Abstract
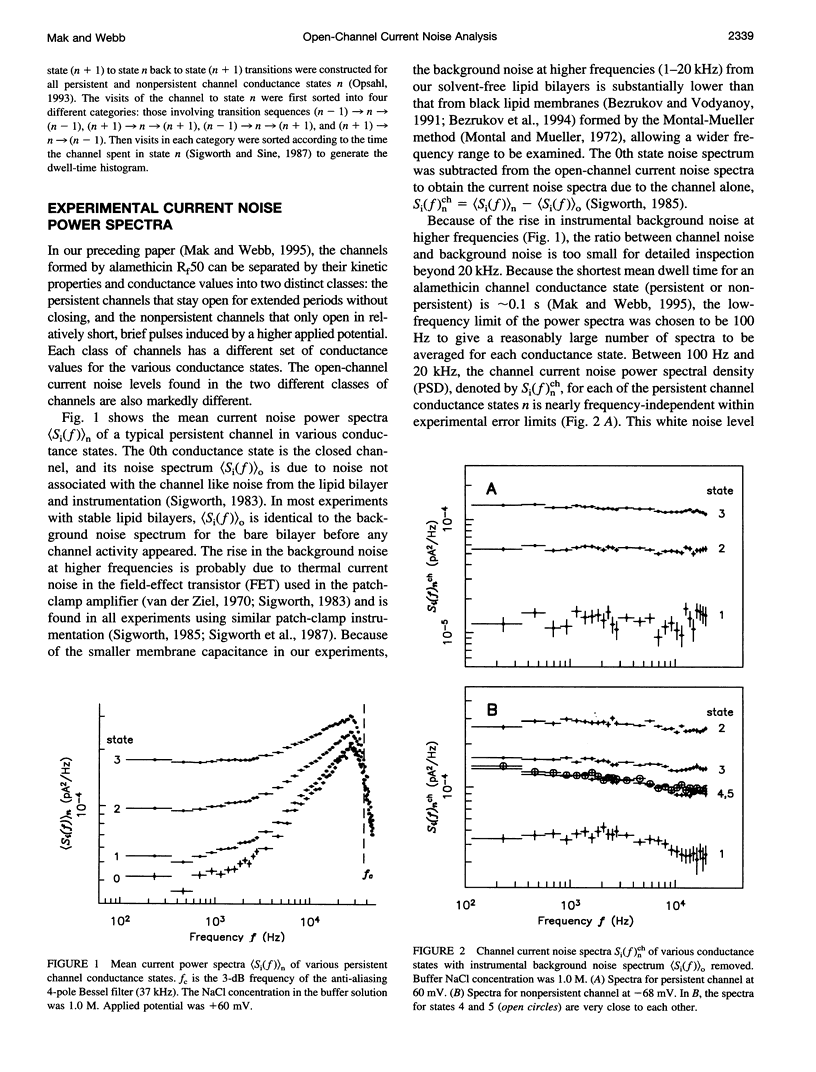
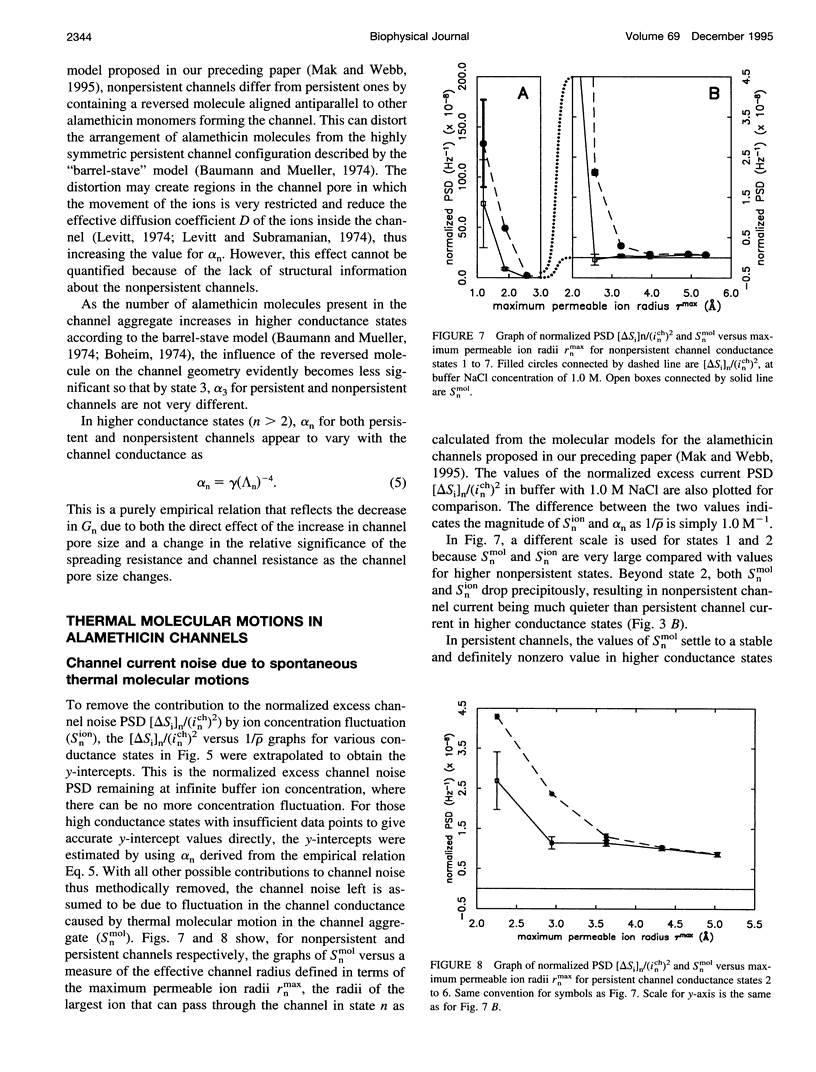
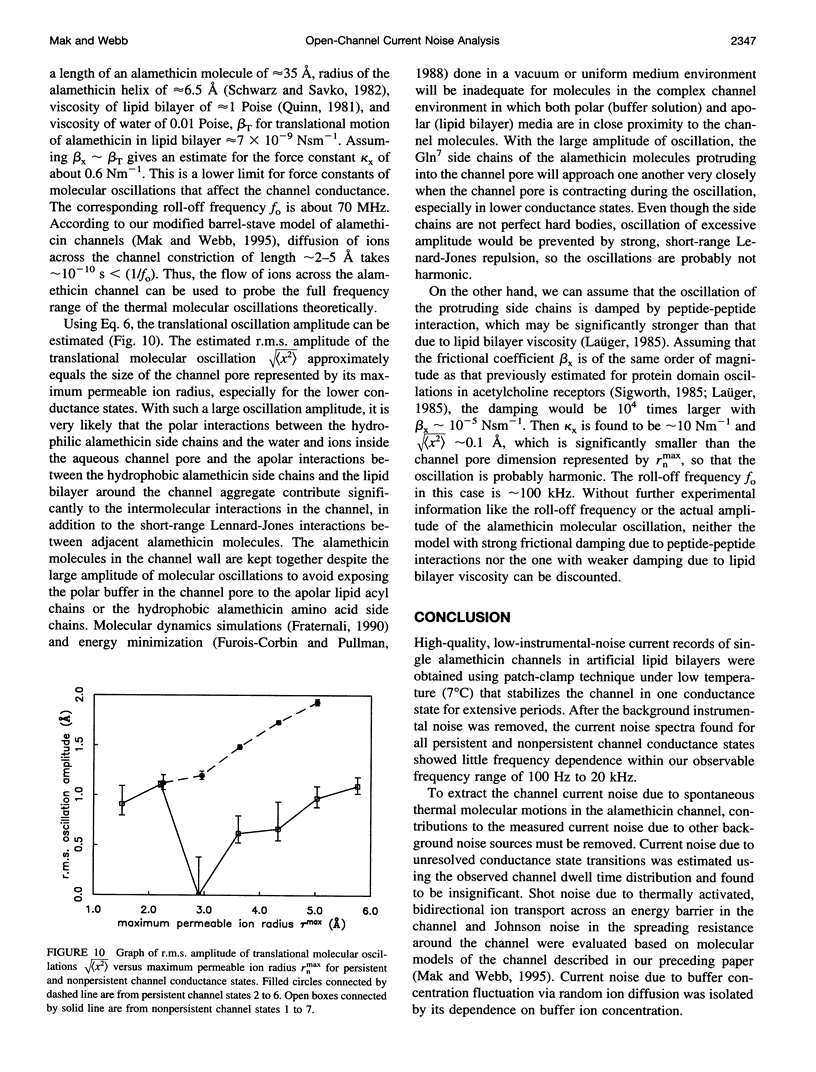
Conductance noise measurement of the open states of alamethicin transmembrane channels reveals excess noise attributable to cooperative low-frequency molecular dynamics that can generate fluctuations approximately 1 A rms in the effective channel pore radius. Single-channel currents through both persistent and nonpersistent channels with multiple conductance states formed by purified polypeptide alamethicin in artificial phospholipid bilayers isolated onto micropipettes with gigaohm seals were recorded using a voltage-clamp technique with low background noise (rms noise < 3 pA up to 20 kHz). Current noise power spectra between 100 Hz and 20 kHz of each open channel state showed little frequency dependence. Noise from undetected conductance state transitions was insignificant. Johnson and shot noises were evaluated. Current noise caused by electrolyte concentration fluctuation via diffusion was isolated by its dependence on buffer concentration. After removing these contributions, significant current noise remains in all persistent channel states and increases in higher conductance states. In nonpersistent channels, remaining noise occurs primarily in the lowest two states. These fluctuations of channel conductance are attributed to thermal oscillations of the channel molecular conformation and are modeled as a Langevin translational oscillation of alamethicin molecules moving radially from the channel pore, damped mostly by lipid bilayer viscosity.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer S. J., Ellena J. F., Cafiso D. S. Dynamics and aggregation of the peptide ion channel alamethicin. Measurements using spin-labeled peptides. Biophys J. 1991 Aug;60(2):389–398. doi: 10.1016/S0006-3495(91)82064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann G., Mueller P. A molecular model of membrane excitability. J Supramol Struct. 1974;2(5-6):538–557. doi: 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- Bezrukov S. M., Vodyanoy I., Parsegian V. A. Counting polymers moving through a single ion channel. Nature. 1994 Jul 28;370(6487):279–281. doi: 10.1038/370279a0. [DOI] [PubMed] [Google Scholar]
- Boheim G. Statistical analysis of alamethicin channels in black lipid membranes. J Membr Biol. 1974;19(3):277–303. doi: 10.1007/BF01869983. [DOI] [PubMed] [Google Scholar]
- Cahalan M., Neher E. Patch clamp techniques: an overview. Methods Enzymol. 1992;207:3–14. doi: 10.1016/0076-6879(92)07003-7. [DOI] [PubMed] [Google Scholar]
- Fraternali F. Restrained and unrestrained molecular dynamics simulations in the NVT ensemble of alamethicin. Biopolymers. 1990;30(11-12):1083–1099. doi: 10.1002/bip.360301109. [DOI] [PubMed] [Google Scholar]
- Furois-Corbin S., Pullman A. Conformation and pairing properties of the N-terminal fragments of trichorzianine and alamethicin: a theoretical study. Biochim Biophys Acta. 1988 Oct 20;944(3):399–413. doi: 10.1016/0005-2736(88)90511-1. [DOI] [PubMed] [Google Scholar]
- Gordon L. G., Haydon D. A. Kinetics and stability of alamethicin conducting channels in lipid bilayers. Biochim Biophys Acta. 1976 Jul 1;436(3):541–556. doi: 10.1016/0005-2736(76)90439-9. [DOI] [PubMed] [Google Scholar]
- Gurd F. R., Rothgeb T. M. Motions in proteins. Adv Protein Chem. 1979;33:73–165. doi: 10.1016/s0065-3233(08)60459-3. [DOI] [PubMed] [Google Scholar]
- Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981 Aug;391(2):85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Karplus M., Petsko G. A. Molecular dynamics simulations in biology. Nature. 1990 Oct 18;347(6294):631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- Latorre R., Alvarez O. Voltage-dependent channels in planar lipid bilayer membranes. Physiol Rev. 1981 Jan;61(1):77–150. doi: 10.1152/physrev.1981.61.1.77. [DOI] [PubMed] [Google Scholar]
- Levitt D. G. A new theory of transport for cell membrane pores. I. General theory and application to red cell. Biochim Biophys Acta. 1974 Nov 27;373(1):115–131. doi: 10.1016/0005-2736(74)90111-4. [DOI] [PubMed] [Google Scholar]
- Levitt D. G., Subramanian G. A new theory of transport for cell membrane pores. II. Exact results and computer simulation (molecular dynamics). Biochim Biophys Acta. 1974 Nov 27;373(1):132–140. doi: 10.1016/0005-2736(74)90112-6. [DOI] [PubMed] [Google Scholar]
- Läuger P. Shot noise in ion channels. Biochim Biophys Acta. 1975 Nov 17;413(1):1–10. doi: 10.1016/0005-2736(75)90053-x. [DOI] [PubMed] [Google Scholar]
- Läuger P. Structural fluctuations and current noise of ionic channels. Biophys J. 1985 Sep;48(3):369–373. doi: 10.1016/S0006-3495(85)83793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby K. L., Weiss D. S. Estimating kinetic parameters for single channels with simulation. A general method that resolves the missed event problem and accounts for noise. Biophys J. 1990 Dec;58(6):1411–1426. doi: 10.1016/S0006-3495(90)82487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. R., Williams R. J. The nature and function of alamethicin. Biochem Soc Trans. 1975;3(1):166–167. doi: 10.1042/bst0030166. [DOI] [PubMed] [Google Scholar]
- Millhauser G. L., Salpeter E. E., Oswald R. E. Diffusion models of ion-channel gating and the origin of power-law distributions from single-channel recording. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1503–1507. doi: 10.1073/pnas.85.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal M., Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko G. A., Ringe D. Fluctuations in protein structure from X-ray diffraction. Annu Rev Biophys Bioeng. 1984;13:331–371. doi: 10.1146/annurev.bb.13.060184.001555. [DOI] [PubMed] [Google Scholar]
- Quinn P. J. The fluidity of cell membranes and its regulation. Prog Biophys Mol Biol. 1981;38(1):1–104. doi: 10.1016/0079-6107(81)90011-0. [DOI] [PubMed] [Google Scholar]
- Rinehart K. L., Jr, Cook J. C., Jr, Meng H., Olson K. L., Pandey R. C. Mass spectrometric determination of molecular formulas for membrane-modifying antibiotics. Nature. 1977 Oct 27;269(5631):832–833. doi: 10.1038/269832a0. [DOI] [PubMed] [Google Scholar]
- Saffman P. G., Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom M. S. Alamethicin and related peptaibols--model ion channels. Eur Biophys J. 1993;22(2):105–124. doi: 10.1007/BF00196915. [DOI] [PubMed] [Google Scholar]
- Sansom M. S. The biophysics of peptide models of ion channels. Prog Biophys Mol Biol. 1991;55(3):139–235. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Allosteric regulation of the epidermal growth factor receptor kinase. J Cell Biol. 1986 Dec;103(6 Pt 1):2067–2072. doi: 10.1083/jcb.103.6.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G., Savko P. Structural and dipolar properties of the voltage-dependent pore former alamethicin in octanol/dioxane. Biophys J. 1982 Aug;39(2):211–219. doi: 10.1016/S0006-3495(82)84510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F. J. Open channel noise. I. Noise in acetylcholine receptor currents suggests conformational fluctuations. Biophys J. 1985 May;47(5):709–720. doi: 10.1016/S0006-3495(85)83968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F. J. Open channel noise. II. A test for coupling between current fluctuations and conformational transitions in the acetylcholine receptor. Biophys J. 1986 May;49(5):1041–1046. doi: 10.1016/S0006-3495(86)83732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F. J., Sine S. M. Data transformations for improved display and fitting of single-channel dwell time histograms. Biophys J. 1987 Dec;52(6):1047–1054. doi: 10.1016/S0006-3495(87)83298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F. J., Urry D. W., Prasad K. U. Open channel noise. III. High-resolution recordings show rapid current fluctuations in gramicidin A and four chemical analogues. Biophys J. 1987 Dec;52(6):1055–1064. doi: 10.1016/S0006-3495(87)83299-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C. F. Inferences about membrane properties from electrical noise measurements. Biophys J. 1972 Aug;12(8):1028–1047. doi: 10.1016/S0006-3495(72)86141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. J., de Levie R. "Reversed" alamethicin conductance in lipid bilayers. Biophys J. 1991 Apr;59(4):873–879. doi: 10.1016/S0006-3495(91)82299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin P. N., Zampighi G. Structure of the junction between communicating cells. Nature. 1980 Feb 7;283(5747):545–549. doi: 10.1038/283545a0. [DOI] [PubMed] [Google Scholar]
- Vogel H., Nilsson L., Rigler R., Meder S., Boheim G., Beck W., Kurth H. H., Jung G. Structural fluctuations between two conformational states of a transmembrane helical peptide are related to its channel-forming properties in planar lipid membranes. Eur J Biochem. 1993 Mar 1;212(2):305–313. doi: 10.1111/j.1432-1033.1993.tb17663.x. [DOI] [PubMed] [Google Scholar]
- Woolley G. A., Wallace B. A. Model ion channels: gramicidin and alamethicin. J Membr Biol. 1992 Aug;129(2):109–136. doi: 10.1007/BF00219508. [DOI] [PubMed] [Google Scholar]


