Abstract
BAP31 is an integral protein of the endoplasmic reticulum membrane and a substrate of caspase-8. Here, we describe the procaspase-8 isoform, procaspase-8L, which is ubiquitously expressed and selectively recruited to the BAP31 complex in response to apoptotic signaling by E1A. Procaspase-8L is characterized by the N-terminal extension (Nex) domain, which extends procaspase-8/a at the N terminus and is required for selective association of procaspase-8L with the BAP31 complex. Gene deletion identified BAP31 and related BAP29 as required for processing of procaspase-8L in response to E1A, by a FADD-independent mechanism that was blocked by BCL-2. Further, Bap29,31 deletion, as well as a Nex-domain dominant-negative mutant, curtailed the activation of downstream caspases (IETDase and DEVDase) and cell death in response to E1A. Preferential recruitment of procaspase-8L by the BAP31 complex at the endoplasmic reticulum suggests an additional pathway for regulating initiator caspase-8 during apoptosis.
Physiological cell death occurs in response to diverse signals, which initiate pathways that ultimately are coupled to complexes that activate the death machinery, typically comprised of caspases. To date, two caspase activation complexes have been identified: death-inducing signaling complexes (DISCs) of the tumor necrosis factor (TNF)-family of death receptors that activate initiator procaspase-8, via the Fas-associated death domain (FADD) adapter, in response to extrinsic cytokine signals, and a mitochondrial-dependent complex, the apoptosome, that activates initiator procaspase-9 in response to multiple cell intrinsic death signals (1, 2). After activation, caspase-8 and caspase-9 stimulate a downstream cascade of events by directly processing effector procaspases such as procaspase-3. The activated effector enzymes then cleave several hundred cellular proteins, causing apoptotic cell suicide (2). An additional target of caspase-8, BID, is cleaved to tBID, which mediates cross talk between death receptors at the plasma membrane and the apoptosome by inducing mitochondria to release a critical cofactor, cytochrome c (3).
BAP31 is a 28-kDa polytopic integral membrane protein that is ubiquitously expressed (refs. 4–6; our unpublished data) and highly enriched at the endoplasmic reticulum (ER) (7, 8), where it forms both a homo-oligomer and a hetero-oligomer with the closely related BAP29 (6, 8). A role for BAP31 as a potential regulator of apoptosis derived from its discovery as a predicted BCL-2/BCL-XL-associated protein (8). The 14-kDa cytosolic tail of human BAP31 contains a weak death effector and overlapping coiled-coil (DECC) domain, flanked on either side by identical caspase-8 recognition sites, and terminating at the C terminus with a canonical KKXX ER retention signal (8, 9). In addition to being a preferred caspase-8 substrate in vitro, the DECC domain of BAP31 can associate, albeit weakly, with procaspase-8 in overexpressing cotransfected 293T cells (8, 9). Of note, two predominant protein isoforms of procaspase-8 have been identified to date, procaspase-8/a and procaspase-8/b, that differ only in a short sequence within the pro-domain that derives from the alternative splicing of CASP8 exon 6 (10). The two isoforms are typically expressed ubiquitously at equivalent levels, and appear to function interchangeably (10).
Here, we describe the cellular isoform of procaspase-8, procaspase-8L, which is procaspase-8/a containing an N-terminal extension (Nex domain) of 59 aa. This domain allows selective recruitment of procaspase-8L to the BAP31 complex in response to apoptotic signaling by the model oncogene, adenovirus E1A. Gene deletion indicates that the BAP proteins are required for E1A-induced processing of procaspase-8L and contribute to downstream activation of caspases and cell death.
Materials and Methods
Antibodies and Reagents.
Calnexin monoclonal antibody was purchased from BD Biosciences (Mississauga, ON, Canada), anti-FLAG M2 monoclonal antibody and M2 FLAG gel were from Sigma, and anti-hemagglutinin (HA) 12CA5 monoclonal antibody was from Babco (Richmond, CA). Rabbit antibody against the p18 catalytic subunit of human caspase-8 was from S. Roy and D. Nicholson (Merck Frosst Canada, Kirkland, QC), anti-Apo-1 was from P. Krammer (German Cancer Research Center, Heidelberg), and anti-procaspase-8 N2 monoclonal antibody (raised against the caspase-8 prodomain) from M. Peter (University of Chicago). Rabbit anti-mouse caspase-8 (p18) was from Santa Cruz Biotechnology. Rabbit anti-BAP31 and anti-Bap29 antibodies have been described (6, 8). The procaspase-8L Nex domain peptide-specific antibody was generated at Research Genetics (Huntsville, AL). Rabbits were immunized with the peptide NH2-EHVELGRLGDSETA-COOH, conjugated at its amino terminus to KLH, and serum was affinity purified against the immunizing peptide. A second anti-Nex antibody generated in our lab against a recombinant GST-Nex domain fusion protein and absorbed on a GST column was used in Fig. 2D.
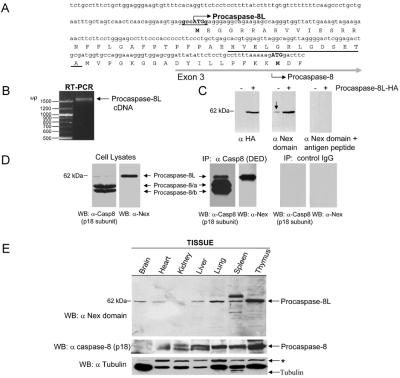
Figure 2.
Characterization and cloning of procaspase-8L. (A) Deduced amino acid sequence of the predicted N-terminal extension (Nex) domain of procaspase-8L. The predicted procaspase-8L upstream start codon and Kozak translation initiation consensus sequence are shown in bold. The positions of exon 3 and the canonical procaspase-8/a and -8/b start site are indicated. The sequence of the peptide used for the procaspase-8L Nex domain antibody production is underlined in black. (B) PCR of first-strand cDNA derived from KB cell mRNA by using primers specific for the 5′ end of the Nex domain and the 3′ end of the canonical caspase-8 p10 subunit demonstrate that the Nex domain of procaspase-8L is connected to the entire procaspase-8/a ORF. The reverse transcription (RT)-PCR product is indicated (arrow). (C) Affinity-purified rabbit polyclonal antibodies raised to the Nex domain of procaspase-8L detect an endogenous protein of ≈62 kDa in human H1299 lung carcinoma cells (arrow, Center) and ectopic expressed procaspase-8L-HA. Cells were transfected with vector (−) or with vector expressing HA-tagged procaspase-8L (+) in the presence of 50 μM zVAD-fmk and, after 24 h, total cell lysates were analyzed by Western blotting with the indicated antibodies, in the presence or absence of 10 μM immunizing peptide. (D) Cell lysates were subjected to immunoprecipitation with an anti-caspase-8 monoclonal antibody recognizing the prodomain (10). Lysates and precipitates were analyzed by blotting with anti-caspase-8 p18 subunit-specific antibody or anti-Nex domain antibody. (E) Tissue distribution of procaspase-8L. The indicated murine tissue lysates were analyzed by immuno blotting as in D. The blot was reprobed with antitubulin monoclonal antibody for loading control. *, Cross-reacting product.
Cell Culture, Virus Infection, and Apoptosis Assays.
Human KB epithelial and H1299 lung carcinoma cells were maintained as described (8). H9 lymphocytes were cultured in RPMI medium 1640 including 10 mM Hepes and supplemented as above. KB cells stably expressing HA-BCL-2, GFP-Flag, BAP31-Flag, or crBAP31-Flag have been described (11, 12) and were grown as above in the presence of 350 mg/ml geneticin (GIBCO/BRL). KB cells stably expressing both crBAP31-Flag and HA-BCL-2 were constructed by transfecting HA-BCL-2 stable transfectants with crBAP31-Flag in pcDNA3.1/Hygro(−) and selecting for clones resistant to both geneticin and hygromycin. Wild-type and Bap29,31-null embryonic stem (ES) cells were grown at 37°C and 7.5% CO2 on embryonic fibroblast feeders in KNOCKOUT D-MEM (GIBCO/BRL) supplemented with 100 units/ml streptomycin sulfate and penicillin, 15% FBS, 2 mM l-glutamine, nonessential amino acids, and 1 mM β-mercaptoethanol, in the presence of leukemia inhibitory factor (LIF). ES cells were differentiated into embryoid epithelial and fibroblast-like cells as follows; cells were propagated on gelatin-coated dishes for three passes (8 days) in the absence of LIF to dilute out feeder cells, and then replated on nongelatinized dishes, allowing monolayers of differentiated cells to form. Infection of cells with Ad5 dl520E1B−, which expresses the 243-residue form (12 S) of E1A and no E1B products, was performed as described (8) and ≥98% of cells were found to be expressing E1A 20 h postinfection. IETD-amc and DEVD-amc caspase activity assays (Upstate Biotechnology, Lake Placid, NY) and Annexin V staining (BioVision, Mountain View, CA) were conducted according to the manufacturer's protocols
Rapid Amplification of cDNA Ends (RACE) Analysis and Expression Constructs.
PolyA+ mRNA from KB cells was isolated using the Oligotex Direct mRNA Kit (Qiagen). Both 5′ and 3′ RACE were performed using the SMART RACE cDNA Amplification Kit (CLONTECH). Primers 5′-CGGGATCCGATTCTGCCTTTCTGCTGG and 5′-GCCAAGCTTTCAATCAGAAGGGAAGAC-3′ were used to PCR-amplify the procaspase-8L reading frame. All PCR products were cloned using the TOPO TA Cloning Kit (Invitrogen), and at least three independent clones of each product were sequenced on both strands. Procaspase-8 and procaspase-8L, each tagged at their carboxy terminus with HA, were created using standard PCR techniques and cloned into pcDNA3. For the Nex-EGFP construct, standard PCR techniques were used to create the Nex domain with a C-terminal Flag tag; this product was then cloned into the pEGFP-N3 Vector so that it was upstream and in-frame with EGFP. Construction of pcDNA3 vectors expressing crBAP31-Flag and BAP31-Flag has been described (12). pcDNA3 Vector expressing Flag-tagged PAIP-I or 12 S E1A were gifts from N. Sonenberg, (McGill University) and R. Marcellus (Gemin X Biotechnologies, Montreal), respectively.
Immunoprecipitation.
For immunoprecipitation of the BAP31 complex, 1–2 × 107 cells per sample were washed once in PBS and lysed for 30 min at 4°C in 2 ml of lysis buffer (50 mM Hepes, pH 7.4/150 mM NaCl/0.5 mM EDTA/1% Triton X-100/10% glycerol/1 mM phenylmethylsulfonyl fluoride/1 μg/ml aprotinin/1 μg/ml leupeptin/1 μg/ml pepstatin). The cell lysate was centrifuged at 12,000 × g for 15 min to remove insoluble membrane and debris, and subsequently precleared with protein G Sepharose (Pharmacia) for 1 h. crBAP31-Flag or BAP31-Flag was then precipitated for 4 h at 4°C with FLAG M2 Gel, and washed five times with lysis buffer. The BAP31 complex was eluted from the beads by Flag peptide competition or by boiling in SDS sample buffer. DISC analysis was done exactly as described (13). All samples were analyzed by SDS/PAGE and Western blotting.
Subcellular Fractionation.
KB cells were fractionated as described by Goping et al. (14) with minor modifications.
Transient Transfection.
For Fig. 5B, 3 × 105 H1299 cells were transfected with Lipofectamine Plus (GIBCO/BRL) and 1 μg of pcDNA3 vector, or this vector expressing either procaspase-8/a-HA or procaspase-8L-HA, together with 0.1 μg of vector expressing luciferase. Three hours after transfection, the cells were incubated in medium containing or lacking 50 μM zVAD-fmk for 24 h and adherent and floating cells were then collected, washed, lysed, and total luciferase activity was measured as described (8). In Fig. 5A, 7 × 106 H1299 cells were transfected as described above with 5 μg of each of the indicated constructs in the presence of zVAD-fmk. After 24 h, the cells were lysed and BAP31-Flag and PAIP-1-Flag were immunoprecipitated as described above. In Fig. 6, 2 × 105 KB cells in 24-well clusters were transfected with the indicated plasmids using Lipofectamine 2000 (GIBCO/BRL).
Figure 5.
Procaspase-8L selectively associates with the BAP31 complex. (A) Anti-Flag immunoprecipitates from H1299 cells cotransfected with the indicated expression constructs in the presence of 50 μM zVAD-fmk were analyzed by SDS/PAGE and Western blotting with anti-HA or anti-Nex domain antibodies. Note that only procaspase-8L-HA immunoprecipitated with BAP31-Flag. *, A nonspecific protein detected by anti-HA antibody in lysates. (B) Procaspase-8L induces loss of cell viability when ectopically expressed. H1299 cells were transfected with a luciferase expression construct and pcDNA3 vector expressing either procaspase-8/a-HA or -8L-HA, in the absence or presence of 50 μM zVAD-fmk, and luciferase activity was measured 24 h later. (C) Procaspase-8L is not detected in the Fas DISC. The DISC was immunoprecipitated following stimulation of H9 lymphocytes with anti-Apo-1/Fas (19) and the presence of procaspase-8L and procaspase-8 in the precipitates was assessed by immunoblotting as in Fig. 4C. IgG, Ig heavy chain. (D) Wt or Fadd-deficient mouse embryo fibroblasts (20) were infected with Ad5 dl520E1B− for 50 h and procaspase-8L cleavage was analyzed as in Fig. 4A.
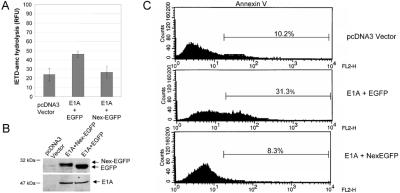
Figure 6.
E1A-induced caspase-8 activity and apoptosis is inhibited by a procaspase-8L DN mutant. (A) KB cells were transiently cotransfected with plasmids encoding 12S E1A and Nex-Flag-EGFP or 12S E1A and Flag-EGFP and 36 h post transfection equivalent amounts of cell lysate were tested for their ability to hydrolyze the caspase-8-preferred substrate IETD-amc. Shown is the mean of four independent experiments. (B) Cell lysates from A were analyzed by SDS/PAGE and immunoblotting with antibodies against E1A or Flag. (C) As in A, except that cells were collected, stained with Annexin V, and analyzed by flow cytometry. Transfection efficiency was estimated to be 20–30% by analyzing EGFP-positive cells by immunofluorescence (not shown).
Targeted Disruption of Bap29 and Bap31.
To target the X-linked Bap31 gene, a targeting construct comprising 1.2 kb of intron 1, exon 2 (containing the start codon) fused in-frame with the full-length Bap31 coding sequence, an internal ribosome entry site (IRES), a promoterless neomycin phosphotransferase coding sequence, a polyadenylation signal, and 4.7 kb of intron 3 was created. This targeting construct contained three loxP sites; the first in the intron 1-derived region, the second between the Bap31 coding sequence and the IRES, and the third between the neomycin cassette and the polyadenylation signal. C57BL/6 ES cells were electroporated with the linearized targeting vector and G418-resistant colonies were screened for homologous recombinants by PCR and Southern blotting. Bap31 protein expression in the gene-targeted cells was indistinguishable from parental cells. A complete disruption of the Bap31 gene was achieved by transient expression of Cre recombinase. Clones with only a Cre-mediated deletion of the IRES and neo cassette were isolated and targeted with a linearized targeting construct composed of a 250-bp homology region, a neomycin phosphotransferase coding sequence fused in-frame to the Bap29 translation initiation codon, a polyadenylation signal, and a 3.5-kb homology region. In this case, homologous recombination resulted in loss of Bap29 expression from the targeted allele. To generate cells negative for both Bap29 and Bap31, Bap29−/− clones underwent another round of transient Cre expression.
Results and Discussion
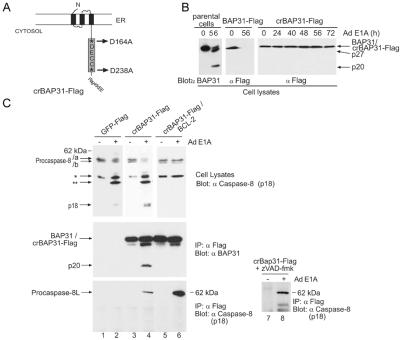
The adenovirus type 5 vector, Ad5 dl520E1B− (15), encodes 12 S E1A mRNA and no E1B products, generating the 243-aa E1A oncoprotein as the only vector expression product. Infection of human KB epithelial cells with this Ad E1A vector stimulates activation of caspases including caspase-8 and -3, cleavage of the caspase-8 preferred substrate BAP31 and the caspase-3 preferred substrate, PARP, and apoptosis (8, 11, 16). To observe the recruitment of procaspase-8 to the caspase-sensitive BAP31 complex during E1A-induced apoptosis, therefore, a KB cell line was created that stably expresses an epitope-tagged caspase-resistant mutant, crBAP31-Flag (Fig. 1A), in which the mutant was expressed at levels only slightly higher than that of endogenous BAP31 (12). As expected, crBAP31, but not endogenous BAP31 or wt BAP31-Flag, remained structurally intact during E1A-induced apoptosis (Fig. 1B).
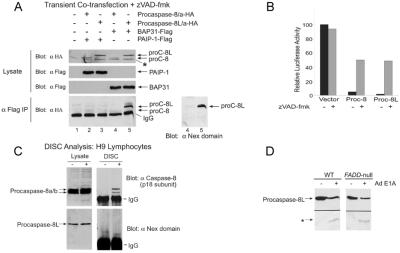
Figure 1.
Procaspase-8L is recruited to the BAP31 complex during E1A-induced apoptosis. (A) Schematic of crBAP31-Flag in the ER. Asp-164 and Asp-238 were mutated to Ala, and a Flag epitope was inserted directly in front of the KKEE ER retrieval sequence. *, Caspase recognition Asp residues. (B) crBAP31-Flag is not cleaved during E1A-induced apoptosis. Parental KB cells or KB cells stably expressing wt BAP31-Flag or crBAP31-Flag (at levels slightly higher than that of endogenous BAP31) were treated with adenovirus type 5 (Ad5) dl520E1B− (expressing only 12 S E1A and no E1B products) for the indicated times and endogenous BA, 31, BAP31-Flag, or crBAP31-Flag cleavage was analyzed by SDS/PAGE and immunoblotting with anti-BAP31 or anti-Flag antibodies. (C) A ≈62-kDa form of procaspase-8, designated procaspase-8L, is recruited to the BAP31 complex in response to E1A signaling. KB cells stably expressing GFP-Flag, crBAP31-Flag, or crBAP31-Flag and BCL-2 were mock infected or infected (E1A) with Ad5 dl520E1B− for 40 h and immunoprecipitation (IP) was conducted with anti-Flag M2 gel followed by SDS/PAGE and Western blotting with anti-BAP31 (Middle) or anti-caspase-8 antibody specific for the p18 subunit (Bottom). Where indicated, cells were cultured in the presence of zVAD-fmk (50 μm). Immunoblots of the corresponding total cell lysate (5% of the input used for immunoprecipitation) are also shown (Top). The positions of the conventional procaspase-8 isoforms, designated/a and/b, their processing intermediates (asterisks), the caspase-8 catalytic subunit (p18), procaspase-8L, crBAP31-Flag, endogenous BAP31, and its p20 cleavage product, are shown.
In Fig. 1C, crBAP31-Flag cells, with or without stable coexpression of BCL-2, were mock-infected or infected with Ad E1A for 40 h, and cell lysates subjected to precipitation with anti-Flag antibody and analysis by Western (immuno) blotting with antibodies either against the p18 catalytic subunit of caspase-8 or against BAP31. An approximately 62-kDa product (labeled procaspase-8L) that reacted with antibody against p18 was observed in crBAP31-Flag precipitates in the presence (Fig. 1C Bottom, lane 4) but not absence (lane 3) of E1A signaling. In contrast, the conventional 55- and 53-kDa procaspase-8/a and procaspase-8/b isoforms observed in cell lysates (Fig. 1C Top) were not detected in the crBAP31-Flag precipitates. Cells expressing a control protein, GFP-Flag, did not yield any anti-p18 reactive product in the anti-Flag precipitate (Fig. 1C Bottom). In KB cells stably expressing both crBAP31-Flag and antiapoptotic BCL-2, processing of the conventional forms of procaspase-8/a and -8/b in response to E1A was inhibited (Fig. 1C Top, lanes 5 and 6), endogenous BAP31 was not cleaved (as judged by the lack of appearance of the p20 product derived from caspase cleavage of endogenous BAP31; Fig. 1C Middle), and apoptosis was blocked (ref. 11; data not shown). BCL-2, however, did not interfere with the E1A-induced recruitment of the 62-kDa product to the crBAP31 complex; the apparent increase in levels of this product that associated with crBAP31 in the presence of BCL-2 (compare lanes 4 and 6 in Fig. 1C Bottom) may arise because BCL-2 blocks its processing (see below) and, therefore, preserves the pool of intact precursor protein. In addition, procaspase-8L recruitment was not blocked by the pan-caspase inhibitor zVAD-fmk (Fig. 1C Bottom, lanes 7 and 8), although caspase activation and cell death were inhibited (data not shown). As expected, the p20 caspase cleavage product of endogenous BAP31 that was generated in the absence of BCL-2 (12) coprecipitated with crBAP31-Flag (Fig. 1C Middle, lane 4) because it forms a stable oligomer with intact BAP31 (9). Compared with procaspase-8/a and -8/b, the 62-kDa product was not clearly detectable in whole-cell lysates with anti-p18 antibody, suggesting that it might be a minor product.
Procaspase-8L.
An extensive analysis of the 5′ ends of procaspase-8 mRNA transcripts in KB epithelial cells was conducted using 5′ RACE. Five transcripts were identified that differ in sequence upstream of CASP8 exon 3 (GenBank accession nos. AF422925–AF422929). Exon 3 contains the ATG start site of translation for conventional procaspase-8/a and -8/b (ref. 17; see Fig. 2A). Four of the transcripts contained in-frame stop codons upstream of exon 3, whereas one transcript, accession no. AF422925, contained an upstream ATG within a strong Kozak translational initiation consensus sequence (GCCATGG), which is in-frame with the procaspase-8 ORF initiating within exon 3 (Fig. 2A). This was the most 5′ ATG codon in the derived transcript, which together with the surrounding consensus sequence suggested that it might function as the preferential site of translation initiation in this transcript (18). If so, the predicted N-terminal extended ORF encodes 59 aa upstream of the conventional procaspase-8 translation initiation site, and was designated the Nex domain. The Nex domain was continuous with the entire procaspase-8/a ORF as determined by PCR and sequencing of first-strand KB cell cDNA, using primers specific for the 5′ end of the Nex domain and the 3′ end of the caspase-8 p10 subunit (Fig. 2B). Importantly, the Nex domain could not be connected to any transcript other than procaspase-8/a by 3′ RACE. The extended ORF consisting of the Nex domain linked to the procaspase-8/a ORF codes for a protein with a predicted molecular size of 61.9 kDa, which is consistent with the size of the 62-kDa anti-p18 reactive protein found associated with the BAP31 complex, and was designated procaspase-8L (long).
Antibody raised against a peptide sequence within the Nex domain (solid black underline in Fig. 2A) detected an endogenous 62-kDa protein in H1299 cells (arrow in Fig. 2E Middle), and demonstrated enhanced reactivity toward procaspase-8L protein obtained by overexpression in these cells of HA-tagged procaspase-8L cDNA by transient transfection (Fig. 2C). The specificity of the anti-Nex domain antibody in Western blot was demonstrated by competitive inhibition with the Nex domain antigen peptide (Fig. 2C Right). Moreover, recovery of procaspase-8 isoforms from cell lysate with a precipitating procaspase-8 antibody (10) and blotting of the precipitate with anti-Nex detected the 62-kDa product (Fig. 2D). As predicted, probing the precipitate with antibody against the common p18 subunit revealed that procaspase-8L is expressed at a considerably lower level than the two main isoforms (Fig. 2D). Analysis of murine tissues by immunoblot suggested that procaspase-8L is widely expressed (Fig. 2E). Taken together, these results suggest that procaspase-8L is a procaspase-8 isoform that is expressed at the protein level and contains a unique N-terminal Nex domain.
Subcellular Distribution of Procaspase-8L.
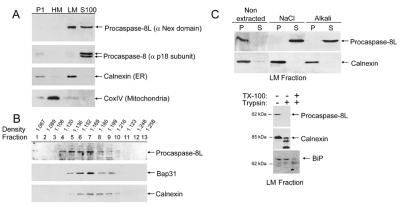
KB cells were fractionated into P1 nuclear, heavy membrane (HM), light membrane (LM), and cytosolic S-100 fractions and the presence of procaspase-8L in each fraction was examined with the anti-Nex domain antibody. As shown in Fig. 3A, procaspase-8L distributed between the S100 cytosolic fraction and the LM fraction enriched in microsomes, as indicated by the ER marker, calnexin. In contrast, procaspase-8/a and -8/b were located exclusively in the S100 fraction. Further investigation using analytical fractionation of rodent liver membranes revealed that procaspase-8L and Bap31 cosedimented with near identical median densities that were slightly lower than the median density of calnexin (Fig. 3B). In contrast to an integral membrane protein like calnexin, procaspase-8L was extracted from the LM fraction with high salt concentrations or alkali (pH 11.5) (Fig. 3C Upper), suggesting that it is peripherally associated with microsomal membrane. Furthermore, treatment of the LM fraction with trypsin showed that procaspase-8L was sensitive to digestion, whereas an ER luminal protein, BiP, was completely resistant (Fig. 3C Lower). Therefore, the initial location of procaspase-8L at the cytosolic face ER may provide a nearby pool for recruitment to the BAP31 complex following E1A stimulation.
Figure 3.
Subcellular distribution of procaspase-8L. (A) KB cells were homogenized in isotonic buffer and the P1 nuclear (500 × g pellet), heavy membrane (HM) (9,000 × g pellet), LM (100,000 × g pellet), and cytosolic S-100 fractions were separated by differential centrifugation. The fractions were analyzed for the presence of procaspase-8L, procaspase-8, the ER marker, calnexin, and the mitochondrial marker, cytochrome c oxidase subunit IV (CoxIV), by immunoblotting with the respective antibodies. (B) Sucrose density gradient fractions of rodent liver membranes were analyzed for the presence of procaspase-8L, BAP31, and calnexin as in A. (C) Procaspase-8L is peripherally associated with the cytosolic face of the light membranes. (Upper) The LM fraction from KB cells was subjected to extraction with 0.5M NaCl or alkali (0.1 M NaCO3, pH 11.5) and, after centrifugation, the membrane pellets (P; lanes 1 and 3) and supernatants (lanes 2 and 4) were analyzed as in A. (Lower) The LM fraction was incubated with (+) or without (−) trypsin and the integrity of procaspase-8L, calnexin (ER transmembrane protein), and BiP (ER luminal protein) assessed by immunoblotting. TritonX-100 was added to 1% in one reaction to demonstrate that BiP was trypsin sensitive following solubilization of the LM microsomal fraction.
Procaspase-8L Recruitment to the BAP31 Complex and Cleavage in Response to E1A Signaling.
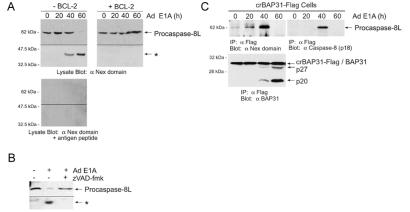
Immunoblot analysis of KB cells with anti-Nex domain antibody detected endogenous procaspase-8L, which was cleaved in response to E1A expression (Fig. 4A). Similar results were observed in cells expressing crBAP31-Flag (not shown) and, in both cases, this apparent procaspase-8L processing was blocked by BCL-2 or by the wide spectrum caspase inhibitor, zVAD-fmk (Fig. 4 A and B; data not shown). Procaspase-8L was maximally recovered in crBAP31-Flag immunoprecipitates 40 h after introducing E1A into cells, as determined by immunoblot analysis with anti-Nex (Fig. 4C Upper Left), and this corresponded to the time when both procaspase-8L processing (Fig. 4A) and caspase cleavage of endogenous BAP31 (Fig. 4C Lower) had been initiated. The anti-Nex blot of the crBAP31-Flag precipitates was then stripped and reprobed with antibody against the p18 subunit of caspase-8. Again, a product was detected that exactly comigrated with the anti-Nex reactive product, showing maximal accumulation with crBAP31-Flag at 40 h postinfection with Ad E1A (Fig. 4C Upper Right). By 60 h, procaspase-8L cleavage was completed (Fig. 4A) and anti-Nex and anti-p18-reactive product was no longer detected in association with crBAP31-Flag (Fig. 4C).
Figure 4.
Endogenous procaspase-8L is recruited to the BAP31 complex and cleaved during E1A-induced apoptosis. (A) Cleavage of procaspase-8L is blocked by BCL-2. Parental KB cells (−BCL-2) or KB cells stably expressing BCL-2 (+BCL-2) were infected with Ad5 dl520E1B− for the indicated times and Western blots were developed with anti-Nex antibody, in the absence (Upper) or presence (Lower) of the Nex immunizing peptide (10 μM). (B) Cleavage of procaspase-8L is inhibited by z-VAD-fmk. Cells were treated as in A in the absence or presence of 50 μM zVAD-fmk. (C) Recruitment of procaspase-8L to the BAP31 complex. KB cells stably expressing crBAP31-Flag were treated as in A and the BAP31 complex was immunoprecipitated (α Flag IP) at the indicated times postinfection. Immuno blots of the precipitates were developed with anti-Nex antibody (Upper Left), then stripped and reprobed with anti-caspase-8 (p18) antibody (Upper Right) or anti-BAP31 antibody (Lower). The anti-Nex domain and anti-caspase-8 (p18) antibodies detected an identical 62-kDa protein. The p20 and p27 cleavage products of endogenous BAP31 (that heterodimerize with crBAP31-Flag) are indicated. A representative experiment is shown.
Selective Recruitment of Procaspase-8L to the BAP31 Complex.
Consistent with the observation that endogenous procaspase-8/a and -8/b were not detectably recruited to crBAP31-Flag following E1A expression (Fig. 1E), overexpression of procaspase-8/a-HA and -8L-HA by transient cotransfection of the respective cDNAs with cDNA encoding BAP31-Flag in H1299 cells showed a marked preference for interactions between the BAP31 complex and procaspase-8L (Fig. 5A, lanes 4 and 5). Neither procaspase exhibited a detectable association with the control protein, PABP Interacting Protein-I-Flag (PAIP-I-Flag). Transfections were conducted in the presence of zVAD-fmk. In the absence of the caspase inhibitor, ectopically expressed procaspase-8/a and -8L underwent processing (data not shown) and potently induced cell death, as assessed by cotransfection with a luciferase reporter and measurement of luciferase activity (Fig. 5B) or by cotransfection with a GFP reporter and measurement of the apoptotic morphology of GFP-transfected cells (not shown).
Procaspase-8/a and -8/b are recruited to DISCs of the tumor necrosis factor (TNF) receptor family on receptor stimulation (10). To determine whether procaspase-8L is also a component of this signaling complex, we stimulated H9 lymphocytes with the agonistic Fas antibody, anti-Apo-1 (19), and immunoprecipitated the DISC. As expected, procaspase-8/a and -8/b were both recruited to the Fas DISC in a stimulation-dependent manner. In the same DISC precipitation, however, no procaspase-8L was detected with the anti-Nex antibody (Fig. 5C), suggesting that if procaspase-8L associates with this complex it is a minor component of it. Moreover, procaspase-8L cleavage was not impaired in Fadd-null primary mouse embryo fibroblasts (20) in response to E1A (Fig. 5D), nor do these same cells exhibit detectable resistance to Ad E1A-induced cell death (11, 20), suggesting that E1A-induced processing of procaspase-8L is FADD-independent.
Nex Domain Dominant-Negative Mutant Inhibits E1A-Induced Apoptosis.
Because the procaspase-8L Nex domain is required for recruitment of the proenzyme to the BAP31 complex in response to E1A, expression of the Nex domain on its own might be expected to exert a dominant-negative influence on E1A death signaling. In Fig. 6, KB cells were transiently cotransfected with a plasmid encoding 12S E1A and plasmid encoding either Nex-EGFP or EGFP. Compared with EGFP, Nex-EGFP significantly inhibited both E1A-induced cleavage of the caspase-8 preferred substrate, IETD-amc (Fig. 6A), and E1A-induced appearance of Annexin V-positive cells (Fig. 6C). Of note, expression of a catalytically inactive full-length procaspase-8L DN, in which the catalytic cys was mutated to ala, also inhibited E1A-induced apoptosis to a similar extent as Nex-EGFP (data not shown). Nex-EGFP was expressed at a similar level as EGFP and did not affect the expression of E1A (Fig. 6B).
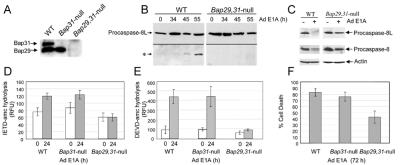
E1A-Induced Cleavage of Procaspase-8L Is Inhibited in Double Bap29- and Bap31-Null Cells.
BAP31 is part of large complex that includes both BAP31 homo-oligomers and hetero-oligomers comprising the closely related BAP29 (ref. 6; our unpublished data). To examine the contribution of these proteins to procaspase-8L processing in response to E1A, mouse ES cells deficient in Bap31 and Bap29 were generated by gene targeting (S.K. and M.R., unpublished data; see Materials and Methods; Fig. 7A), and both wild-type and gene-deleted cells were subjected to growth conditions that favor differentiation into epithelial- and fibroblast-like cells. The levels of procaspase-8L were similar in wild-type and Bap29,31-null cells and E1A protein was expressed at equal levels in all cell types following infection with Ad E1A (Fig. 7B and data not shown). Following expression of E1A in wild-type cells, procaspase-8L was cleaved, generating the N-terminal fragment detected by anti-Nex domain antibody (Fig. 7B). In Bap29,31-null cells, on the other hand, this processing was strongly impaired (Fig. 7B) and the double-null cells exhibited a significant decrease in their ability to hydrolyze the caspase-8 preferred substrate, IETD-amc, and the caspase-3 preferred substrate, DEVD-amc, in response to E1A (Fig. 7 D and E). Procaspase-8L processing was not significantly altered in either Bap31 or Bap29 single knockout cells (data not shown), suggesting that Bap31 and Bap29 can functionally complement one another. Interestingly, procaspase-8/a and -8/b cleavage was also reduced in the Bap29,31-null cells (Fig. 7C), consistent with idea that activated procaspase-8L might directly cleave conventional procaspase-8/a and -8/b. Importantly, E1A-induced cell death in Bap29,31-null cells was reproducibly less than in either wild-type or Bap31-null cells (Fig. 7F).
Figure 7.
Procaspase-8L processing is inhibited in Bap29,31-null cells. (A) Loss of Bap29 and Bap31 expression was confirmed by immunoblotting with anti-BAP31 and anti-Bap29 antibodies. (B) E1A-induced procaspase-8L cleavage is inhibited in Bap29,31-null ES cells. ES cells were differentiated into epithelial- and fibroblast-like cells as described in Materials and Methods, and infected with Ad5 dl520E1B− for the indicated times. Procaspase-8L cleavage was analyzed as in Fig. 3A. (C) As in B, procaspase-8L and procaspase-8/a and -8/b cleavage was analyzed with anti-Nex and anti-caspase-8 (p18) antibodies. (D) Decreased caspase-8 activity in Bap29,31-null cells. Aliquots of lysate from wild-type and Bap29,31-null cells (containing equivalent protein concentrations) stimulated with E1A for 24h (time of maximum caspase activity) were tested for their ability to hydrolyze the preferred caspase-8 substrate IETD-amc. Shown is the average of three independent experiments. RFU, relative fluorescence units. (E) Decreased DEVD-ase activity in Bap29,31-null cells. As in C, except that lysates were tested for their ability to hydrolyze the caspase-3 preferred substrate DEVD-amc. (F) Cell death was measured by trypan blue exclusion 72 h post infection. Shown is a representative of four independent experiments.
Collectively, these results identify the procaspase-8 isoform, procaspase-8L, whose Nex domain allows preferential recruitment to the BAP complex in the ER in response to apoptotic signaling by oncogenic E1A. Although E1A likely triggers several proapoptotic pathways (21, 22), gene deletion identified the BAP proteins as directly contributing to procaspase-8L processing, activation of downstream caspases, and cell death. The involvement of procaspase-8L in E1A-induced apoptosis was further supported by the fact that procaspase-8L DN mutants inhibited caspase-8 activity and cell death. E1A-induced cleavage of procaspase-8L was normal in FADD-null cells but sensitive to the caspase inhibitor zVAD-fmk, consistent with either FADD-independent autoprocessing or cleavage by an upstream caspase. This cleavage was also inhibited by BCL-2, which might relate to observed interactions between BCL-2 and BAP31 (8, 9) and potential functions for BCL-2 at the ER in regulating organelle-specific initiation of cell death (23). The potential activation of caspase-8 at a site other than cell surface death receptors suggests an additional pathway for regulating this important initiator caspase, which could initiate a cascade of caspase activation by processing conventional procaspase-8/a,b (24) and/or downstream effector procaspases, as well as to activate proapoptotic targets such as BID (3). Of note, BAP31 itself is a preferred substrate of caspase-8 and its cleavage contributes directly to apoptosis progression (12). Recruitment of procaspase-8L to the BAP complex, therefore, may also place the enzyme at an important site of action for caspase-8.
Acknowledgments
We are grateful to W.-C. Yeh for Fadd-null cells, P. Krammer, D. Nicholson, and M. Peter for antibodies, J. Mathai for KB cell PolyA+ mRNA, and M. Gravel and J. Pelletier for advice. D.G.B. is the recipient of a Canadian Institutes of Health Research doctoral research award. This work was supported by the National Cancer Institute of Canada, the Canadian Institutes of Health Research, the Deutsche Forschungsgemeinschaft through SFB 388, and by the Leibniz program.
Abbreviations
- DISC
death-inducing signaling complex
- ER
endoplasmic reticulum
- ES
embryonic stem
- HA
hemagglutinin
- LM
light membrane
- Nex
N-terminal extension
- RACE
rapid amplification of cDNA ends
- FADD
Fas-associated death domain
Footnotes
References
- 1.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw W C, Martins L M, Kaufmann S H. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 3.Korsmeyer S J, Wei M C, Saito M, Weiler S, Oh K J, Schlesinger P H. Cell Death Differ. 2000;12:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 4.Kim K M, Adachi T, Nielsen P J, Terashima M, Lamers M C, Kohler G, Reth M. EMBO J. 1994;13:3793–3800. doi: 10.1002/j.1460-2075.1994.tb06690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosser J, Sarde C O, Vicaire S, Yates J R, Mandel J L. Genomics. 1994;22:469–471. doi: 10.1006/geno.1994.1413. [DOI] [PubMed] [Google Scholar]
- 6.Adachi T, Schamel W W, Kim K M, Watanabe T, Becker B, Nielsen P J, Reth M. EMBO J. 1996;15:1534–1541. [PMC free article] [PubMed] [Google Scholar]
- 7.Annaert W G, Becker B, Kistner U, Reth M, Jahn R. J Cell Biol. 1997;139:1397–1410. doi: 10.1083/jcb.139.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng F W, Nguyen M, Kwan T, Branton P E, Nicholson D W, Cromlish J A, Shore G C. J Cell Biol. 1997;139:327–338. doi: 10.1083/jcb.139.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng F W, Shore G C. J Biol Chem. 1998;273:3140–3143. doi: 10.1074/jbc.273.6.3140. [DOI] [PubMed] [Google Scholar]
- 10.Scaffidi C, Medema J P, Krammer P H, Peter M E. J Biol Chem. 1997;272:26953–26958. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen M, Branton P E, Roy S, Nicholson D W, Alnemri E S, Yeh W C, Mak T W, Shore G C. J Biol Chem. 1998;273:33099–33102. doi: 10.1074/jbc.273.50.33099. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen M, Breckenridge D G, Ducret A, Shore G C. Mol Cell Biol. 2000;20:6731–6740. doi: 10.1128/mcb.20.18.6731-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medema J P, Scaffidi C, Kischkel F C, Shevchenko A, Mann M, Krammer P H, Peter M E. EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goping I S, Gross A, Lavoie J N, Nguyen M, Jemmerson R, Roth K, Korsmeyer S J, Shore G C. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd S E, Howe J A, Mymryk J S, Bayley S T. J Virol. 1993;68:2944–2949. doi: 10.1128/jvi.67.5.2944-2949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulakia C A, Chen G, Ng F W, Teodoro J G, Branton P E, Nicholson D W, Poirier G G, Shore G C. Oncogene. 1996;12:529–535. [PubMed] [Google Scholar]
- 17.Grenet J, Teitz T, Wei T, Valentine V, Kidd V J. Gene. 1999;226:225–232. doi: 10.1016/s0378-1119(98)00565-4. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh W C, Pompa J L, McCurrach M E, Shu H B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry W S, Lowe S W, Goeddel D V, Mak T W. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 21.Lowe S W. Endocr Relat Cancer. 1999;6:45–48. doi: 10.1677/erc.0.0060045. [DOI] [PubMed] [Google Scholar]
- 22.Breckenridge D G, Shore G C. Crit Rev Eukaryotic Gene Expression. 2001;10:273–280. doi: 10.1615/critreveukargeneexpr.v10.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 23.Ferri K F, Kroemer G. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 24.Stegh A H, Herrmann H, Lampel S, Weisenberger D, Andra K, Seper M, Wiche G, Krammer P H, Peter M E. Mol Cell Biol. 2000;20:5665–5679. doi: 10.1128/mcb.20.15.5665-5679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]