Abstract
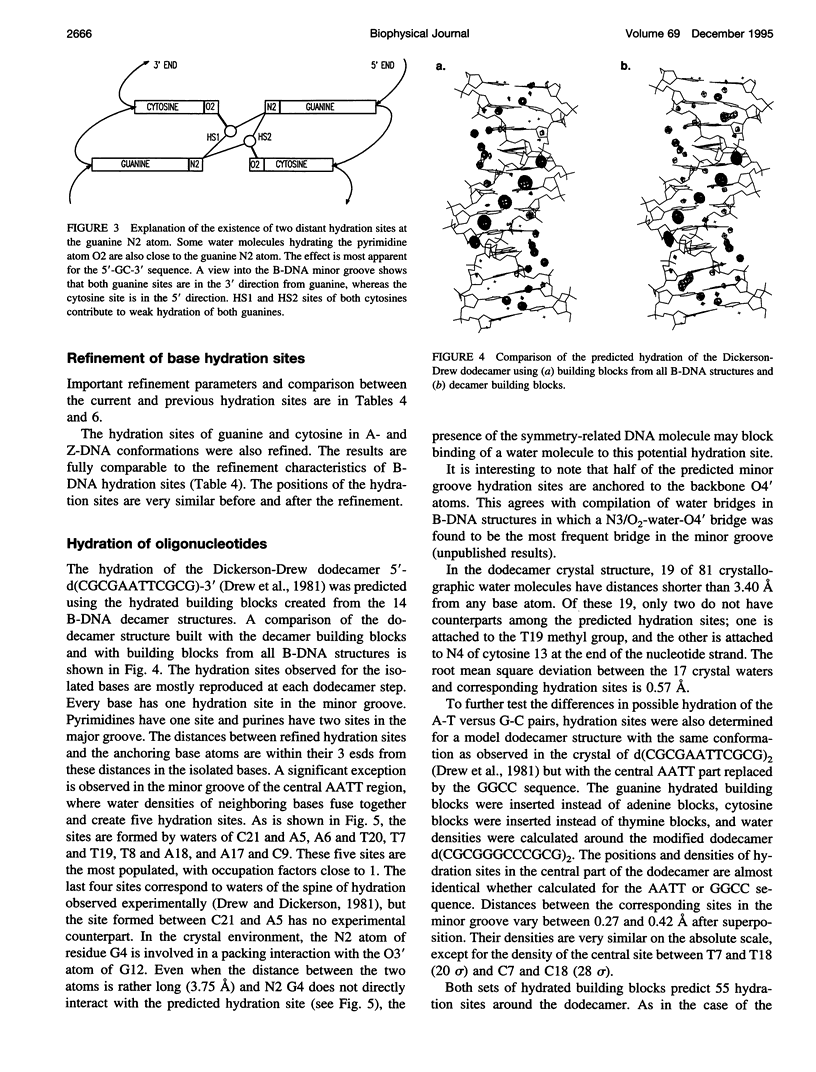
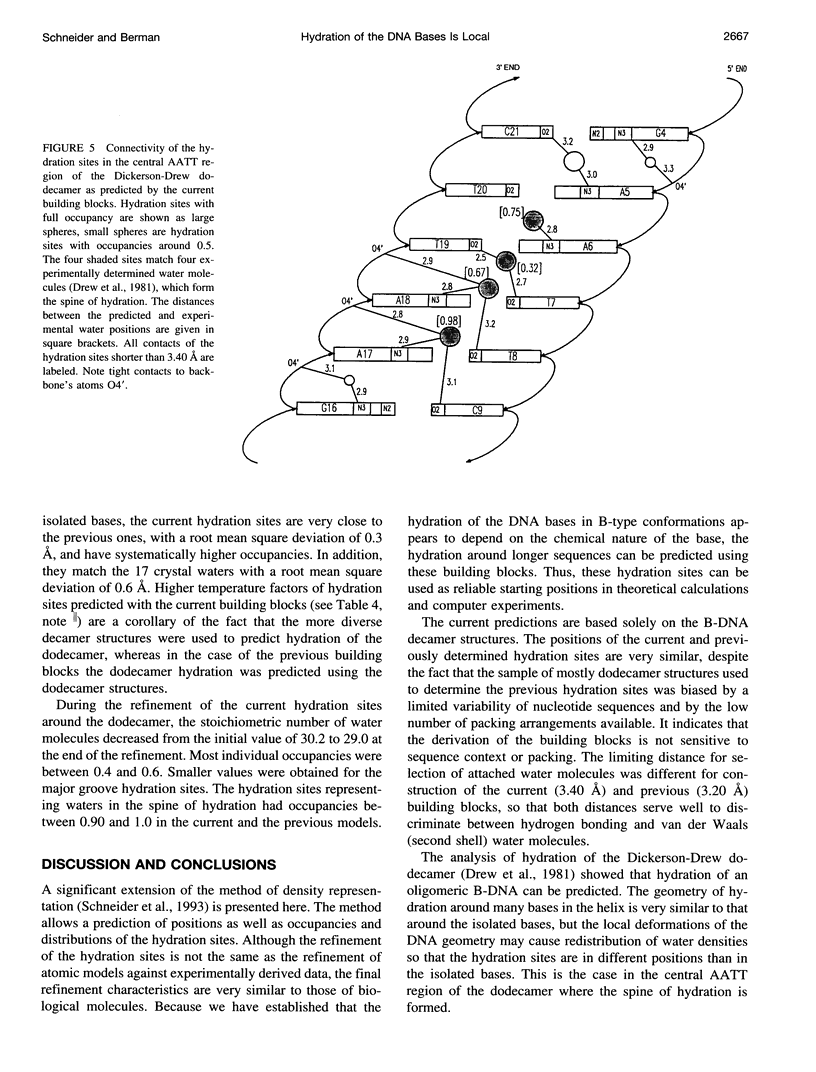
Ordered hydration sites were determined for the nucleotide bases in B-type conformations using the crystal structure data on 14 B-DNA decamer structures. A method of density representation was extended so that positions, occupancies, and distributions of the hydration sites were predicted around a B-DNA double helix by a method analogous to crystallographic refinement. The predicted hydration sites correctly reproduce the main features of hydration around the B-DNA dodecamer. In contrast to the previous observations, the newly available crystal data show the same extent of hydration of guanine and adenine, and of cytosine and thymine.
Full text
PDF

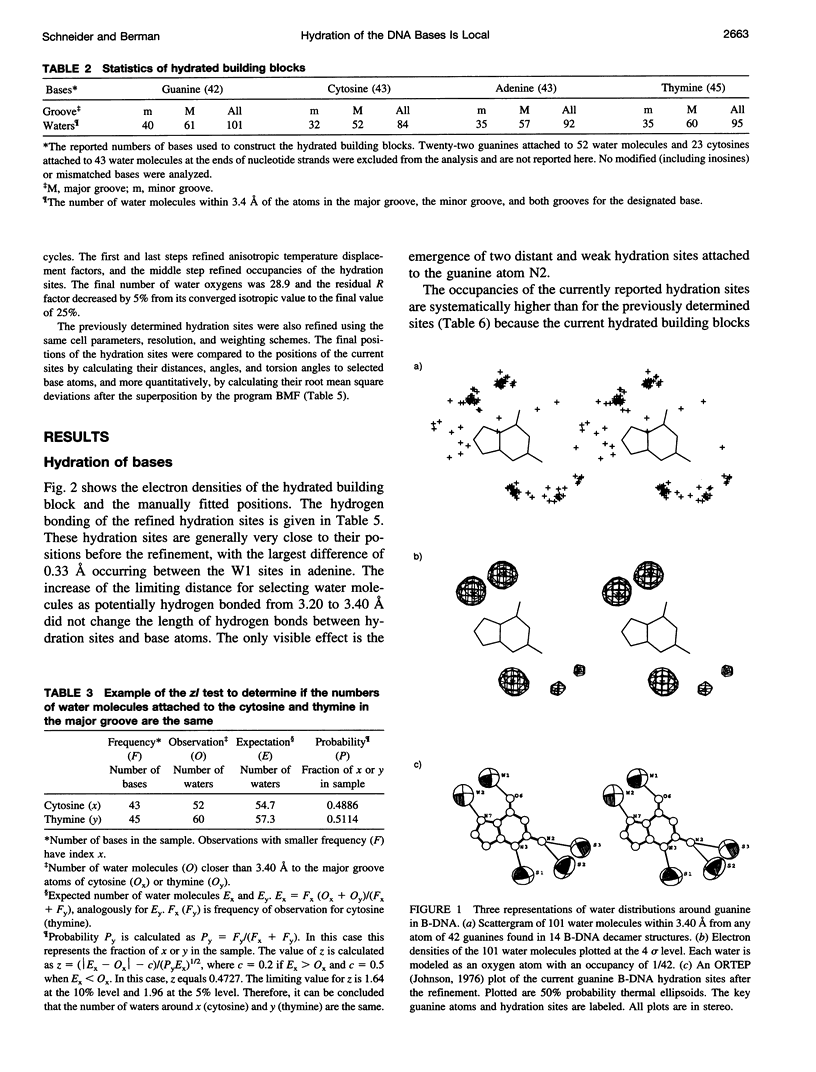
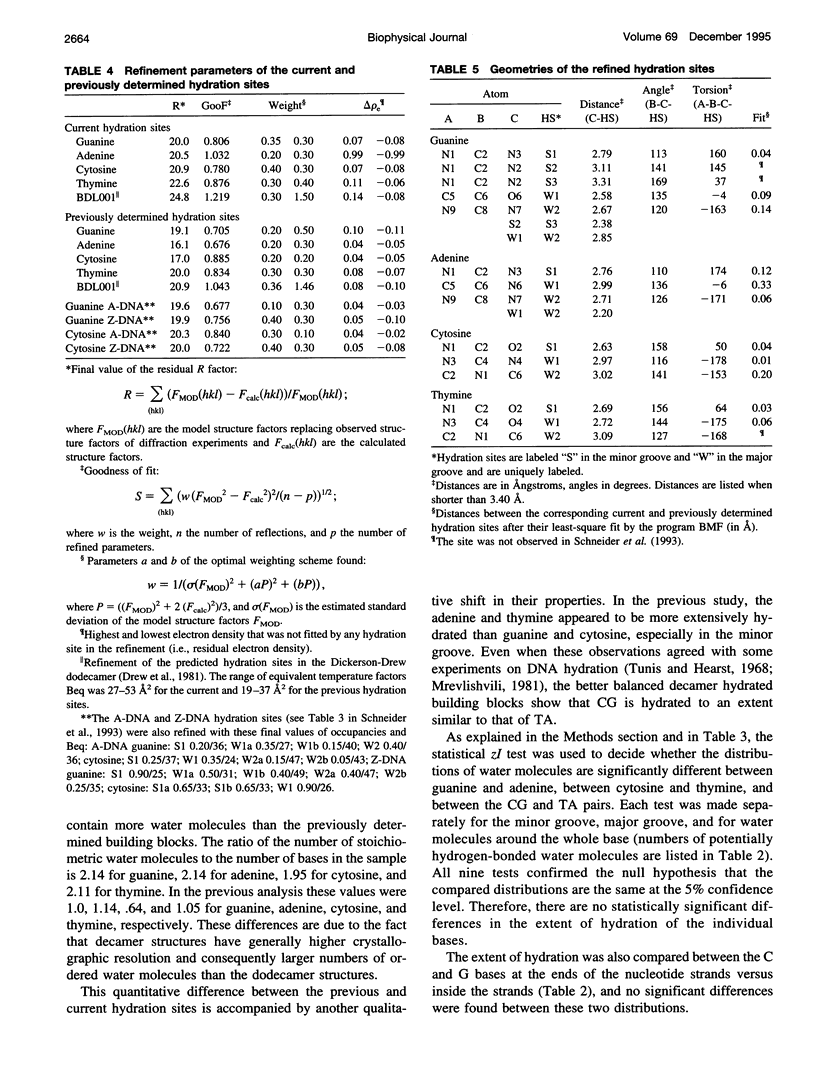
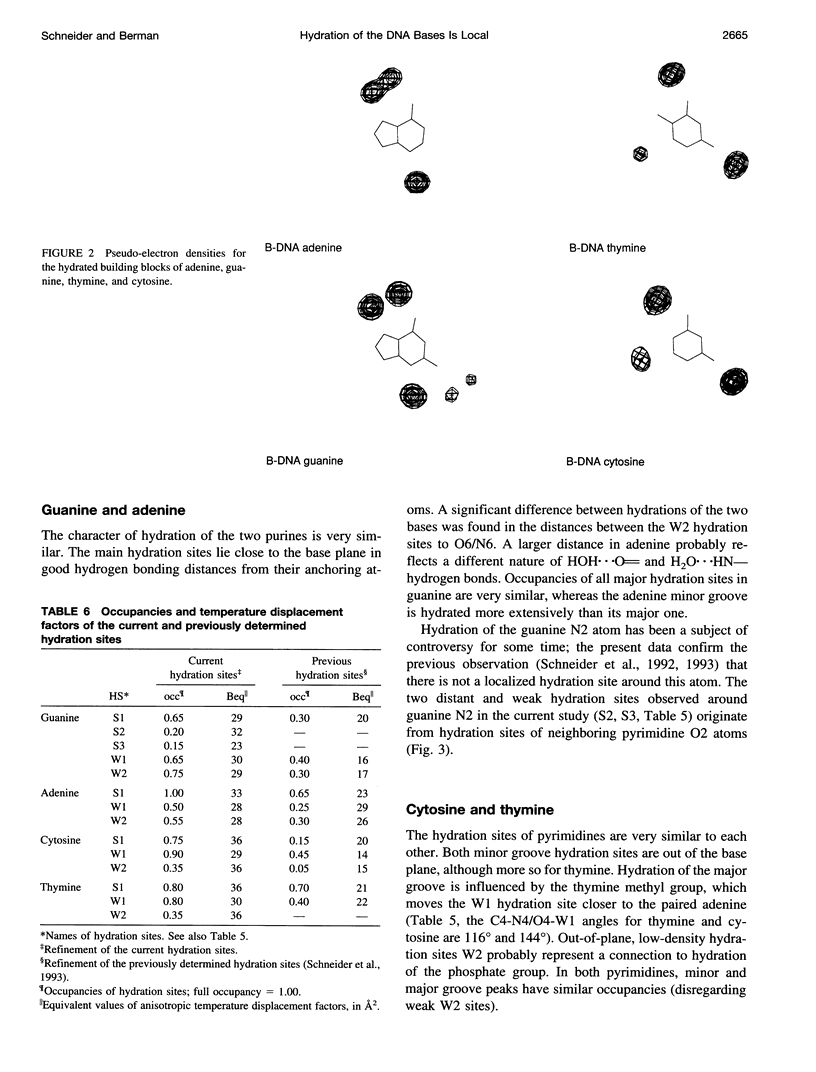


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badger J., Caspar D. L. Water structure in cubic insulin crystals. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):622–626. doi: 10.1073/pnas.88.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikalov I., Grzeskowiak K., Yanagi K., Quintana J., Dickerson R. E. The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn. J Mol Biol. 1993 Jun 5;231(3):768–784. doi: 10.1006/jmbi.1993.1325. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Sowri A., Ginell S., Beveridge D. A systematic study of patterns of hydration in nucleic acids:(I) guanine and cytosine. J Biomol Struct Dyn. 1988 Apr;5(5):1101–1110. doi: 10.1080/07391102.1988.10506451. [DOI] [PubMed] [Google Scholar]
- Chalikian T. V., Sarvazyan A. P., Plum G. E., Breslauer K. J. Influence of base composition, base sequence, and duplex structure on DNA hydration: apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25 degrees C. Biochemistry. 1994 Mar 8;33(9):2394–2401. doi: 10.1021/bi00175a007. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Prohofsky E. W. Synergistic effects in the melting of DNA hydration shell: melting of the minor groove hydration spine in poly(dA).poly(dT) and its effect on base pair stability. Biophys J. 1993 May;64(5):1385–1393. doi: 10.1016/S0006-3495(93)81504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Kopka M. L., Cascio D., Dickerson R. E. Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeskowiak K., Yanagi K., Privé G. G., Dickerson R. E. The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. The effect of base pair reversals. J Biol Chem. 1991 May 15;266(14):8861–8883. doi: 10.2210/pdb1d23/pdb. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C., Bansal M. Double helix conformation, groove dimensions and ligand binding potential of a G/C stretch in B-DNA. EMBO J. 1992 May;11(5):1931–1939. doi: 10.1002/j.1460-2075.1992.tb05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Hahn M. C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J Biol Chem. 1992 Apr 15;267(11):7332–7341. [PubMed] [Google Scholar]
- Langan P., Forsyth V. T., Mahendrasingam A., Pigram W. J., Mason S. A., Fuller W. A high angle neutron fibre diffraction study of the hydration of the A conformation of the DNA double helix. J Biomol Struct Dyn. 1992 Dec;10(3):489–503. doi: 10.1080/07391102.1992.10508664. [DOI] [PubMed] [Google Scholar]
- Lipanov A., Kopka M. L., Kaczor-Grzeskowiak M., Quintana J., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA. Biochemistry. 1993 Feb 9;32(5):1373–1389. doi: 10.1021/bi00056a024. [DOI] [PubMed] [Google Scholar]
- Milton J. G., Galley W. C. Evidence for heterogeneity in DNA-associated solvent mobility from acridine phosphorescence spectra. Biopolymers. 1986 Sep;25(9):1673–1684. doi: 10.1002/bip.360250910. [DOI] [PubMed] [Google Scholar]
- Mrevlishvili G. M. Zavisimost' gidratatsii prirodnykh DNK ot GC-soderzhaniia. Dokl Akad Nauk SSSR. 1981;260(3):761–764. [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Grzeskowiak K., Yanagi K., Dickerson R. E. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. J Mol Biol. 1992 May 20;225(2):379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- Rentzeperis D., Kupke D. W., Marky L. A. Volume changes correlate with entropies and enthalpies in the formation of nucleic acid homoduplexes: differential hydration of A and B conformations. Biopolymers. 1993 Jan;33(1):117–125. doi: 10.1002/bip.360330111. [DOI] [PubMed] [Google Scholar]
- Roe S. M., Teeter M. M. Patterns for prediction of hydration around polar residues in proteins. J Mol Biol. 1993 Jan 20;229(2):419–427. doi: 10.1006/jmbi.1993.1043. [DOI] [PubMed] [Google Scholar]
- Schneider B., Cohen D. M., Schleifer L., Srinivasan A. R., Olson W. K., Berman H. M. A systematic method for studying the spatial distribution of water molecules around nucleic acid bases. Biophys J. 1993 Dec;65(6):2291–2303. doi: 10.1016/S0006-3495(93)81306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B., Cohen D., Berman H. M. Hydration of DNA bases: analysis of crystallographic data. Biopolymers. 1992 Jul;32(7):725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- Schreiner L. J., MacTavish J. C., Pintar M. M., Rupprecht A. NMR spin grouping and correlation exchange analysis. Application to low hydration NaDNA paracrystals. Biophys J. 1991 Jan;59(1):221–234. doi: 10.1016/S0006-3495(91)82213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T., Kuwabara S., Mashimo S., Yagihara S. Dielectric study on hydration of B-, A-, and Z-DNA. Biopolymers. 1990;30(7-8):649–656. doi: 10.1002/bip.360300702. [DOI] [PubMed] [Google Scholar]
- Unis M. J., Hearst J. E. On the hydration of DNA. II. Base composition dependence of the net hydration of DNA. Biopolymers. 1968;6(9):1345–1353. doi: 10.1002/bip.1968.360060909. [DOI] [PubMed] [Google Scholar]
- Yuan H., Quintana J., Dickerson R. E. Alternative structures for alternating poly(dA-dT) tracts: the structure of the B-DNA decamer C-G-A-T-A-T-A-T-C-G. Biochemistry. 1992 Sep 1;31(34):8009–8021. [PubMed] [Google Scholar]