Abstract
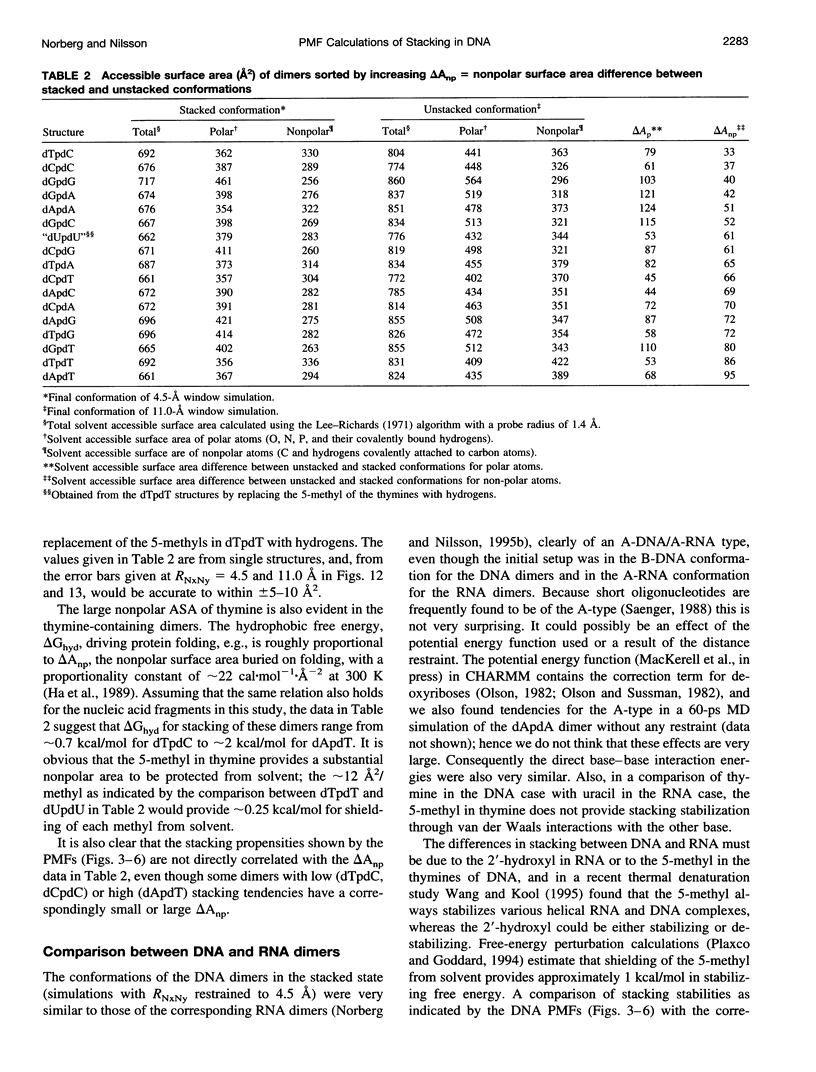
The free energy of the stacking-unstacking process of deoxyribodinucleoside monophosphates in aqueous solution has been investigated by potential of mean force calculations along a reaction coordinate, defined by the distance between the glycosidic nitrogen atoms of the bases. The stacking-unstacking process of a ribodinucleoside monophosphate was observed to be well characterized by this coordinate, which has the advantage that it allows for a dynamical backbone and flexible bases. All 16 naturally occurring DNA dimers composed of the adenine, cytosine, guanine, or thymine bases in both the 5' and the 3' positions were studied. From the free-energy profiles we observed the deepest minima for the stacked states of the purine-purine dimers, but good stacking was also observed for the purine-pyrimidine and pyrimidine-purine dimers. Substantial stacking ability was found for the dimers composed of a thymine base and a purine base and also for the deoxythymidylyl-3',5'-deoxythymidine dimer. Very poor stacking was observed for the dCpdC dimer. Conformational properties and solvent accessibility are discussed for the stacked and unstacked dimers. The potential of mean force profiles of the stacking-unstacking process for the DNA dimers are compared with the RNA dimers.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aida M. An ab initio molecular orbital study on the sequence-dependency of DNA conformation: an evaluation of intra- and inter-strand stacking interaction energy. J Theor Biol. 1988 Feb 7;130(3):327–335. doi: 10.1016/s0022-5193(88)80032-8. [DOI] [PubMed] [Google Scholar]
- Alden C. J., Kim S. H. Solvent-accessible surfaces of nucleic acids. J Mol Biol. 1979 Aug 15;132(3):411–434. doi: 10.1016/0022-2836(79)90268-7. [DOI] [PubMed] [Google Scholar]
- Beveridge D. L., DiCapua F. M. Free energy via molecular simulation: applications to chemical and biomolecular systems. Annu Rev Biophys Biophys Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- Cheng D. M., Sarma R. H. Intimate details of the conformational characteristics of deoxyribodinucleoside monophosphates in aqueous solution. J Am Chem Soc. 1977 Oct 26;99(22):7333–7348. doi: 10.1021/ja00464a038. [DOI] [PubMed] [Google Scholar]
- Frechet D., Ehrlich R., Remy P., Gabarro-Arpa J. Thermal perturbation differential spectra of ribonucleic acids. II. Nearest neighbour interactions. Nucleic Acids Res. 1979 Dec 11;7(7):1981–2001. doi: 10.1093/nar/7.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. A., Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J. 1995 Oct;69(4):1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Livingstone J. R., Spolar R. S., Record M. T., Jr Contribution to the thermodynamics of protein folding from the reduction in water-accessible nonpolar surface area. Biochemistry. 1991 Apr 30;30(17):4237–4244. doi: 10.1021/bi00231a019. [DOI] [PubMed] [Google Scholar]
- Nakano N. I., Igarashi S. J. Molecular interactions of pyrimidines, purines, and some other heteroaromatic compounds in aqueous media. Biochemistry. 1970 Feb 3;9(3):577–583. doi: 10.1021/bi00805a019. [DOI] [PubMed] [Google Scholar]
- Norberg J., Nilsson L. Stacking-unstacking of the dinucleoside monophosphate guanylyl-3',5'-uridine studied with molecular dynamics. Biophys J. 1994 Aug;67(2):812–824. doi: 10.1016/S0006-3495(94)80541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxco K. W., Goddard W. A., 3rd Contributions of the thymine methyl group to the specific recognition of poly- and mononucleotides: an analysis of the relative free energies of solvation of thymine and uracil. Biochemistry. 1994 Mar 15;33(10):3050–3054. doi: 10.1021/bi00176a038. [DOI] [PubMed] [Google Scholar]
- Ravishanker G., Swaminathan S., Beveridge D. L., Lavery R., Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: "curves", dials and windows. J Biomol Struct Dyn. 1989 Feb;6(4):669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- Wang S., Kool E. T. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2'-hydroxyl effects. Biochemistry. 1995 Mar 28;34(12):4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]


