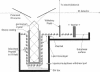
Abstract
Polarized attenuated total internal reflectance techniques were applied to study the infrared dichroism of the amide I transition moment in two membrane-bound peptides that are known to form oriented transmembrane helices: gramicidin A in a supported phospholipid monolayer and Ac-Lys2-Leu24-Lys2-amide (L24) in oriented multibilayers. These studies were performed to test the ability of these techniques to determine the orientation of these peptides, to verify the value of optical parameters used to calculate electric field strengths, to examine the common assumptions regarding the amide I transition moment orientation, and to ascertain the effect of surface imperfections on molecular disorder. The two peptides exhibit marked differences in the shape and frequency of their amide I absorption bands. Yet both peptides are highly ordered and oriented with their helical axes perpendicular to the membrane surface. In the alpha-helix formed by L24, there is evidence for a mode with type E1 symmetry contributing to amide I, and the amide I transition moment must be more closely aligned with the peptide C=O (< 34 degrees) than earlier studies have suggested. These results indicate that long-standing assumptions about the orientation of amide I in a peptide require some revision, but that in general, infrared spectroscopy yields reliable information about the orientation of membrane-bound helical peptides.
Full text
PDF
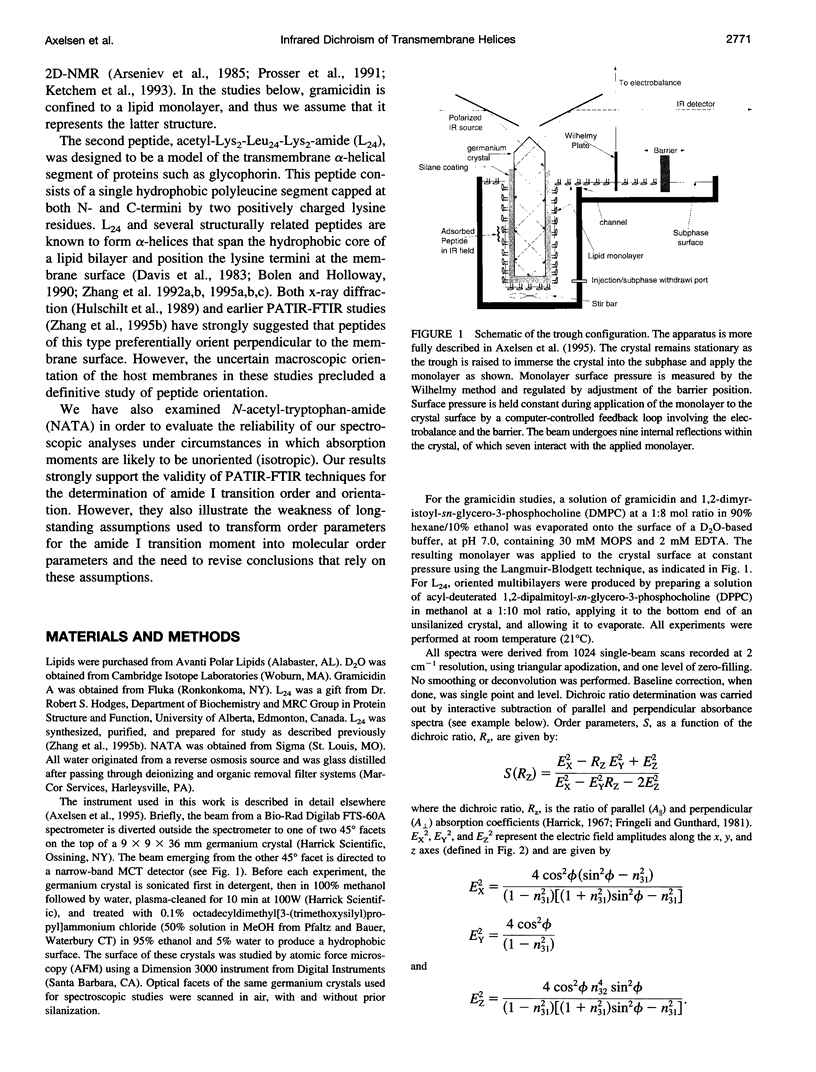
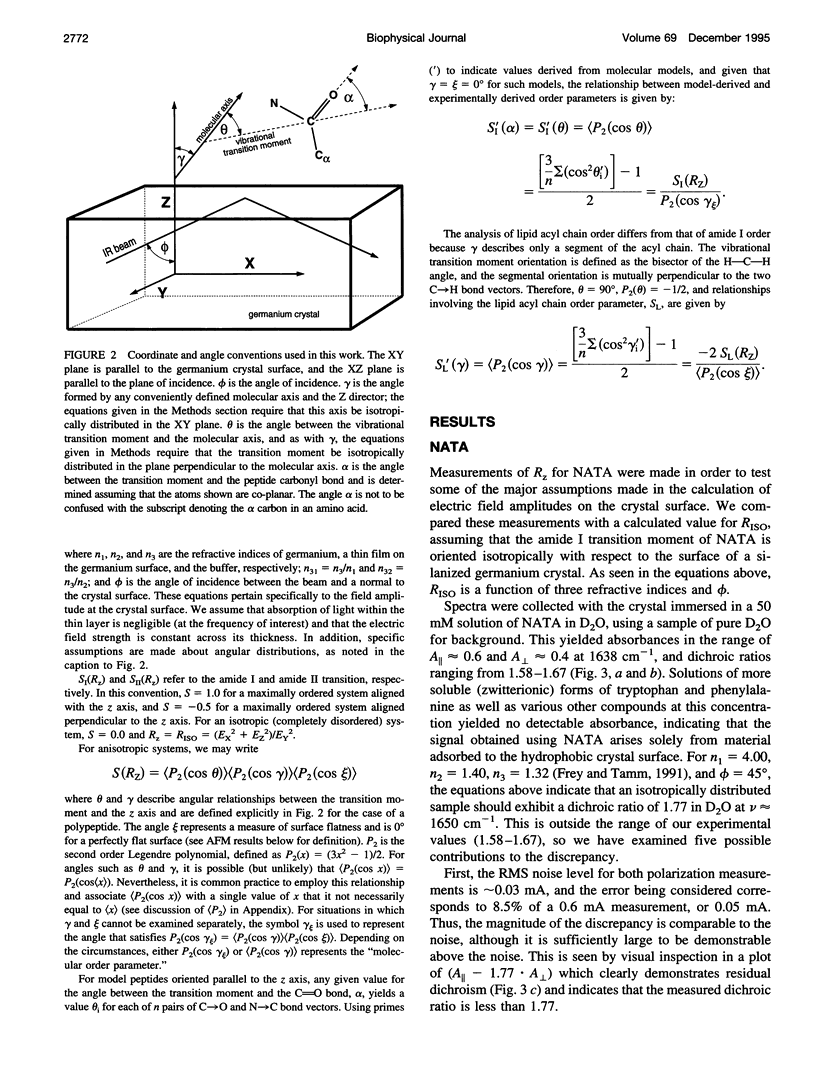
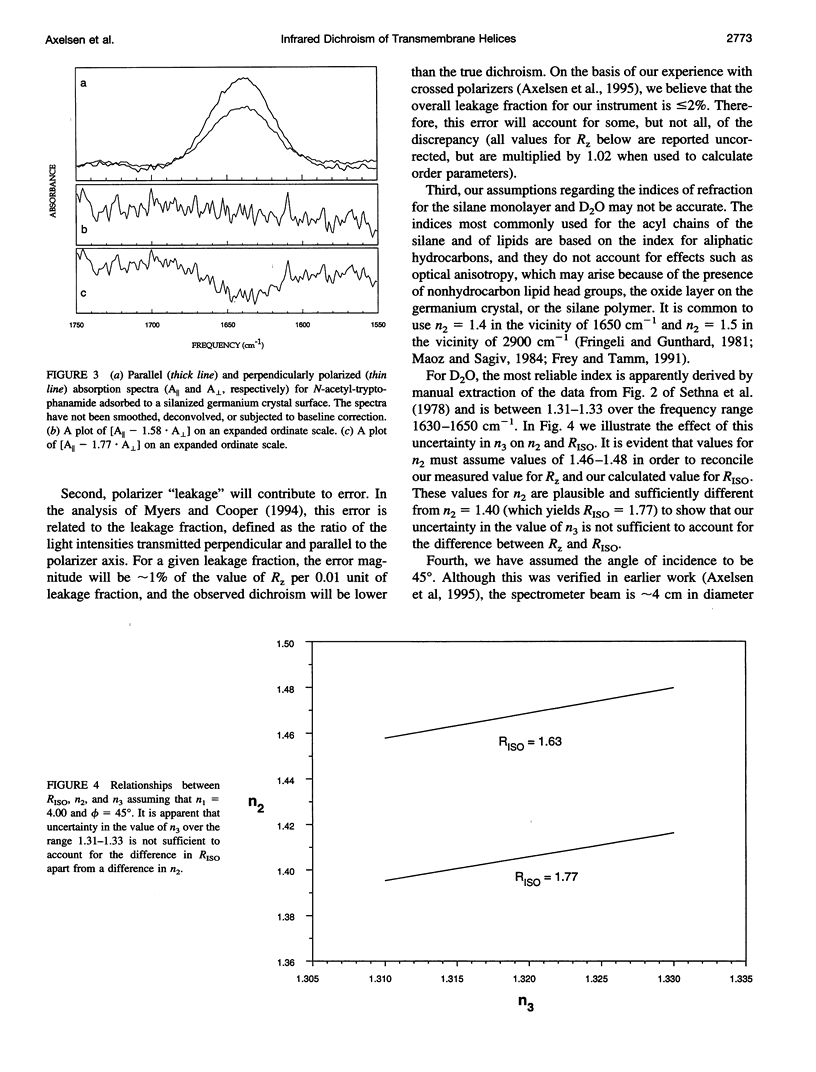



Images in this article
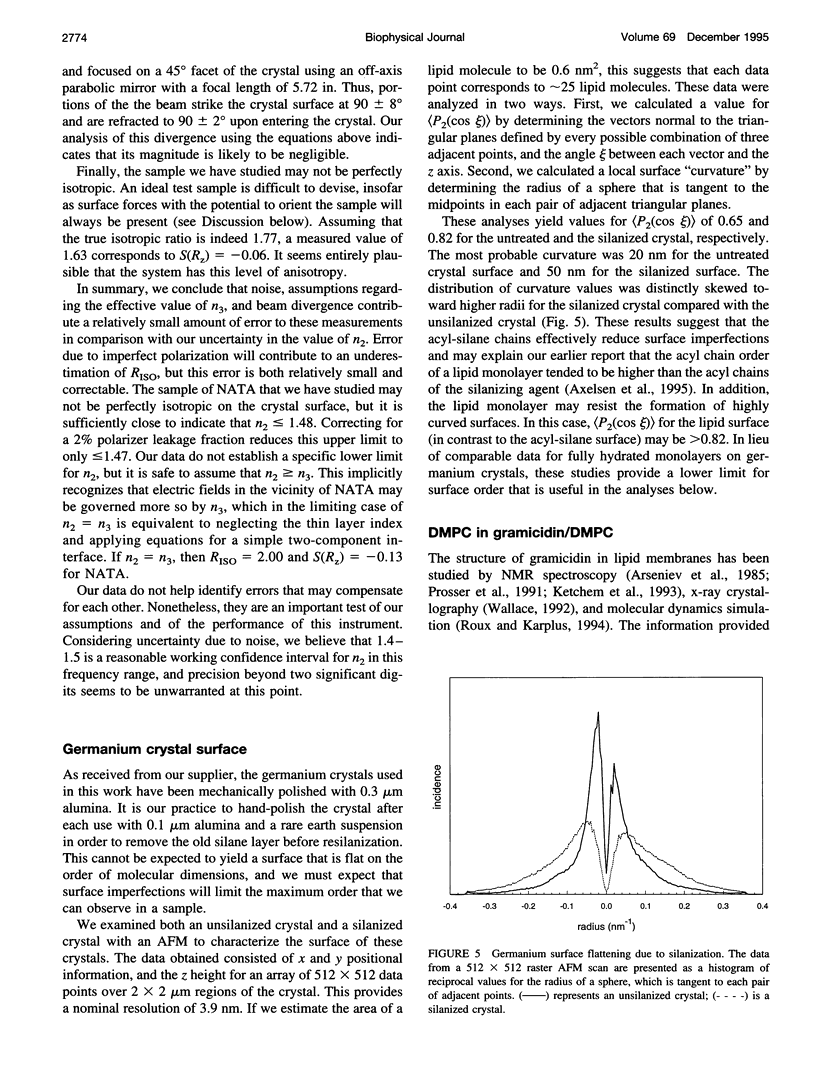
Selected References
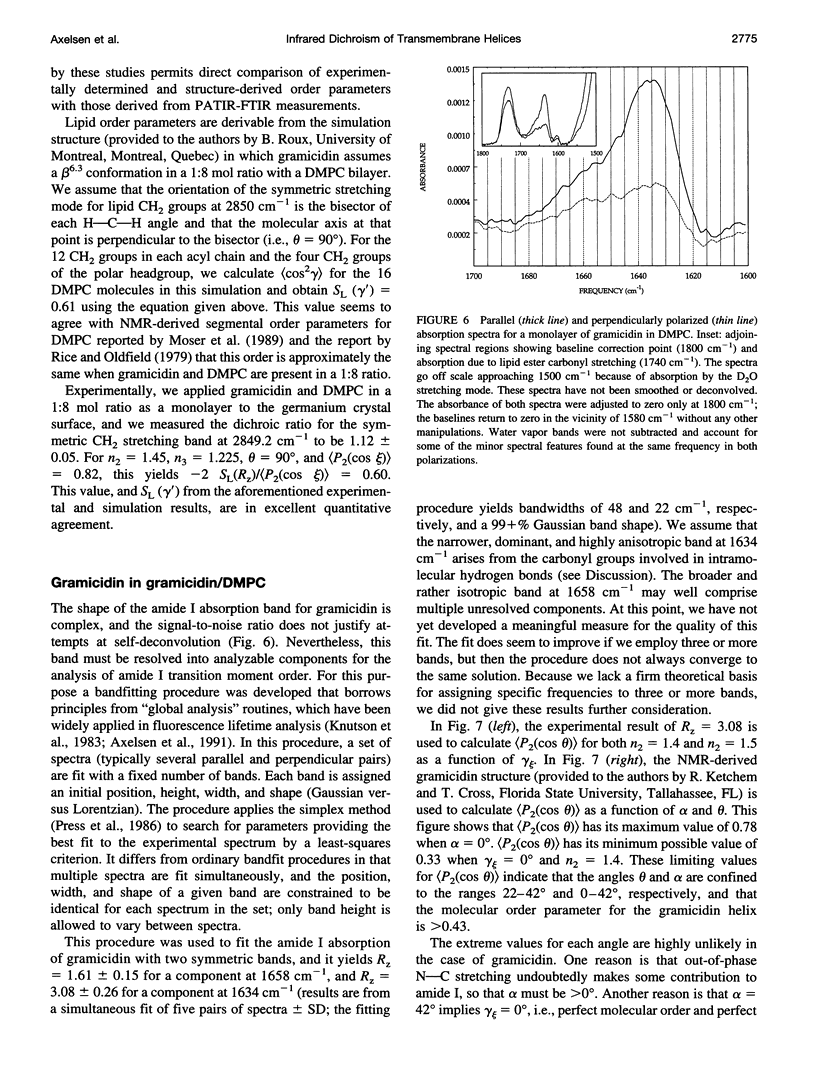
These references are in PubMed. This may not be the complete list of references from this article.
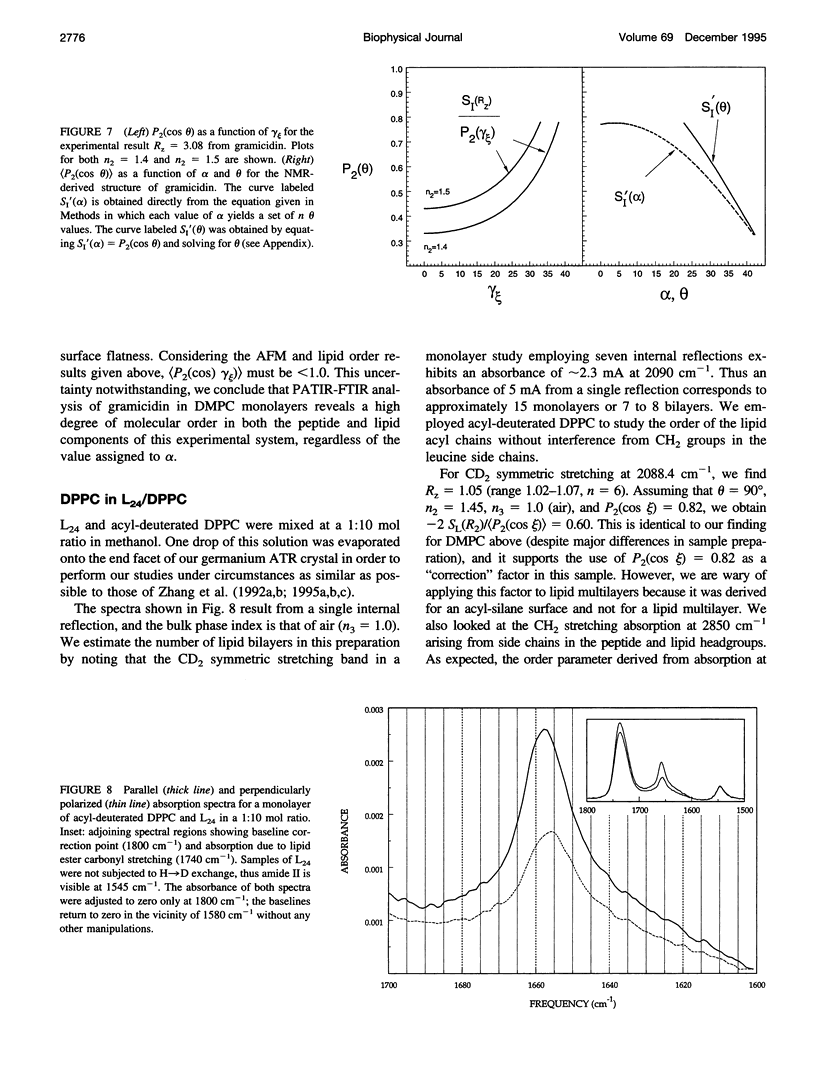
- Arrondo J. L., Muga A., Castresana J., Goñi F. M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog Biophys Mol Biol. 1993;59(1):23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
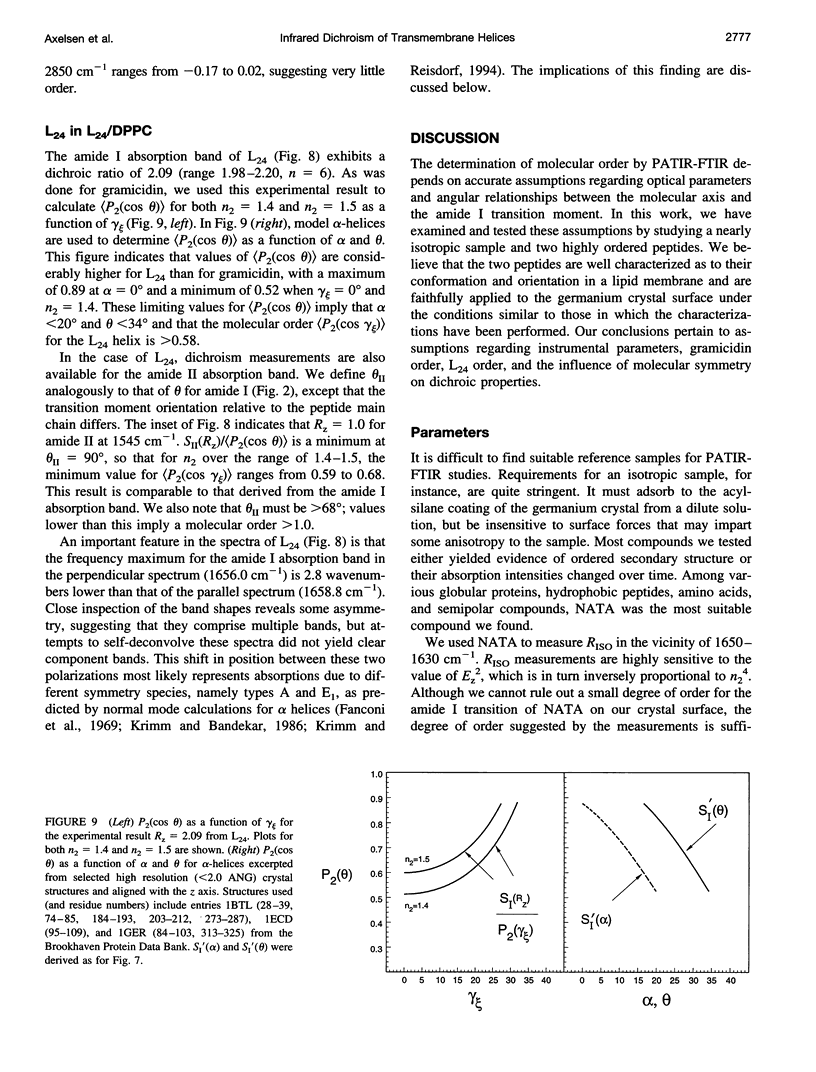
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Axelsen P. H., Bajzer Z., Prendergast F. G., Cottam P. F., Ho C. Resolution of fluorescence intensity decays of the two tryptophan residues in glutamine-binding protein from Escherichia coli using single tryptophan mutants. Biophys J. 1991 Sep;60(3):650–659. doi: 10.1016/S0006-3495(91)82094-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRADBURY E. M., BROWN L., DOWNIE A. R., ELLIOTT A., FRASER R. D., HANBY W. E. The structure of the omegaform of poly-Beta-benzyl-L-aspartate. J Mol Biol. 1962 Aug;5:230–247. doi: 10.1016/s0022-2836(62)80086-2. [DOI] [PubMed] [Google Scholar]
- Bolen E. J., Holloway P. W. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 1990 Oct 16;29(41):9638–9643. doi: 10.1021/bi00493a019. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Carpenter B. G., Stephens R. M. Conformational studies of polymers and copolymers of L-aspartate ester. II. Infrared studies and the factors involved in the formation of the omega-helix. Biopolymers. 1968;6(7):905–915. doi: 10.1002/bip.1968.360060702. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Rothschild K. J. Fourier transform infrared techniques for probing membrane protein structure. Annu Rev Biophys Biophys Chem. 1988;17:541–570. doi: 10.1146/annurev.bb.17.060188.002545. [DOI] [PubMed] [Google Scholar]
- Deinum G., van Langen H., van Ginkel G., Levine Y. K. Molecular order and dynamics in planar lipid bilayers: effects of unsaturation and sterols. Biochemistry. 1988 Feb 9;27(3):852–860. doi: 10.1021/bi00403a003. [DOI] [PubMed] [Google Scholar]
- FRASER R. D. B., PRICE W. C. Infra-red dichroism and protein structure. Nature. 1952 Sep 20;170(4325):490–491. doi: 10.1038/170490a0. [DOI] [PubMed] [Google Scholar]
- Fanconi B., Tomlinson B., Nafie L. A., Small W., Peticolas W. L. Polarized laser Raman studies of biological polymers. J Chem Phys. 1969 Nov 1;51(9):3993–4005. doi: 10.1063/1.1672620. [DOI] [PubMed] [Google Scholar]
- Frey S., Tamm L. K. Orientation of melittin in phospholipid bilayers. A polarized attenuated total reflection infrared study. Biophys J. 1991 Oct;60(4):922–930. doi: 10.1016/S0006-3495(91)82126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huschilt J. C., Millman B. M., Davis J. H. Orientation of alpha-helical peptides in a lipid bilayer. Biochim Biophys Acta. 1989 Feb 13;979(1):139–141. doi: 10.1016/0005-2736(89)90534-8. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- Moser M., Marsh D., Meier P., Wassmer K. H., Kothe G. Chain configuration and flexibility gradient in phospholipid membranes. Comparison between spin-label electron spin resonance and deuteron nuclear magnetic resonance, and identification of new conformations. Biophys J. 1989 Jan;55(1):111–123. doi: 10.1016/S0006-3495(89)82784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabedryk E., Gingold M. P., Breton J. Orientation of gramicidin A transmembrane channel. Infrared dichroism study of gramicidin in vesicles. Biophys J. 1982 Jun;38(3):243–249. doi: 10.1016/S0006-3495(82)84555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of gramicidin A. I. Normal mode analysis. Biophys J. 1986 Jun;49(6):1131–1145. doi: 10.1016/S0006-3495(86)83742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of gramicidin A. II. Vibrational spectra. Biophys J. 1986 Jun;49(6):1147–1154. doi: 10.1016/S0006-3495(86)83743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser R. S., Davis J. H., Dahlquist F. W., Lindorfer M. A. 2H nuclear magnetic resonance of the gramicidin A backbone in a phospholipid bilayer. Biochemistry. 1991 May 14;30(19):4687–4696. doi: 10.1021/bi00233a008. [DOI] [PubMed] [Google Scholar]
- Rice D., Oldfield E. Deuterium nuclear magnetic resonance studies of the interaction between dimyristoylphosphatidylcholine and gramicidin A'. Biochemistry. 1979 Jul 24;18(15):3272–3279. doi: 10.1021/bi00582a012. [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Clark N. A. Polarized infrared spectroscopy of oriented purple membrane. Biophys J. 1979 Mar;25(3):473–487. doi: 10.1016/S0006-3495(79)85317-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Molecular dynamics simulations of the gramicidin channel. Annu Rev Biophys Biomol Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Jonas R., Braiman M., Bormann B. J. Structure and orientation of the transmembrane domain of glycophorin A in lipid bilayers. Biochemistry. 1994 May 24;33(20):6334–6341. doi: 10.1021/bi00186a037. [DOI] [PubMed] [Google Scholar]
- Susi H., Timasheff S. N., Stevens L. Infrared spectra and protein conformations in aqueous solutions. I. The amide I band in H2O and D2O solutions. J Biol Chem. 1967 Dec 10;242(23):5460–5466. [PubMed] [Google Scholar]
- Wallace B. A. Crystallographic studies of a transmembrane ion channel, gramicidin A. Prog Biophys Mol Biol. 1992;57(2):59–69. doi: 10.1016/0079-6107(92)90004-p. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Henry G. D., Sykes B. D., Hodges R. S., McElhaney R. N. Peptide models of helical hydrophobic transmembrane segments of membrane proteins. 1. Studies of the conformation, intrabilayer orientation, and amide hydrogen exchangeability of Ac-K2-(LA)12-K2-amide. Biochemistry. 1995 Feb 21;34(7):2348–2361. doi: 10.1021/bi00007a031. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N. FTIR spectroscopic studies of the conformation and amide hydrogen exchange of a peptide model of the hydrophobic transmembrane alpha-helices of membrane proteins. Biochemistry. 1992 Nov 24;31(46):11572–11578. doi: 10.1021/bi00161a041. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N. Interaction of a peptide model of a hydrophobic transmembrane alpha-helical segment of a membrane protein with phosphatidylcholine bilayers: differential scanning calorimetric and FTIR spectroscopic studies. Biochemistry. 1992 Nov 24;31(46):11579–11588. doi: 10.1021/bi00161a042. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N. Interaction of a peptide model of a hydrophobic transmembrane alpha-helical segment of a membrane protein with phosphatidylethanolamine bilayers: differential scanning calorimetric and Fourier transform infrared spectroscopic studies. Biophys J. 1995 Mar;68(3):847–857. doi: 10.1016/S0006-3495(95)80261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N. Peptide models of helical hydrophobic transmembrane segments of membrane proteins. 2. Differential scanning calorimetric and FTIR spectroscopic studies of the interaction of Ac-K2-(LA)12-K2-amide with phosphatidylcholine bilayers. Biochemistry. 1995 Feb 21;34(7):2362–2371. doi: 10.1021/bi00007a032. [DOI] [PubMed] [Google Scholar]