Abstract
The misregulation of programmed cell death, or apoptosis, contributes to the pathogenesis of many diseases. We used Nomarski microscopy to screen for mutants containing refractile cell corpses in a C. elegans strain in which all programmed cell death is blocked and such corpses are absent. We isolated a mutant strain that accumulates refractile bodies resembling irregular cell corpses. We rescued this mutant phenotype with the C. elegans mucolipidosis type IV (ML-IV) homolog, the recently identified cup-5 (coelomocyte-uptake defective) gene. ML-IV is a human autosomal recessive lysosomal storage disease characterized by psychomotor retardation and ophthalmological abnormalities. Our null mutations in cup-5 cause maternal-effect lethality. In addition, cup-5 mutants contain excess lysosomes in many and possibly all cell types and contain lamellar structures similar to those observed in ML-IV cell lines. The human ML-IV gene is capable of rescuing both the maternal-effect lethality and the lysosome-accumulation abnormality of cup-5 mutants. cup-5 mutants seem to contain excess apoptotic cells as detected by staining with terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling. We suggest that the increased apoptosis seen in cup-5 mutants is a secondary consequence of the lysosomal defect, and that abnormalities in apoptosis may be associated with human lysosomal storage disorders.
Programmed cell death or apoptosis regulates cell number during metazoan development (1). The misregulation of programmed cell death, resulting in either too many or too few dying cells, contributes to the pathogenesis of many human diseases (2). For example, animal models of human retinitis pigmentosa indicate that retinal degeneration occurs by apoptotic death, and that blocking this death can prevent retinal degeneration and restore vision (3–5). Mutations that disrupt Fas-mediated apoptosis in the immune systems of mice and humans lead to lymphoproliferative disorders (6, 7). The inhibition of apoptosis caused by the misexpression of the proto-oncogene Bcl-2 is a primary cause of human follicular lymphoma (8), a cancer of the immune system.
Studies of the nematode Caenorhabditis elegans have played an important role in defining the components that regulate and execute programmed cell death (9). To seek new genes that can affect programmed cell death, we screened for suppressors of a gain-of-function (gf) allele of the C. elegans Bcl-2-like gene ced-9. The ced-9 gene normally protects cells that should survive from undergoing programmed cell death (10), and the ced-9(gf) allele n1950 causes all programmed cell death to be blocked (11). In this paper, we describe one ced-9(gf) suppressor, n3194, which proved to be a mutation in the C. elegans counterpart of the human mucolipidosis type IV (ML-IV) gene (12, 13), cup-5 (coelomocyte-uptake defective; ref. 14).
Materials and Methods
Genetics and Strains.
Strains were cultured as described (15) at 20°C. The wild-type parental strain was C. elegans Bristol N2. Mutagenesis with ethyl methanesulfonate and genetic mapping were performed by standard methods (16). Mutations used were: linkage group I (LGI): ced-1(e1735), sem-4(n1378); LGIII: unc-36(e251), dpy-17(e164), ncl-1(e1865), sma-3(e491), ced-9(n1950), ced-9(n2812), ced-4(n1162), unc-79(e1068), qC1 [dpy-19(e1259) glp-1(q339)]; LGIV: ced-3(n717), unc-31(e928), ced-5(n1812). We used three-factor mapping to localize n3194 between sma-3 and ncl-1: sma-3 (10/14) n3194 (4/14) ncl-1.
Cloning and Molecular Characterization.
Cosmids from the region between sma-3 and ncl-1 were injected into unc-36 cup-5(n3194)/qC1 animals at 20 μg/ml with pRF4, which contains the rol-6(su1006) allele, and at 80 μg/ml as a coinjection marker. Rescue of maternal-effect lethality was tested in Rol Unc animals from stably transmitting transgenic lines. cDNA clones for cup-5 were provided by Y. Kohara (National Institute of Genetics, Japan). We determined the 5′ end of the cup-5 message by 5′ rapid amplification of cDNA ends (5′-RACE System, GIBCO/BRL).
Heat-Shock Experiments.
To test rescue of the cup-5 mutant phenotype by cup-5 and human ML-IV (hML-IV) cDNAs, full-length cDNAs were cloned into the heat-shock vectors pPD49.78 and pPD49.83 (17). These clones were injected at 50 μg/ml each with pRF4 into unc-36 cup-5(n3194)/qC1 animals. At least six Rol Unc animals from stable transgenic lines were used for each experiment. Animals were allowed to lay eggs at 20°C for 24 h before heat shock (−HS). Adults then were transferred to new plates, incubated at 33°C for 1 h, and allowed to lay eggs at 20°C for an additional 24 h (+HS). Adults were removed, and the number of embryos on each plate was determined. The number of embryos that had hatched was counted 1 day after the removal of Rol Unc adults. The number of embryos that had reached adulthood was determined 4 days after removal of adults.
TUNEL, Acridine Orange (AO), and LysoTracker Staining.
Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining was performed as described (18). cup-5(n3194) embryos for TUNEL staining were obtained from cup-5(n3194) hermaphrodites.
For AO staining, embryos were isolated by treating adults with 0.8 M NaOH/8% hypochlorite solution until adults had been dissolved and embryos were released. Embryos were incubated in 50 μg/ml AO in embryo buffer (113 mM NaCl/40 mM KCl/3.4 mM CaCl2/3.4 mM MgCl2/10 mM Na-Hepes, pH 7.5) for 1 h before observation by using epifluorescence microscopy.
For LysoTracker staining, NGM agar plates (16) were supplemented with LysoTracker Red (Molecular Probes) at 2 μM. L4 stage animals were placed on LysoTracker plates and incubated in the dark at 20°C. Embryos from these plates were observed with confocal microscopy (Zeiss LSM 510) at an excitation wavelength of 543 nm.
Electron Microscopy.
Electron microscopy was performed as described (19). L1 larvae were obtained from either cup-5(n3264) or cup-5(n3194) homozygous hermaphrodites.
Results
Isolation of n3194.
ced-9(n1950) is a gain-of-function mutation that blocks all somatic programmed cell deaths that occur during normal C. elegans development (11). To seek mutations that can cause cells to undergo programmed cell death even in the presence of the ced-9(n1950) mutation, we performed a screen for suppressors of ced-9(n1950). We used Nomarski optics microscopy to look for cell corpses in progeny of mutagenized ced-1(e1735) sem-4(n1378); ced-9(n1950) animals. Mutations in the ced-1 gene cause the persistence of unengulfed apoptotic corpses because of defects in a receptor that recognizes and promotes the engulfment of dying cells (20). Mutations in the sem-4 gene disrupt the development of the hermaphrodite sex muscles and therefore prevent egg-laying (21), such that the progeny of a single hermaphrodite hatch and begin development within that individual; the progeny can be easily screened, and siblings of animals carrying homozygous lethal mutations can be obtained.
We performed two screens, one of approximately 10,000 mutagenized haploid genomes for mutations with a zygotic suppression phenotype and one of approximately 6,000 haploid genomes for mutations conferring a maternal-effect suppression phenotype. From the maternal-effect screen we isolated three mutations that conferred a recessive maternal-effect lethal phenotype: n3196, n3213, and n3194.
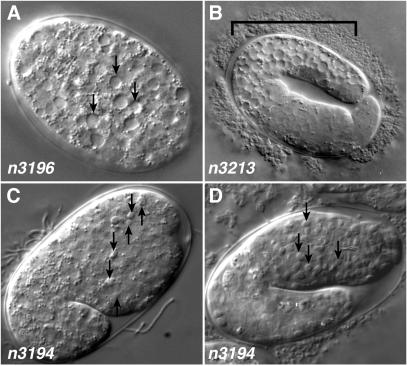
The n3196 mutation resulted in embryos containing apparently vacuolated cells, beginning at about the 50-cell stage (Fig. 1A). The n3213 mutation resulted in a similar degenerative vacuolarization defect, but the effect was limited to the posterior portion of the embryo (Fig. 1B). Neither of these mutations restored the presence of refractile, unengulfed corpses as seen in ced-1(e1735) embryos, so they likely represent defects unrelated to the well characterized pathway of programmed cell death; n3196 and n3213 might affect nonapoptotic degenerative deaths.
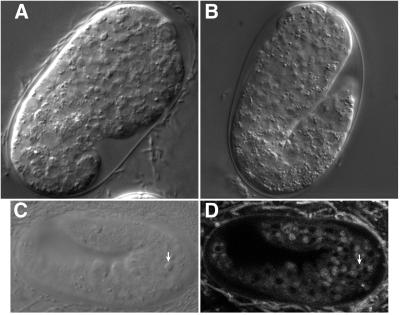
Figure 1.
Mutants isolated from a screen for suppressors of ced-9(n1950). (A) Approximately 50-cell stage embryo carrying the n3196 mutation, which confers a maternal-effect lethal phenotype characterized by vacuolization (arrows) and degeneration of the embryo. (B) Two-fold stage embryo carrying the n3213 mutation, which confers a phenotype similar to that of n3196 embryos but is restricted to the posterior portion (bracket) of the embryo. The abnormality first appears in approximately comma-staged embryos. (C) Comma-stage cup-5(n3194) embryo derived from a homozygous cup-5(n3194) mother showing early appearance of refractile bodies (arrows). (D) cup-5(n3194) embryo arrested at approximately the two-fold stage filled with refractile bodies.
The third mutation, n3194, seemed to affect programmed cell death. Embryos derived from homozygous n3194 hermaphrodites accumulated refractile corpse-like bodies beginning at the comma stage (Fig. 1C), approximately the time at which corpses begin to accumulate in ced-1(e1735) embryos. By the two-fold to three-fold stages of embryogenesis (Fig. 1D), n3194 embryos were filled with refractile bodies, and the embryos arrested development shortly thereafter. Rare animals that survived to hatching arrested as L1 larvae.
By using the maternal-effect lethal phenotype, we mapped the n3194 mutation to chromosome III (data not shown). ced-9 maps to chromosome III, and loss-of-function mutations in ced-9 both suppress ced-9(n1950gf) and cause maternal-effect lethality with an accumulation of corpses (10). However, n3194 was unlikely to be an allele of ced-9, because n3194 complemented ced-9(n2812), a putative null allele of ced-9 (22). n3194 seemed to allow cell death to occur in ced-9(n1950) animals, in which cells that normally die instead survive. To determine if n3194 suppresses this effect of ced-9(n1950), we counted the number of cells in the anterior pharynx of homozygous n3194 ced-9(n1950) animals derived from n3194 ced-9(n1950)/+ ced-9(n1950) mothers. These animals contained 12.0 ± 1.6 extra cells in the anterior pharynx, as compared with 11.9 ± 1.1 for ced-9(n1950gf) animals. Thus, either n3194 does not suppress the survival of extra cells caused by ced-9(n1950gf), or a maternal contribution of the n3194 gene product is sufficient for function. As embryos derived from homozygous n3194 mothers do not survive until cells in the pharynx can be counted, this latter possibility cannot be directly tested.
Two additional mutations that failed to complement n3194 were identified in unrelated screens. The mutation zu223 was isolated in a screen for mutations with a maternal-effect lethal phenotype and observed to have excess refractile corpses in arrested embryos (J. Priess, personal communication). The mutation n3264 was isolated in a screen for mutants defective in cell-corpse engulfment (Z. Zhou and H.R.H., unpublished data). We found that n3264 and zu223 failed to complement n3194 for both maternal-effect lethality and the accumulation of refractile bodies. n3264 seems to be a partial loss-of-function allele, because unlike zu223 or n3194, it does not confer a maternal-effect lethal phenotype; n3264 does cause accumulation of refractile bodies in embryos.
n3194 Is a Mutation in the C. elegans ML-IV Gene.
We further mapped n3194 to a region of chromosome III between sma-3 and ncl-1 (see Materials and Methods). We injected cosmids from this region and rescued the maternal-effect lethality conferred by n3194 with cosmid C30C5. We subcloned this cosmid and identified an 8.6-kb KpnI-HpaI rescuing fragment that contained only one complete ORF, R13A5.1. Deleting a portion of this reading frame while retaining the two partial ORFs 5′ and 3′ of R13A5.1 eliminated rescuing ability. This minimal rescuing fragment also completely rescued the accumulation of refractile corpses in n3194 embryos.
A mutation causing inappropriate accumulation of green fluorescent protein (GFP) in coelomocytes, cells of unknown function in the C. elegans pseudocoelom, was recently found by Fares and Greenwald (14) to be a missense mutation in R13A5.1. This mutation, ar465, defined the gene cup-5. Thus, n3194, n3264, and zu223 are alleles of cup-5. cup-5(ar465) animals have endocytic trafficking defects but no defects in viability.
Two classes of expressed sequence tag (EST) clones isolated by the C. elegans EST project correspond to the predicted gene R13A5.1 and differ slightly from each other and from the GENEFINDER (23) prediction for the structure of this gene. These two classes differ specifically in the length of exon 8. Three clones (yk517f10, yk611f2, and yk280c8) are in-frame with exon 9 and encode a predicted protein of 668 amino acids, which we refer to as CUP-5L (L, long). Three other clones (yk253a6, yk643f6, and yk561c9) extend exon 8 by four nucleotides, shifting the reading frame with respect to exon 9 and encode a predicted protein of 611 amino acids, which we refer to as CUP-5S (S, short) and which correspond to the structure described by Fares and Greenwald (14). The two predicted proteins are identical through the first 603 amino acids. The cup-5S transcript corresponds to the previously reported cup-5 sequence (14), but the longer cup-5L transcript was not previously reported. Draft genome sequence from the nematode Caenorhabditis briggsae (Sanger Institute, Hinxton, U.K., and Genome Sequencing Center, Washington University, St. Louis) encodes a 665 amino acid CUP-5L protein. Nonfunctional regions of C. elegans and C. briggsae are typically not highly conserved (24), so the conservation of the CUP-5L-specific portion of the sequence suggests this sequence is likely functional. The C. briggsae sequence also contains the four extra bps and splice site that could generate CUP-5S. Neither class of C. elegans cDNA contains the first exon predicted by GENEFINDER. Therefore, we performed 5′-RACE analysis on mixed-stage N2 RNA to determine the starting point of the transcript. The cDNA amplified in this manner contained an SL1 leader sequence (25) followed immediately 3′ by the in-frame ATG codon at the start of the GENEFINDER-predicted second exon, consistent with the starting point we observed in the longest of the EST clones.
We expressed cup-5L and cup-5S individually under the control of the C. elegans heat-shock promoters to test their abilities to rescue the maternal-effect lethality caused by cup-5(n3194). cup-5(n3194) mutant animals bearing either a Phspcup-5L or a Phspcup-5S transgenic array were subjected to heat shock, and embryos derived from these animals were assayed for viability. The Phspcup-5L and Phspcup-5S constructs each effectively rescued the maternal-effect lethality (Table 1), indicating that both forms are functional. The vector alone did not rescue.
Table 1.
cup-5L, cup-5S, and hML-IV rescue cup-5(n3194) lethality
| Transgene | Heat shock | Total no. eggs laid | Total no. eggs hatched | Eggs hatched, % | No. of adults |
|---|---|---|---|---|---|
| Phsp alone | − | 178 | 0 | 0 | 0 |
| Phsp alone | + | 360 | 3 | 0.8 | 1 |
| Phspcup-5L | − | 232 | 28 | 12.1 | 24 |
| Phspcup-5L | + | 301 | 127 | 42.2 | 118 |
| Phspcup-5S | − | 178 | 26 | 14.6 | 24 |
| Phspcup-5S | + | 271 | 135 | 49.8 | 121 |
| PhsphML-IV | − | 136 | 0 | 0 | 0 |
| PhsphML-IV | + | 263 | 24 | 9.1 | 21 |
| PhsphML-IV(fs) | − | 243 | 0 | 0 | 0 |
| PhsphML-IV(fs) | + | 314 | 1 | 0.3 | 0 |
Transgenic extrachromosomal arrays carried by unc-36(e251) cup-5(n3194)/qC1 animals were tested for rescue as described in Materials and Methods. Animals were removed from Petri plates 24 h following heat-shock treatment, and the number of eggs present on each plate was counted. Hatching was assayed 2 days after heat-shock treatment, and the presence of adult animals was assayed 4 days after heat-shock treatment. At least three transgenic lines were tested for each construct. Data for a single representative line are shown here. fs, frameshift.
The CUP-5 protein is similar to the product of the hML-IV gene (refs. 12 and 13; Fig. 2). hML-IV is a lysosomal storage disorder found most often in Ashkenazic Jews and is characterized by lipid accumulation in lysosomes and consequent psychomotor retardation, impairment of cognitive development, corneal clouding, and retinal degeneration (26, 27). Cells derived from ML-IV patients are not defective in the lysosomal hydrolases that participate in lipid catabolism but seem to have a block in transport or sorting of late endosome-lysosome vesicles (28, 29). CUP-5 also is similar to the predicted Drosophila protein CG8743 and the predicted human protein FLJ11006.
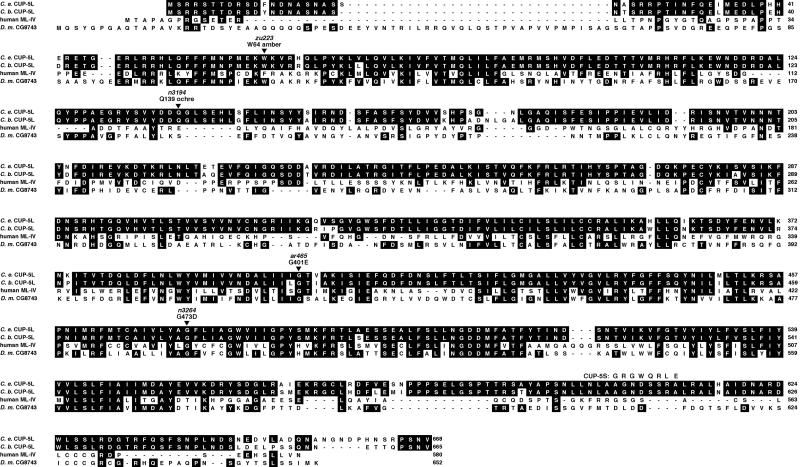
Figure 2.
CUP-5L sequence and alignment. Alignment of C. elegans CUP-5L with predicted C. briggsae CUP-5L, the product of the hML-IV gene (12, 13) and the predicted Drosophila protein CG8743. Identities to the CUP-5L protein are shaded. The divergent portion of CUP-5S following amino acid 603 is indicated, and the positions of the three cup-5 mutants alleles described in the text as well as cup-5(ar465) are shown.
We identified mutations in the coding region of cup-5 in all three of the alleles we studied (Fig. 2). n3194 and zu223, which confer a maternal-effect lethal phenotype, introduce stop codons early in the gene. The weaker allele, n3264, converts a conserved glycine to an aspartate. Both zu223 and n3194 are candidate molecular-null alleles, and cup-5(n3194) homozygotes are similar in the penetrance of maternal-effect lethality (Table 2) and refractility (data not shown) to cup-5(n3194)/Df animals. Therefore, the phenotype of cup-5(n3194) and cup-5(zu223) animals likely represents the null phenotype of this gene.
Table 2.
Mutations that block programmed cell death slightly improve cup-5 viability
| Genotype | Avg no. eggs laid | Avg no. eggs hatched | Eggs hatched, % | n |
|---|---|---|---|---|
| cup-5(n3194) unc-36 | 216 | 2.4 | 1.1 | 11 |
| cup-5(n3194) unc-36/nDf16 | 187 | 0.2 | 0.1 | 13 |
| cup-5(zu223) unc-36 | 206 | 0.4 | 0.2 | 7 |
| cup-5(zu223) unc-36/nDf16 | 246 | 0.6 | 0.2 | 10 |
| sma-3 cup-5(n3194) | 110 | 0.6 | 0.5 | 12 |
| unc-79 ced-4(n1162) sma-3 cup-5(n3194) | 114 | 10.0 | 8.8 | 10 |
| sma-3 cup-5(n3194); unc-31 | 56 | 0.6 | 1.1 | 10 |
| sma-3 cup-5(n3194); unc-31 ced-3(n717) | 64 | 7.7 | 12.1 | 9 |
Effects of mutations blocking programmed cell death on the viability of cup-5 embryos. The number of eggs laid and number of eggs hatched was determined for n individual animals, and the mean value is indicated. Hatched % was calculated by dividing the total number of eggs hatched by the total number of eggs laid for each indicated genotype.
hML-IV Can Substitute for cup-5.
We obtained hML-IV cDNA from G. Borsani (Telethon Institute of Genetics and Medicine, Milan, Italy) and placed it under the control of the C. elegans heat-shock promoters. The expression of hML-IV rescued the maternal-effect lethality associated with cup-5(n3194), although not as strongly as did either of the C. elegans cDNA clones (Table 1). Most animals that survived to hatching progressed to become healthy adults. A 5-bp frameshift introduced into the hML-IV cDNA (BstEII site, nucleotide position 460) abolished rescue, demonstrating that functional ML-IV gene product is required for rescue.
cup-5 Mutants Contain Excess Lysosomes.
As a single missense allele of cup-5 had been previously identified on the basis of accumulated GFP in coelomocytes (14); because this allele seems not to be a null allele, we tested whether the null alleles of cup-5 we identified also conferred such a defect. A secreted GFP expressed under the control of the myo-3 promoter normally is diffuse through the C. elegans pseudocoelom but accumulated in the coelomocytes of both cup-5(n3194) homozygotes and cup-5(n3264) animals (data not shown), as it does in cup-5(ar465) animals (14).
We also tested whether cup-5 mutants contain an increased number of lysosomes, as occurs in patients with ML-IV. We stained embryos with AO, an acidophilic dye that stains acidified lysosomal and endosomal compartments in protists, plants, insects, nematodes, and cultured mammalian cells (30) but also stains apoptotic cells in Drosophila and C. elegans (31, 32). Thus, an increase in AO staining could indicate an increase in either the number of lysosomes or programmed cell deaths or both. Embryos derived from homozygous cup-5(n3194) mothers contained greatly increased numbers of AO+ bodies compared with embryos derived from wild-type mothers (Fig. 3 A and B).
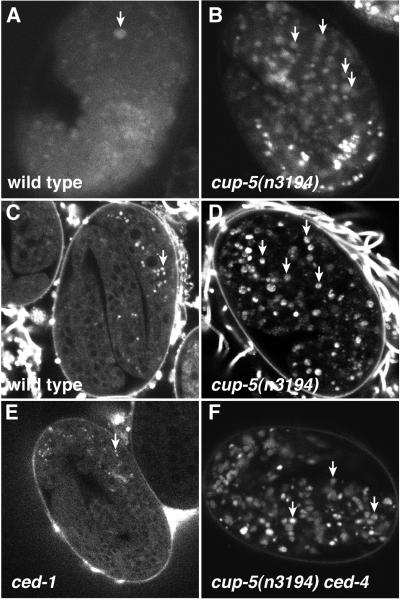
Figure 3.
cup-5 mutants accumulate excess lysosomes. (A and B) AO staining in embryos. (A) Wild-type embryo. Apoptotic corpse is indicated by arrow. (B) cup-5(n3194) embryo. Arrows indicate a few of the many additional AO+ bodies. (C–F) LysoTracker staining in embryos. (C) Late wild-type embryos with staining restricted to intestinal granules (arrow). (D) Increased accumulation of LysoTracker in cup-5(n3194) embryos. (E) ced-1 embryo, which accumulates apoptotic corpses, did not accumulate LysoTracker. (F) ced-4 cup-5(n3194) embryo accumulated LysoTracker.
To distinguish whether the AO+ bodies in cup-5 mutants represent lysosomes or corpses, we grew animals on media containing the acidophilic dye LysoTracker Red, which accumulates in highly acidified lysosomal compartments (33). We found that persistent cell corpses do not accumulate LysoTracker, because ced-1 and ced-5 mutants, both of which have persistent cell corpses (34), did not accumulate LysoTracker in corpses; both mutants displayed a distribution of LysoTracker indistinguishable from that of the wild type (Fig. 3E and data not shown). In early wild-type embryos, LysoTracker had a granular cytoplasmic distribution that became restricted to intestinal granules in later embryos (Fig. 3C). By contrast, all three of our cup-5 mutants accumulated LysoTracker in most and possibly all tissues of the embryo (Fig. 3D). By using polarized-light microscopy, which illuminates intestinal granules in C. elegans embryos and allows visualization of intestinal cells, we found that cup-5 mutants did not have excess intestinal cells that might have been responsible for the broader distribution of LysoTracker. Furthermore, cup-5 mutants showed normal distribution of the mitochondrial marker MitoTracker Red (35), indicating that the accumulation of LysoTracker is not a consequence of a nonspecific general accumulation of dye in an unspecified intracellular compartment in these animals (data not shown). We conclude that cup-5 mutant animals contain excess acidified intracellular compartments that most likely are lysosomes.
We analyzed the cellular ultrastructure of cup-5 mutant animals with electron microscopy (Fig. 4). cup-5 mutants were disorganized and displayed irregular cellular morphologies. Specifically, cup-5 mutant animals contained many cells with enlarged vacuoles or lysosomes and membranous lamellar structures that are not seen in wild-type animals. We observed these structures both in the weak cup-5(n3264) mutant and in the strong cup-5(n3194) mutant. Cells derived from hML-IV patients have enlarged vacuolar spaces and characteristic membranous cytoplasmic bodies (26, 36) that are similar to the subcellular structures present in cup-5 mutants.
Figure 4.
Accumulation of vacuoles and lamellar material in cup-5 mutants. Electron micrographs of (A) wild-type and (B) cup-5(n3194) mutant larva at the level of the pharynx(ph). Cellular structure in the cup-5 mutant appears disorganized. Multiple cells with enlarged vacuolar spaces (arrows) that may represent enlarged lysosomes can be seen.
cup-5 Mutants Display Increased Programmed Cell Death.
As the null allele cup-5(n3194) seems to cause the accumulation of refractile bodies similar to cell corpses, we examined the role of programmed cell death in the phenotype of cup-5 mutant animals. The activities of ced-3 and ced-4 are required for essentially all programmed cell deaths in C. elegans (37). ced-3 encodes a caspase (38), and ced-4 encodes a protein similar to the human caspase activator Apaf-1 (39, 40). Introducing either a ced-3 or ced-4 mutation into a cup-5 mutant background suppressed only slightly the lethality of cup-5 mutants (Table 2) and did not obviously suppress the accumulation of refractile bodies. Thus, cup-5 maternal-effect lethality either is not primarily a consequence of too much programmed cell death and the refractile bodies are not cell corpses, or programmed cell death is being activated independently of ced-3 and ced-4 in cup-5(n3194) animals. In addition, mutations in neither ced-3 nor ced-4 blocked the accumulation of LysoTracker in cup-5 mutants (Fig. 3F and data not shown), indicating that this accumulation does not depend on ced-4 and ced-3-mediated programmed cell death.
Blocking programmed cell death did slightly increase the viability of cup-5 mutants. Compared with cup-5(n3194) embryos, greater numbers of both cup-5(n3194) ced-4(n1162) and cup-5(n3194); ced-3(n717) double mutants survived to hatching (Table 2). Double-mutant embryos that hatched typically did not survive to adulthood: 1/42 cup-5(n3194); ced-3(n717), 7/72 cup-5(n3194) ced-4(n1162), and 4/24 cup-5(n3194) mutants survived to adulthood. Blocking cell death by expressing the baculovirus caspase inhibitor p35 (41, 42) similarly slightly increased viability but did not completely rescue lethality (Table 3).
Table 3.
Overexpression of the baculoviral anti-apoptotic gene p35 partially rescues viability of cup-5 embryos
| Transgene | Heat-shock | Total no. eggs laid | Total no. hatched | Hatched, % |
|---|---|---|---|---|
| Phsp p35 | − | 253 | 2 | 0.8 |
| Phsp p35 | + | 351 | 27 | 7.7 |
| Phsp p35 | + 2x | 187 | 16 | 8.6 |
At least 10 cup-5(n3194) unc-36(e251) animals carrying a transgenic array of p35 were subjected to a single pulse of 33°C heat-shock (+) or two pulses of heat-shock separated by 1 hr recovery at 20°C (+ 2x). The total numbers of eggs laid and hatched were counted.
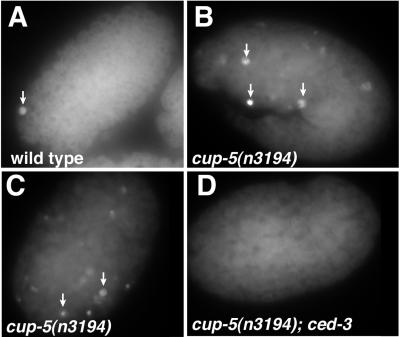
To examine further whether programmed cell death was increased in cup-5 mutants, we performed the TUNEL assay, which labels nicked DNA ends generated during programmed cell death (18). Whereas wild-type animals averaged 0.8 ± 0.9 (n = 15) TUNEL+ cells per embryo, cup-5 mutants averaged 7.7 ± 4.6 (n = 10) TUNEL+ cells per embryo (Fig. 5). We tested whether this increase in TUNEL+ cells depended on programmed cell death by performing the TUNEL assay with cup-5(n3194) ced-4(n1162), cup-5(n3194); ced-3(n717), cup-5(n3194) ced-9(n1950), and cup-5(n3194); egl-1(n1084n3082) double mutants. In these double mutants, TUNEL+ cells were completely absent. Therefore, the increase in TUNEL+ cells in cup-5 mutants depends on programmed cell death mediated by egl-1, ced-9, ced-4, and ced-3. We conclude that cup-5 mutants have an increase in programmed cell death, but that this increased cell death is not primarily responsible for their lethal phenotype. Alternatively, some aspects of programmed cell death in cup-5 mutants may be induced downstream of or parallel to ced-3 and may contribute to lethality, but such cell death apparently is not capable of generating TUNEL reactivity in the absence of ced-3.
Figure 5.
Increased numbers of TUNEL+ cells in cup-5(n3194) embryos. (A) Wild-type embryo with very few TUNEL+ cells (arrows). (B and C) cup-5(n3194) embryos derived from homozygous cup-5(n3194) animals contain many TUNEL+ cells. (D) cup-5(n3194); ced-3(n717) embryos contain no TUNEL+ cells.
cup-5 Refractile Bodies Represent both Lysosomes and Cell Corpses.
We attempted to determine whether the refractile bodies in cup-5 mutants represent lysosomes or programmed cell death corpses or both. Some of the refractile bodies in cup-5 mutants were smaller and more irregular in shape than corpses generated by programmed cell death (Fig. 1D). Mutations in ced-8 delay the appearance of programmed cell death corpses (43). In cup-5(n3194);ced-8 animals, we observed a delay in the appearance of the larger but not of the smaller irregular refractile bodies in cup-5 mutants (Fig. 6 A and B). This result suggests that the larger refractile bodies in cup-5 mutants represent cell corpses. In addition, we determined that some, but not all, of the refractile bodies label with Lyso Tracker (Fig. 6 C and D), which we showed above does not label cell corpses. Thus, the refractile bodies in cup-5 mutants seem to represent both enlarged lysosomes and programmed cell deaths.
Figure 6.
cup-5 refractile bodies represent both lysosomes and cell corpses. (A and B) cup-5(n3194); ced-8 animals contain smaller refractile bodies but have a delay in the appearance of cell corpses (compare with Fig. 1C, in which a number of cell corpses are obvious). (C) Nomarski image of cup-5(n3194)embryo with large refractile body indicated (arrow). (D) LysoTracker image of embryo in C indicating that refractile body did not stain with LysoTracker.
Discussion
From a screen for mutations affecting programmed cell death, we isolated a null allele of the C. elegans homolog of the hML-IV gene. A recently identified missense mutation in this gene, cup-5, indicated that it regulates the uptake and disposal of extracellular material by coelomocytes (14), scavenger cells of poorly understood function in the C. elegans pseudocoelom. Our analyses have demonstrated that the function of cup-5 is not limited to coelomocytes, and that cup-5 is likely broadly involved in lysosomal regulation in C. elegans.
What is the relationship between the effects of cup-5 mutants on lysosomes and on programmed cell death? Both larger and smaller refractile bodies in cup-5 mutants appear during development at about the same time as cell corpses. We suggest that there may be two classes of refractile bodies in cup-5 mutant animals, corresponding to enlarged lysosomes and to corpses generated by programmed cell death. Distinctions between these classes may be possible at the molecular level (e.g., LysoTracker vs. TUNEL staining) but are not unambiguous at the level of morphological detail visible by light microscopy.
We also studied other markers of programmed cell death in cup-5 mutants. There is an increase in the number of TUNEL+ cells in cup-5 mutants, and the presence of these cells depends on ced-3 and ced-4 function. These findings indicate that the increase in TUNEL staining involves the known genetic pathway for programmed cell death (9). One simple possibility is that the increased number of TUNEL+ cells in cup-5 mutants is a consequence of an increased number of programmed cell deaths. If so, because these deaths occur in a ced-9(n1950) mutant, they may resemble radiation-induced deaths (44) in that their activation seems to occur via a pathway independent of ced-9(n1950) and egl-1. Alternatively, it is conceivable that the apparent increase in TUNEL-reactivity is a consequence of a decreased rate of cell death (43) or DNA degradation (18), such that TUNEL-reactivity persists and accumulates. That the kinetics of DNA degradation seemed to be normal, based upon 4′,6-diamidino-2-phenylindole (DAPI) visualization of nuclei in cup-5 mutants argues against this possibility (B.H. and H.R.H., unpublished observations). Regardless of whether it is the rate of DNA degradation or the amount of programmed cell death that is altered in C. elegans cup-5 mutants, some aspect of programmed cell death is clearly abnormal in these animals.
Follicular lymphoma, autoimmune disease, and retinal degeneration all involve abnormalities in apoptosis (5–8). In addition, human neurodegenerative diseases, including Alzheimer's Disease, Huntington's Disease, and amyotrophic lateral sclerosis, are associated with increased levels of apoptotic cell death (45). Our studies of the C. elegans homolog of the hML-IV gene suggest that this disease, and by extension, other lysosomal storage disorders, may also involve abnormalities in programmed cell death. Indeed, in neuronal tissue of Hexb−/− mice, which provide a model for Sandhoff disease (46), and in spinal cord sections of Tay-Sachs and Sandhoff disease patients, apoptotic cells are present (47). Many lysosomal storage disorders seem to dramatically affect neuronal function (48). It is possible that neurons are particularly sensitive to disruptions in lysosome function. Alternatively, neurons may be more sensitive to the activation of programmed cell death. If lysosomal storage diseases indeed involve abnormalities in programmed cell death, it may be possible to alleviate aspects of these diseases by blocking the abnormal programmed cell deaths.
We have shown that null mutations in the C. elegans homolog of the ML-IV gene cause maternal-effect lethality, the accumulation of excess lysosomes, and abnormalities in programmed cell death. We suggest that abnormalities in programmed cell death may be secondary consequences of lysosomal storage disorders. cup-5 may provide a good model for ML-IV and may allow a detailed genetic analysis of the mechanisms involved in the disease pathology. For example, the isolation of suppressors of the C. elegans maternal-effect lethal phenotype could allow the identification of genes that act with cup-5/hML-IV to regulate lysosome function and to initiate programmed cell death. Such genes could define targets for therapeutic intervention in hML-IV.
Acknowledgments
We thank Z. Zhou for the n3264 mutation, J. Priess for the zu223 mutation, and G. Borsani for the hML-IV cDNA. We thank C. Ceol, P. Reddien, and Z. Zhou for critical reading of the manuscript. B.M.H. was supported by a Howard Hughes Medical Institute predoctoral fellowship. H.R.H. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- ML-IV
mucolipidosis type IV
- hML-IV
human ML-IV
- gf
gain-of-function
- RACE
rapid amplification of cDNA ends
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- AO
acridine orange
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF482952).
References
- 1.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Chang G Q, Hao Y, Wong F. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- 4.Portera-Cailliau C, Sung C H, Nathans J, Adler R. Proc Natl Acad Sci USA. 1994;91:974–978. doi: 10.1073/pnas.91.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson F F, Steller H. Nature (London) 1998;391:587–591. doi: 10.1038/35385. [DOI] [PubMed] [Google Scholar]
- 6.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts I A, Debatin K M, Fischer A, de Villartay J P. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 7.Fisher G H, Rosenberg F J, Straus S E, Dale J K, Middleton L A, Lin A Y, Strober W, Lenardo M J, Puck J M. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 8.Vaux D L, Cory S, Adams J M. Nature (London) 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 9.Metzstein M M, Stanfield G M, Horvitz H R. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 10.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 11.Hengartner M O, Horvitz H R. Nature (London) 1994;369:318–320. doi: 10.1038/369318a0. [DOI] [PubMed] [Google Scholar]
- 12.Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G, et al. Nat Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 13.Bassi M T, Manzoni M, Monti E, Pizzo M T, Ballabio A, Borsani G. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fares H, Greenwald I. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 15.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood W B the Community of C. elegans Researchers. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 17.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y C, Stanfield G M, Horvitz H R. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- 19.Bargmann C I, Hartwieg E, Horvitz H R. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Hartwieg E, Horvitz H R. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 21.Basson M, Horvitz H R. Genes Dev. 1996;10:1953–1965. doi: 10.1101/gad.10.15.1953. [DOI] [PubMed] [Google Scholar]
- 22.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 23.The C. elegans Genome Sequencing Consortium. Science. 1998;282:2012–2018. [Google Scholar]
- 24.Fitch D H A, Thomas W K. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 815–850. [Google Scholar]
- 25.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amir N, Zlotogora J, Bach G. Pediatrics. 1987;79:953–959. [PubMed] [Google Scholar]
- 27.Berman E R, Livni N, Shapira E, Merin S, Levij I S. J Pediatr. 1974;84:519–526. doi: 10.1016/s0022-3476(74)80671-2. [DOI] [PubMed] [Google Scholar]
- 28.Chen C S, Bach G, Pagano R E. Proc Natl Acad Sci USA. 1998;95:6373–6378. doi: 10.1073/pnas.95.11.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bargal R, Bach G. J Inherit Metab Dis. 1997;20:625–632. doi: 10.1023/a:1005362123443. [DOI] [PubMed] [Google Scholar]
- 30.Allison A C, Young M R. In: Lysosomes in Biology and Pathology. Dingle J T, Fell H B, editors. Vol. 2. Amsterdam: North–Holland; 1969. pp. 600–628. [Google Scholar]
- 31.Abrams J M, White K, Fessler L I, Steller H. Development (Cambridge, UK) 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- 32.Gumienny T L, Lambie E, Hartwieg E, Horvitz H R, Hengartner M O. Development (Cambridge, UK) 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 33.Haller T, Dietl P, Deetjen P, Volkl H. Cell Calcium. 1996;19:157–165. doi: 10.1016/s0143-4160(96)90084-6. [DOI] [PubMed] [Google Scholar]
- 34.Ellis R E, Jacobson D M, Horvitz H R. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, Hersh B M, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz H R. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 36.Schiffmann R, Dwyer N K, Lubensky I A, Tsokos M, Sutliff V E, Latimer J S, Frei K P, Brady R O, Barton N W, Blanchette-Mackie E J, et al. Proc Natl Acad Sci USA. 1998;95:1207–1212. doi: 10.1073/pnas.95.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis H M, Horvitz H R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 38.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J, Horvitz H R. Development (Cambridge, UK) 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 40.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 41.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayzur T, Li P, et al. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 42.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 43.Stanfield G M, Horvitz H R. Mol Cell. 2000;5:423–433. doi: 10.1016/s1097-2765(00)80437-2. [DOI] [PubMed] [Google Scholar]
- 44.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner M O. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 45.Nijhawan D, Honarpour N, Wang X. Annu Rev Neurosci. 2000;23:73–87. doi: 10.1146/annurev.neuro.23.1.73. [DOI] [PubMed] [Google Scholar]
- 46.Kolter T, Sandhoff K. J Inherited Metab Dis. 1998;21:548–563. doi: 10.1023/a:1005419122018. [DOI] [PubMed] [Google Scholar]
- 47.Huang J Q, Trasler J M, Igdoura S, Michaud J, Hanal N, Gravel R A. Hum Mol Genet. 1997;6:1879–1885. doi: 10.1093/hmg/6.11.1879. [DOI] [PubMed] [Google Scholar]
- 48.Gieselmann V. Biochim Biophys Acta. 1995;1270:103–136. doi: 10.1016/0925-4439(94)00075-2. [DOI] [PubMed] [Google Scholar]