Abstract
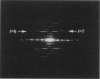
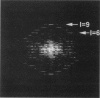
The Escherichia coli RecA protein has served as a model for understanding protein-catalyzed homologous recombination, both in vitro and in vivo. Although RecA proteins have now been sequenced from over 60 different bacteria, almost all of our structural knowledge about RecA has come from studies of the E. coli protein. We have used electron microscopy and image analysis to examine three different structures formed by the RecA protein from the thermophilic bacterium Thermus aquaticus. This protein has previously been shown to catalyze an in vitro strand exchange reaction at an optimal temperature of about 60 degrees C. We show that the active filament formed by the T. aquaticus RecA on DNA in the presence of a nucleotide cofactor is extremely similar to the filament formed by the E. coli protein, including the extension of DNA to a 5.1-A rise per base pair within this filament. This parameter appears highly conserved through evolution, as it has been observed for the eukaryotic RecA analogs as well. We have also characterized bundles of filaments formed by the T. aquaticus RecA in the absence of both DNA and nucleotide cofactor, as well as hexameric rings of the protein formed under all conditions examined. The bundles display a very large plasticity of mass within the RecA filament, as well as showing a polymorphism in filament-filament contacts that may be important to understanding mutations that affect surface residues on the RecA filament.
Full text
PDF

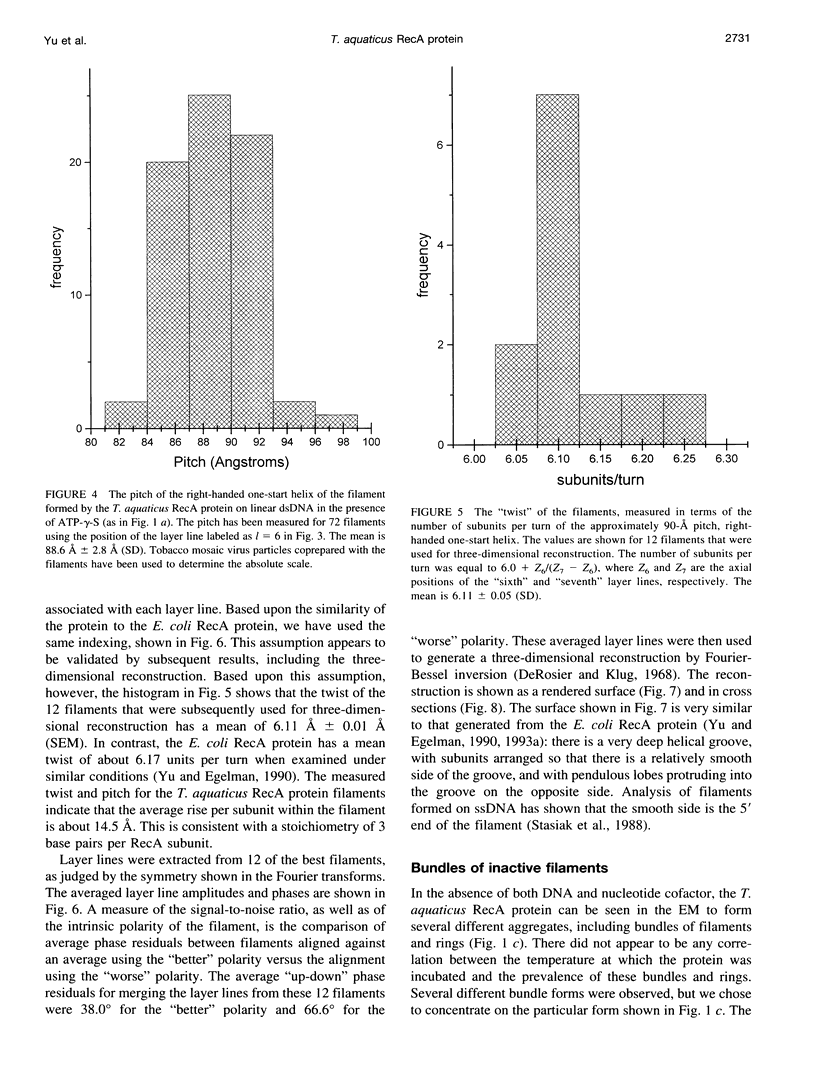
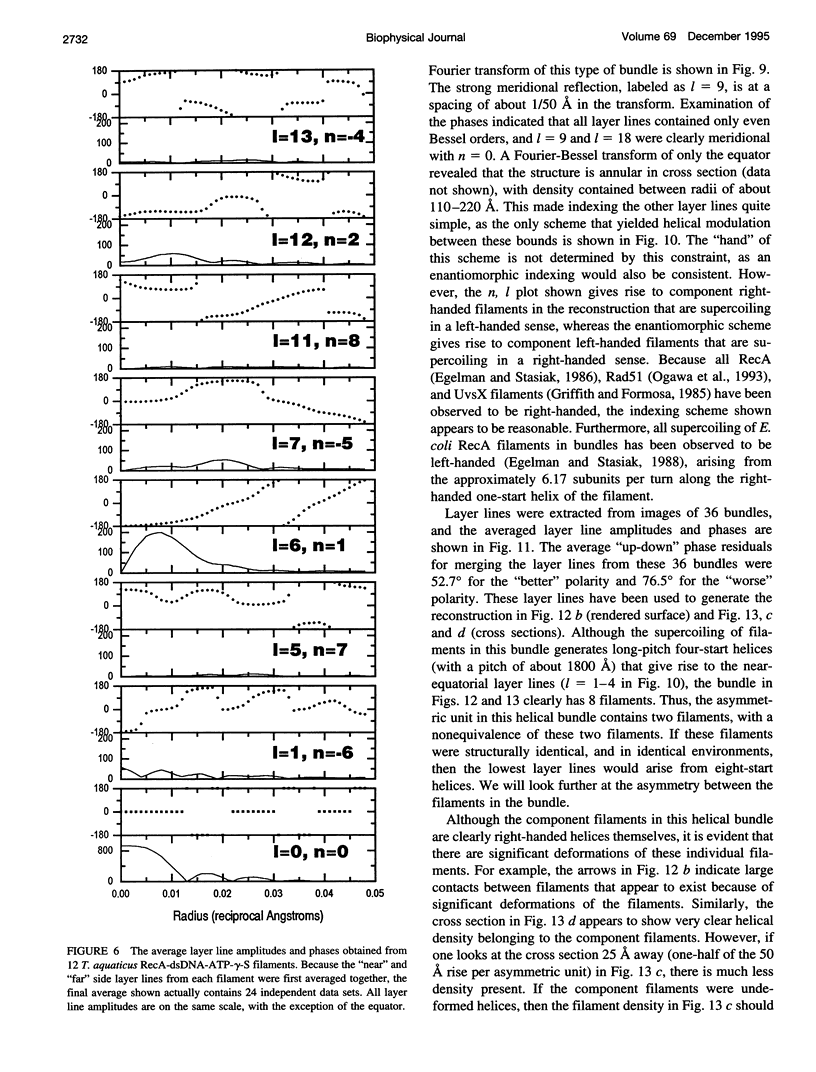
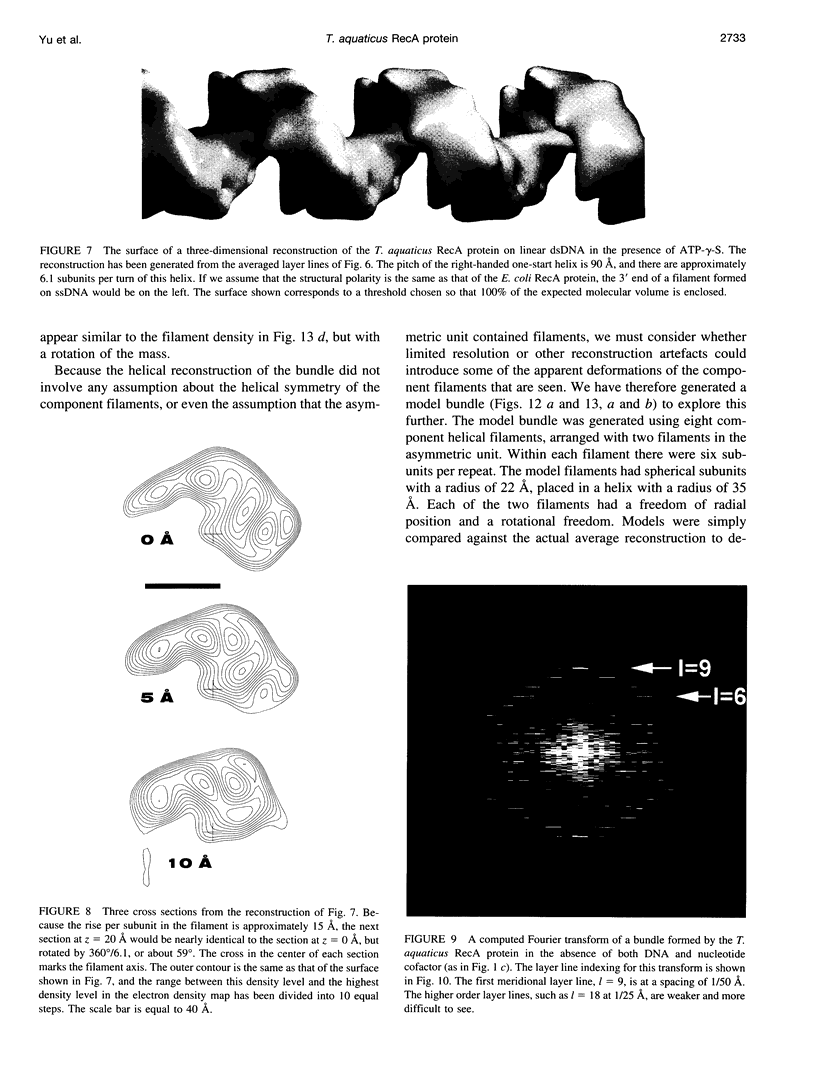
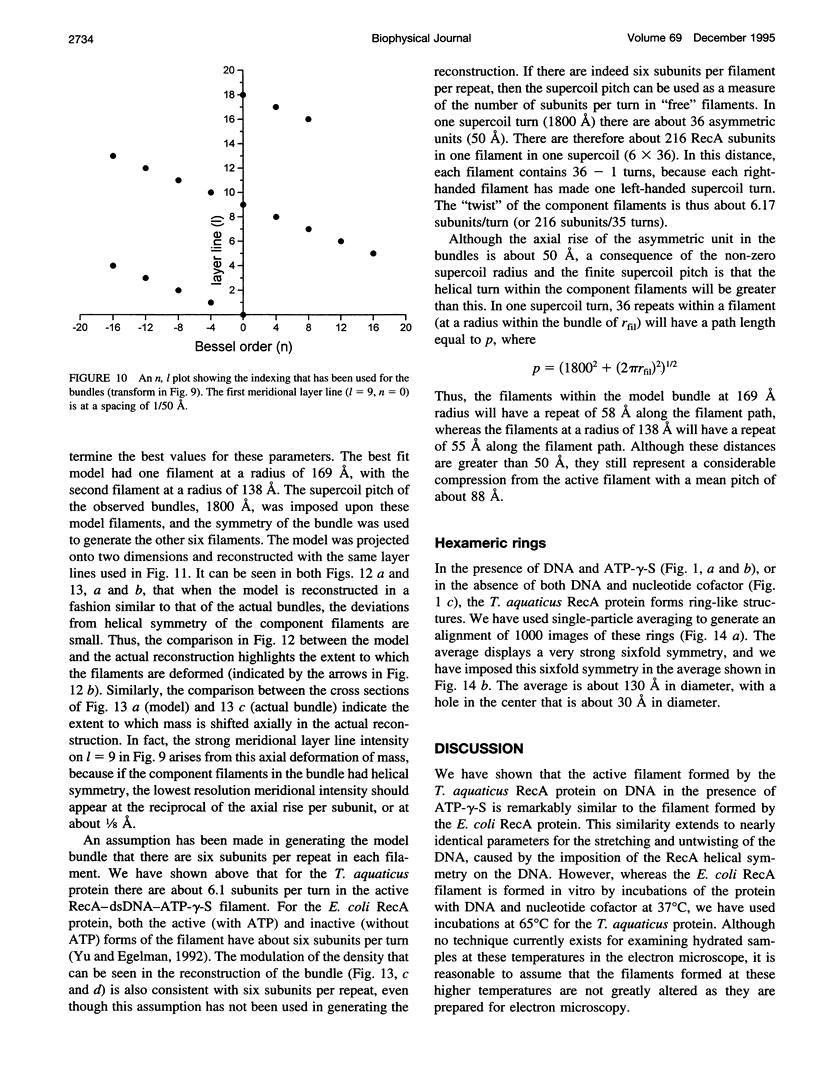


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angov E., Camerini-Otero R. D. The recA gene from the thermophile Thermus aquaticus YT-1: cloning, expression, and characterization. J Bacteriol. 1994 Mar;176(5):1405–1412. doi: 10.1128/jb.176.5.1405-1412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benight A. S., Wilson D. H., Budzynski D. M., Goldstein R. F. Dynamic light scattering investigations of RecA self-assembly and interactions with single strand DNA. Biochimie. 1991 Feb-Mar;73(2-3):143–155. doi: 10.1016/0300-9084(91)90197-9. [DOI] [PubMed] [Google Scholar]
- Benson F. E., Stasiak A., West S. C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994 Dec 1;13(23):5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. L., Zlotnick A., Griffith J. D. RecA protein self-assembly. Multiple discrete aggregation states. J Mol Biol. 1988 Dec 20;204(4):959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- Brenner S. L., Zlotnick A., Stafford W. F., 3rd RecA protein self-assembly. II. Analytical equilibrium ultracentrifugation studies of the entropy-driven self-association of RecA. J Mol Biol. 1990 Dec 20;216(4):949–964. doi: 10.1016/S0022-2836(99)80013-8. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Hsieh P. Parallel DNA triplexes, homologous recombination, and other homology-dependent DNA interactions. Cell. 1993 Apr 23;73(2):217–223. doi: 10.1016/0092-8674(93)90224-e. [DOI] [PubMed] [Google Scholar]
- Egelman E. H. An algorithm for straightening images of curved filamentous structures. Ultramicroscopy. 1986;19(4):367–373. doi: 10.1016/0304-3991(86)90096-3. [DOI] [PubMed] [Google Scholar]
- Egelman E. H., Stasiak A. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J Mol Biol. 1986 Oct 20;191(4):677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- Egelman E. H., Stasiak A. Structure of helical RecA-DNA complexes. II. Local conformational changes visualized in bundles of RecA-ATP gamma S filaments. J Mol Biol. 1988 Mar 20;200(2):329–349. doi: 10.1016/0022-2836(88)90245-8. [DOI] [PubMed] [Google Scholar]
- Egelman E. H., Yu X., Wild R., Hingorani M. M., Patel S. S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Formosa T. The uvsX protein of bacteriophage T4 arranges single-stranded and double-stranded DNA into similar helical nucleoprotein filaments. J Biol Chem. 1985 Apr 10;260(7):4484–4491. [PubMed] [Google Scholar]
- Heuser J., Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J Mol Biol. 1989 Dec 5;210(3):473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Eggleston A. K. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Lee J. W., Cox M. M. Inhibition of recA protein promoted ATP hydrolysis. 1. ATP gamma S and ADP are antagonistic inhibitors. Biochemistry. 1990 Aug 21;29(33):7666–7676. doi: 10.1021/bi00485a016. [DOI] [PubMed] [Google Scholar]
- Lee J. W., Cox M. M. Inhibition of recA protein promoted ATP hydrolysis. 2. Longitudinal assembly and disassembly of recA protein filaments mediated by ATP and ADP. Biochemistry. 1990 Aug 21;29(33):7677–7683. doi: 10.1021/bi00485a017. [DOI] [PubMed] [Google Scholar]
- Liu S. K., Eisen J. A., Hanawalt P. C., Tessman I. recA mutations that reduce the constitutive coprotease activity of the RecA1202(Prtc) protein: possible involvement of interfilament association in proteolytic and recombination activities. J Bacteriol. 1993 Oct;175(20):6518–6529. doi: 10.1128/jb.175.20.6518-6529.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo I. A., Hough P. V., Wall J. S., Dodson M., Dean F. B., Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989 Apr 20;338(6217):658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E. H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993 Mar 26;259(5103):1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Penczek P., Radermacher M., Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992 Jan;40(1):33–53. [PubMed] [Google Scholar]
- Rao B. J., Chiu S. K., Bazemore L. R., Reddy G., Radding C. M. How specific is the first recognition step of homologous recombination? Trends Biochem Sci. 1995 Mar;20(3):109–113. doi: 10.1016/s0968-0004(00)88976-8. [DOI] [PubMed] [Google Scholar]
- Ruigrok R. W., Bohrmann B., Hewat E., Engel A., Kellenberger E., DiCapua E. The inactive form of recA protein: the 'compact' structure. EMBO J. 1993 Jan;12(1):9–16. doi: 10.1002/j.1460-2075.1993.tb05626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Matsuda Y., Ushio N., Ikeo K., Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993 Jul;4(3):239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992 May 1;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Di Capua E., Koller T. Elongation of duplex DNA by recA protein. J Mol Biol. 1981 Sep 25;151(3):557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982 Sep 9;299(5879):185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Egelman E. H., Howard-Flanders P. Structure of helical RecA-DNA complexes. III. The structural polarity of RecA filaments and functional polarity in the RecA-mediated strand exchange reaction. J Mol Biol. 1988 Aug 5;202(3):659–662. doi: 10.1016/0022-2836(88)90293-8. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Tsaneva I. R., West S. C., Benson C. J., Yu X., Egelman E. H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story R. M., Weber I. T., Steitz T. A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992 Jan 23;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Weinstock G. M., McEntee K., Lehman I. R. Interaction of the recA protein of Escherichia coli with adenosine 5'-O-(3-thiotriphosphate). J Biol Chem. 1981 Aug 25;256(16):8850–8855. [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- Williams R. C., Spengler S. J. Fibers of RecA protein and complexes of RecA protein and single-stranded phi X174 DNA as visualized by negative-stain electron microscopy. J Mol Biol. 1986 Jan 5;187(1):109–118. doi: 10.1016/0022-2836(86)90410-9. [DOI] [PubMed] [Google Scholar]
- Yu X., Egelman E. H. DNA conformation induced by the bacteriophage T4 UvsX protein appears identical to the conformation induced by the Escherichia coli RecA protein. J Mol Biol. 1993 Jul 5;232(1):1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- Yu X., Egelman E. H. Image analysis reveals that Escherichia coli RecA protein consists of two domains. Biophys J. 1990 Mar;57(3):555–566. doi: 10.1016/S0006-3495(90)82571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Egelman E. H. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992 Sep 5;227(1):334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- Yu X., Egelman E. H. The LexA repressor binds within the deep helical groove of the activated RecA filament. J Mol Biol. 1993 May 5;231(1):29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]