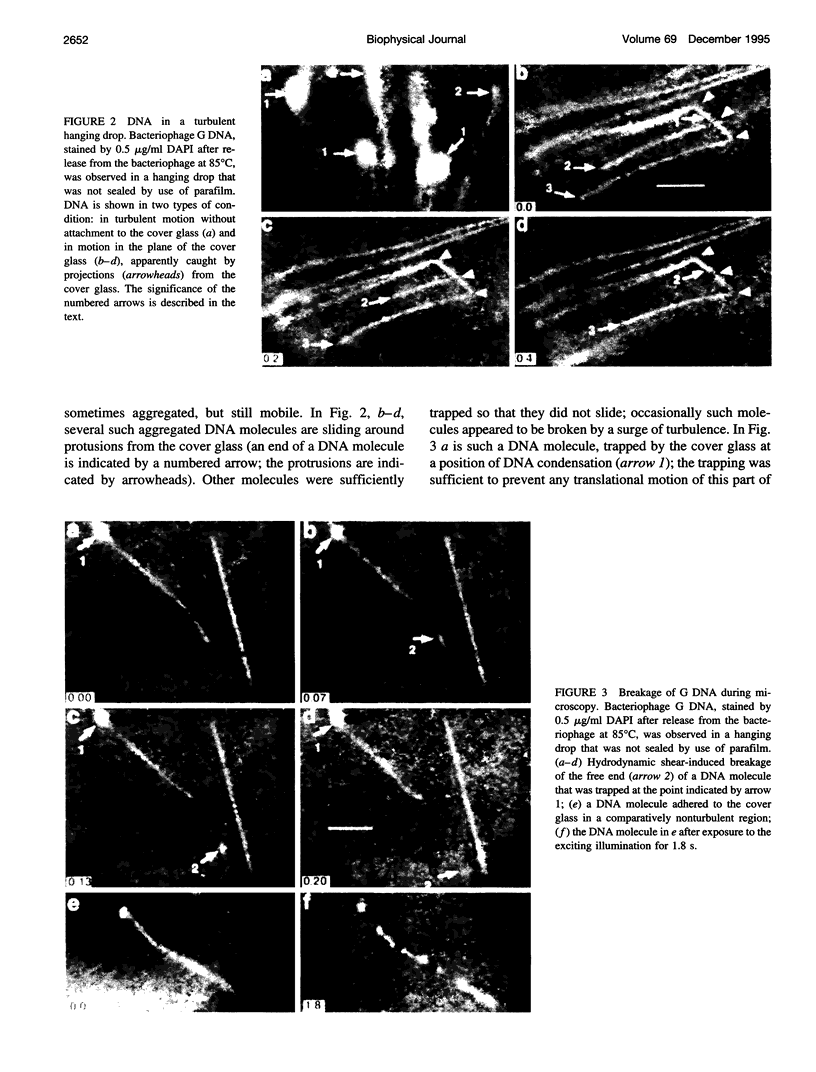
Abstract
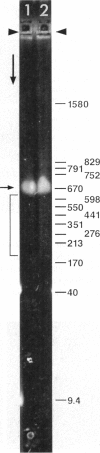
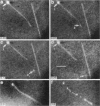
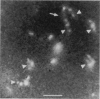
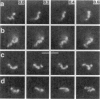
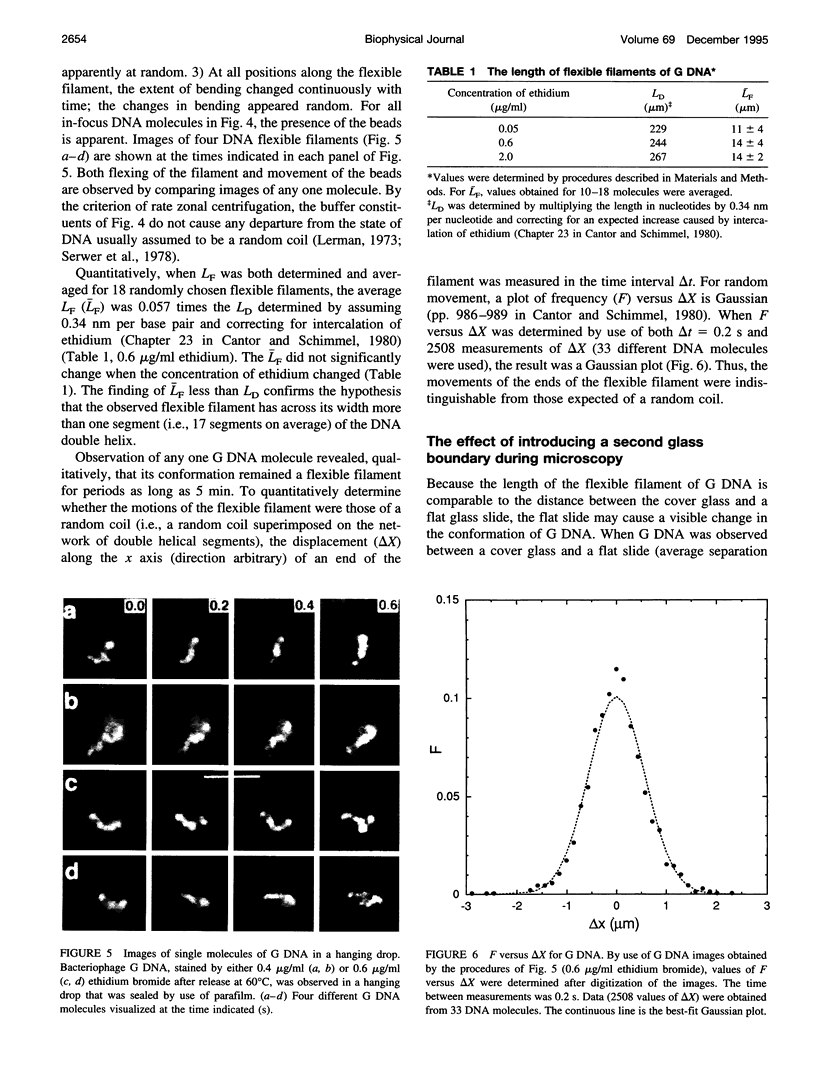
Although its conformation has not been observed directly, double-stranded DNA in solution is usually assumed to be randomly coiled at the level of the DNA double helix. By video light microscopy of ethidium-stained DNA at equilibrium in a nonturbulent hanging drop, in the present study, the 670 kb linear bacteriophage G DNA is found to form a flexible filament that has on average 17 double helical segments across its width. This flexible filament 1) has both asymmetry and dimensions expected of a random coil and 2) has ends that move according to the statistics expected of a random walk. After unraveling the flexible filament-associated DNA double helix near the surface of a hanging drop, recompaction occurs without perceptible rotation of the DNA. Both conformational change and intermolecular tangling of the DNA are observed when G DNA undergoes nondiffusive motion in a hanging drop. The characteristics of the G DNA flexible filament are explained by the assumption that the flexible filament is a random coil of double helical segments that are unperturbed by motion of the suspending medium.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arshad M. F., Dunn F. J., Vega R., Valvano J. W., Serwer P. Progress in developing improved programs for pulsed field agarose gel electrophoresis of DNA. Electrophoresis. 1993 Apr;14(4):344–348. doi: 10.1002/elps.1150140158. [DOI] [PubMed] [Google Scholar]
- Bensimon A., Simon A., Chiffaudel A., Croquette V., Heslot F., Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994 Sep 30;265(5181):2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- Bustamante C. Direct observation and manipulation of single DNA molecules using fluorescence microscopy. Annu Rev Biophys Biophys Chem. 1991;20:415–446. doi: 10.1146/annurev.bb.20.060191.002215. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Fangman W. L. Separation of very large DNA molecules by gel electrophoresis. Nucleic Acids Res. 1978 Mar;5(3):653–665. doi: 10.1093/nar/5.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griess G. A., Guiseley K. B., Serwer P. The relationship of agarose gel structure to the sieving of spheres during agarose gel electrophoresis. Biophys J. 1993 Jul;65(1):138–148. doi: 10.1016/S0006-3495(93)81072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griess G. A., Khan S. A., Serwer P. Variation of the permeability of bacteriophage T4: analysis by use of a protein-specific probe for the T4 interior. Biopolymers. 1991 Jan;31(1):11–21. doi: 10.1002/bip.360310103. [DOI] [PubMed] [Google Scholar]
- Gurrieri S., Rizzarelli E., Beach D., Bustamante C. Imaging of kinked configurations of DNA molecules undergoing orthogonal field alternating gel electrophoresis by fluorescence microscopy. Biochemistry. 1990 Apr 3;29(13):3396–3401. doi: 10.1021/bi00465a036. [DOI] [PubMed] [Google Scholar]
- Houseal T. W., Bustamante C., Stump R. F., Maestre M. F. Real-time imaging of single DNA molecules with fluorescence microscopy. Biophys J. 1989 Sep;56(3):507–516. doi: 10.1016/S0006-3495(89)82697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie D., Serwer P. Effects of temperature on excluded volume-promoted cyclization and concatemerization of cohesive-ended DNA longer than 0.04 Mb. Nucleic Acids Res. 1991 Jun 11;19(11):3047–3054. doi: 10.1093/nar/19.11.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S., Morikawa K., Yanagida M. Light microscopic structure of DNA in solution studied by the 4',6-diamidino-2-phenylindole staining method. J Mol Biol. 1981 Oct 25;152(2):501–516. doi: 10.1016/0022-2836(81)90255-2. [DOI] [PubMed] [Google Scholar]
- Rudnick J., Gaspari G. The shapes of random walks. Science. 1987 Jul 24;237(4813):384–389. doi: 10.1126/science.237.4813.384. [DOI] [PubMed] [Google Scholar]
- Saxton M. J. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys J. 1993 Jun;64(6):1766–1780. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Koval M. Conformational dynamics of individual DNA molecules during gel electrophoresis. Nature. 1989 Apr 6;338(6215):520–522. doi: 10.1038/338520a0. [DOI] [PubMed] [Google Scholar]
- Serwer P., Graef P. R., Garrison P. N. Use of ethidium bromide fluorescence enhancement to detect duplex DNA and DNA bacteriophages during zone sedimentation in sucrose gradients: molecular weight of DNA as a function of sedimentation rate. Biochemistry. 1978 Apr 4;17(7):1166–1170. doi: 10.1021/bi00600a005. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Aldridge P. K., Callis J. B. Observation of individual DNA molecules undergoing gel electrophoresis. Science. 1989 Jan 13;243(4888):203–206. doi: 10.1126/science.2911733. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Bendich A. J. Electrophoretic charge density and persistence length of DNA as measured by fluorescence microscopy. 1990 Jul-Aug 5Biopolymers. 29(8-9):1167–1173. doi: 10.1002/bip.360290807. [DOI] [PubMed] [Google Scholar]
- Sobel E. S., Harpst J. A. Effects of Na+ on the persistence length and excluded volume of T7 bacteriophage DNA. Biopolymers. 1991 Nov;31(13):1559–1564. doi: 10.1002/bip.360311311. [DOI] [PubMed] [Google Scholar]
- Yanagida M., Hiraoka Y., Katsura I. Dynamic behaviors of DNA molecules in solution studied by fluorescence microscopy. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):177–187. doi: 10.1101/sqb.1983.047.01.023. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Levene S. D. Problems and prospects in the theory of gel electrophoresis of DNA. Q Rev Biophys. 1992 May;25(2):171–204. doi: 10.1017/s0033583500004662. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Reese H. R. The degradation of T7 DNA in converging flow. Nucleic Acids Res. 1990 Aug 11;18(15):4469–4470. doi: 10.1093/nar/18.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]