Abstract
Metallo-β-lactamases (MBLs) represent the latest generation of β-lactamases. The structural diversity and broad substrate profile of MBLs allow them to confer resistance to most β-lactam antibiotics. To explore the evolutionary potential of these enzymes, we have subjected the Bacillus cereus MBL (BcII) to a directed evolution scheme, which resulted in an increased hydrolytic efficiency toward cephalexin. A systematic study of the hydrolytic profile, substrate binding, and active-site features of the evolved lactamase reveal that directed evolution has shaped the active site by means of remote mutations to better hydrolyze cephalosporins with small, uncharged C-3 substituents. One of these mutations is found in related enzymes from pathogenic bacteria and is responsible for the increase in that enzyme's hydrolytic profile. The mutations lowered the activation energy of the rate-limiting step rather than improved the affinity of the enzyme toward these substrates. The following conclusions can be made: (i) MBLs are able to expand their substrate spectrum without sacrificing their inherent hydrolytic capabilities; (ii) directed evolution is able to mimic mutations that occur in nature; (iii) the metal-ligand strength is tuned by second-shell mutations, thereby influencing the catalytic efficiency; and (iv) changes in the position of the second Zn(II) ion in MBLs affect the substrate positioning in the active site. Overall, these results show that the evolution of enzymatic catalysis can take place by remote mutations controlling reactivity.
Keywords: Co(II)-substitution, directed evolution, zinc enzymes, antibiotic resistance
The β-lactam antibiotics account for more than half of the world's antibiotic market and are one of the cornerstones of antibacterial chemotherapy (1). The increasing use of these drugs, both in the clinical setting and in animal feed, induces the development of different resistance mechanisms in pathogenic and opportunistic microorganisms (2, 3). Among them, the expression of β-lactamases is predominant. In the last decade, there has been growing concern about the dissemination of genes coding for metallo-β-lactamases (MBLs) among infectious microorganisms (4–7). MBLs are zinc-dependent enzymes that hydrolyze β-lactams by favoring the deprotonation of a metal-bound water molecule, which (as a hydroxide) is a powerful nucleophile (8–11). Much recent literature has been devoted to the elucidation of the structure and mechanism of action of MBLs (8–14). Parallel to these efforts, the significant growth of new MBL sequences has revealed an initially unforeseen molecular and structural diversity in this family of enzymes (6). This diversity encompasses changes in the coordination features of the Zn(II) ions as well as in the topology of the active site. This fact has hitherto thwarted the successful development of clinically useful inhibitors. The potential danger of MBLs is further enhanced by the broad substrate spectrum displayed by these enzymes, as compared with the better-characterized Ser-dependent β-lactamases.
MBLs have been classified into three subclasses (B1, B2, and B3) based on their sequence similarities (6). So far, only B1 lactamases have been found encoded in plasmids and transposons transmitted among pathogens. The prototype B1 enzyme BcII from Bacillus cereus has been thoroughly studied (8, 15–24). The recent finding of an MBL with 94% sequence identity to BcII in Bacillus anthracis has renewed the interest in this enzyme (25). BcII, as do most MBLs, exhibits a broad substrate spectrum, albeit with a lower catalytic efficiency toward some substrates than that of other enzymes from pathogenic bacteria. Based on this finding, and taking into account that BcII is not produced by a clinically relevant pathogen, several groups have proposed that BcII could be a precursor of the more efficient MBLs present in bacteria exposed to increasing amounts of antibiotics (16, 17). Thus, BcII provides a unique template to explore the potential improvement of its catalytic efficiency and substrate spectrum and, hence, the generation of enhanced levels of resistance.
There is no information yet on mutations that are known to increase resistance by MBLs because these represent a relatively recent clinical problem. Here, we employ directed evolution, a technique that has been put forward as a powerful tool to predict antibiotic resistance (26–31), to explore the effect of challenging MBLs with different antibiotics. We have performed in vitro evolution experiments on BcII by DNA shuffling and have selected cephalexin (Fig. 1), a cephalosporin that is not efficiently hydrolyzed by BcII (kcat/KM = 2.7 × 104 M–1·s–1, mostly due to a low kcat value of 3.3 s–1). The directed molecular evolution experiment resulted in an expanded substrate spectrum in the evolved enzyme, without sacrificing its hydrolytic efficiency toward the classical substrates of BcII. Interestingly, one of the mutations found (Gly-262 → Ser) has been associated with increased resistance in MBLs from pathogenic bacteria (32, 33) and corresponds to a residue acting as second-shell ligand of one of the Zn(II) ions. Spectroscopic data confirm that this mutation is able to influence the metal–ligand binding features, suggesting that it controls the structure of the active site and, ultimately, the activity of the enzyme.
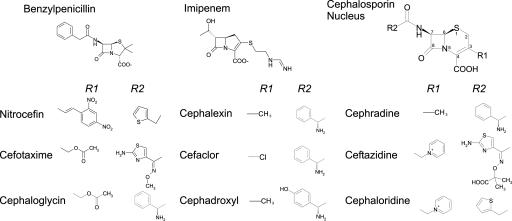
Fig. 1.
Chemical structures of the β-lactam substrates used in this work. Side chains R1 and R2 are specified for cephalosporins.
Experimental Procedures
Materials. Tetracyclin, ampicillin, chloramphenicol, kanamycin, benzylpenicillin, cephalexin, cefotaxime, cefaclor, cephaloglycin, ceftazidime, cephaloridine, 4-(2-pyridylazo)-resorcinol (PAR), and DNase I were purchased from Sigma; nitrocefin was purchased from Calbiochem; and imipenem was a kind gift from Merck, Sharp & Dohme. Escherichia coli XL1 Blue MRF cells (Stratagene) were used as the cloning and recipient strain for plasmids and as the host used to screen for susceptibility to antibiotics. E. coli BL21 (DE3) pLysS′ cells (Stratagene) were used for protein production. Luria–Bertani (LB) medium or Mueller–Hinton broth was used as growth medium for all bacterial strains. The enzymes used for DNA manipulation were purchased from Promega. TaqDNA polymerase was from Invitrogen. Oligonucleotides were synthesized by Biosynthesis (Lewisville, Texas).
DNA Techniques and Cloning Procedure. DNA preparation and related techniques were performed according to standard protocols (34). Plasmid DNA was isolated by using the Wizard Plus SV Minipreps kit (Promega). DNA was extracted from agarose gels by using the QIAEX II kit (Qiagen, Valencia, CA) or GFX columns (Amersham Pharmacia).
The plasmid pKP [a derivative of the pBluescript II vector (Stratagene, CA) with a kanamycin resistance cassette] was designed to have a phenotypic detection system to select β-lactamase mutants conferring increased resistance toward antibiotics. The plasmid pKPBcII contains the bcII gene corresponding to the MBL from B. cereus fused with the leader sequence pelB (from pectate lyase) to target the enzyme to the periplasm. Plasmid pKP derivatives with the native and/or mutant bcII genes were used for the DNA shuffling and for the screening procedure. The plasmid pET-Term is a derivative of the PetGEX vector used to overexpress the bcII gene to yield a fusion protein with GST (16).
Directed Evolution. DNA shuffling was performed as described by Stemmer (27) with some modifications as described below. The substrate DNA for the initial shuffling reaction was a 1,024-bp dsDNA obtained from a PCR amplification of the bcII gene by using the universal forward and reverse primers and plasmid pKPBcII as template. The standard PCR program used was 94°C 5′/30× (94°C 50″, 52°C 50″, 72°C 1′ 30″)/72°C 5′. In the next rounds, a mixture of selected colonies (100 μl of cells grown in LB broth, boiled for 10 min) was used as the template for a colony PCR with the same program. After the initial PCR, the primers were removed by elution through GFX columns. Two to four micrograms of the substrate DNA were digested with 0.015 units of DNase I (Sigma) in 50 mM Tris/10 mM MgCl2 (pH 7.4) for 10–20 min at 20°C. The digestion was terminated by heating at 90°C for 5 min. Fragments of <300 bp were purified from a 2% agarose gel by using the QIAEX II kit (Qiagen) and resuspended in a PCR mix (0.2 mM dNTP/2.2 mM MgCl2/Taq buffer/2 units of TaqDNA polymerase). No primers were added at this point. Thermocycling was performed with a PCR program of 94°C 1′/40× (94°C 30″, 50°C 30″, 72°C 30″) for the reassembly. After a 50-fold dilution of the previous solution, a new PCR [94°C 5′/30× (94°C 50″, 52°C 50″, 72°C 1′ 30″)/72°C 5′] was run, using the universal primers T7 and T3, located closer to the coding region, yielding an amplification product of 940 bp. The reassembled product was digested sequentially with BamHI and XhoI. A fragment of 717 bp (the coding DNA for mature BcII) was purified from a 1% agarose gel and ligated back into the pKP plasmid.
Electrocompetent E. coli cells were transformed with the DNA ligation mixture and plated out on LB agar with 35 μg/ml tetracycline and 25 μg/ml kanamycin. Libraries of 5 × 105 transformants were typically obtained. Plasmids obtained from these libraries were purified and used to transform a new set of E. coli cells. Approximately 1,000 cells per plate were screened on LB agar containing 35 μg/ml tetracycline, 25 μg/ml kanamycin, and 500 μM IPTG, supplemented with increasing concentrations of cephalexin (from 16 to 1,024 μg/ml). After incubation at 37°C for 18 h, several colonies were picked from the selected transformants, separately grown, and the corresponding plasmids were purified from each culture. This protocol was repeated four times iteratively to select colonies with enhanced resistance toward cephalexin. To avoid selection of chromosomal mutations, E. coli cells were transformed after each round with plasmid DNA from the selected clones.
The minimum inhibitory concentrations (MICs) for cephalexin and cefotaxime were determined by the standard agar macrodilution method, and the susceptibilities were determined by the standard disk diffusion method.
Protein Expression and Purification. Plasmids pKPBcII and pKPm5 were digested with BamHI and HindIII, and the resulting 696-bp fragments were cloned into a pET-Term plasmid digested with the same enzymes, giving rise to the plasmids pET-Term-BcII and pET-Term-m5, respectively. Both wild-type (WT) and mutant proteins were expressed in E. coli BL21(DE3)pLysS′ as fusion proteins with glutatione S-transferase, purified, and quantified as described in ref. 16. The metal content was determined in protein samples dialyzed four times against metal-free 10 mM Hepes (pH 7.5)/0.2 M NaCl at 4°C, by using the colorimetric reagent 4-(2-pyridylazo)-resorcinol under denaturing conditions (35). The Co(II) derivative was obtained as described in ref. 16.
Steady-State Kinetic Measurements. β-lactamase activity was followed by monitoring the changes in substrates' absorbance in a Jasco V-550 spectrometer thermostated at 30°C. All kinetic measurements were performed in 15 mM Hepes (pH 7.5), 0.2 M NaCl, 50 μg/ml BSA, and 20 μM Zn(II). The complete reaction time courses or the initial rates were recorded by using 0.1- or 1-cm light path cuvettes depending on the substrate absorbance. The kinetic parameters KM and kcat were estimated by nonlinear fitting of the data to the integrated form of the Michaelis–Menten equation or from the initial hydrolysis rates of reactions by using the Henri–Michaelis equation. The used Δελmax values were as follows: benzylpenicillin Δε235 = –800 M–1·cm–1; nitrocefin Δε485 = +17,400 M–1·cm–1; imipenem Δε300 = –9,000 M–1·cm–1; cefotaxime Δε262 = –7,500 M–1·cm–1; cephalexin Δε266 = –7,800 M–1·cm–1; cefaclor Δε261 = –6,000 M–1·cm–1; cephadroxyl Δε260 = –6,355 M–1·cm–1; cephaloridine Δε260 = –13,600 M–1·cm–1; ceftazidime Δε260 = –9,000 M–1·cm–1; cephradine Δε260 = –13,600 M–1·cm–1; and cephaloglycin Δε263 = –11,000 M–1·cm–1.
Pre-Steady-State Kinetic Measurements and Data Analysis. Experiments were carried out on an SX.18-MVR stopped-flow spectrometer (Applied Photophysics, Surrey, U.K.) as reported in refs. 36 and 37. Enzyme fluorescence measurements were performed by exciting at 280 nm (slit width = 0.5 mm) and detecting through a cutoff filter at >305 nm. The photomultiplier voltage was set to obtain a 4.0- or 0.8-V signal with the free enzyme. Absorbance changes were measured at the corresponding λmax for each substrate by using an absorption photomultiplier. All reactions were performed in 15 mM Hepes (pH 7.5), 0.2 M NaCl, and 20 μM Zn(II) at 6°C.
Kinetic runs under pseudofirst-order conditions to follow substrate binding were performed by mixing 2- or 20-μM enzyme solutions with 10–160 μM substrate (syringe concentrations) and measuring the protein intrinsic fluorescence. Twenty-one runs were averaged for all tested conditions, due to the low amplitude of the signal obtained and the high rate constants observed even at low substrate concentration. The enzyme fluorescence quenching processes were satisfactorily fitted to simple exponential functions. A linear fit of the data points was performed to calculate (k–1 + kcat) and k+1. Subtraction of the kcat values obtained under the same conditions allowed us to estimate k–1.
Results
Libraries of BcII Mutants by DNA Shuffling. The bcII gene from B. cereus was subjected to DNA shuffling. Some directed evolution strategies include mutations in the gene promoter region (27), which we deliberately omitted by cloning the coding DNA sequence for mature BcII in the plasmid used in the phenotypic screening procedure. In this way, the mutations subjected to the screening were only those present in the expressed enzyme. In each round, E. coli transformants with the mutant libraries were challenged with increasing concentrations of cephalexin and compared with the control strain corresponding to WT BcII. The MIC of cephalexin toward E. coli cells transformed with the plasmid pKPBcII (that expresses and targets WT BcII to the periplasmic space) is 16 μg/ml. After four rounds of DNA shuffling, we obtained colonies on agar plates with up to 1,024 μg/ml cephalexin. The upper limit for the concentration of cephalexin was set by the solubility of this antibiotic. None of the libraries obtained during the different rounds exhibited increased MICs toward cefotaxime (used as a control cephalosporin).
After four rounds, 18 clones were selected at cephalexin concentrations of >768 μg/ml and sequenced. Most of them contained three to five mutations each. All of the detected mutations (22 total) corresponded to a single base change and resulted in amino acid changes, i.e., there were no silent mutations. None of these mutations involved any metal ligand or any residue directly involved in substrate recognition. Four mutations occurred with high frequency in the screening process: Asn-70 → Ser (83%), Val-112 → Ala (100%), Leu-250 → Ser (55%), and Gly-262 → Ser (94%). We selected one clone, named m5 hereafter, containing only these four most frequent mutations. Sequencing different clones in the libraries before the selection steps (data not shown) revealed that other mutations present in these libraries were not preserved, suggesting that they may be deleterious. The Val-112 → Ala and Gly-262 → Ser mutations (the most frequent among the selected clones) were already present in the first shuffling round, suggesting that they are able to confer a higher resistance phenotype.
The MICs of cephalexin for E. coli cells harboring the mutant plasmid pKPm5 was 1,024 μg/ml, compared with a value of 16 μg/ml for the plasmid containing the native bcII gene (i.e., a 64-fold increase). Instead, the MIC of cefotaxime for WT and for the mutant m5 was the same (8 μg/ml).
Biochemical Characterization of the M5 Mutant. The metal content of purified WT BcII and M5 was similar [1.6 ± 0.1 and 1.5 ± 0.1 Zn(II)/enzyme for BcII and M5, respectively]. WT BcII is fully active as a dinuclear zinc enzyme. We therefore studied the catalytic performance of WT BcII and M5 toward different substrates in the presence of excess Zn(II) (Table 1) to ensure the formation of a dinuclear center in the active site. The catalytic efficiency of M5 toward cephalexin (the antibiotic used in the screening process of directed evolution) was 7.4-fold higher compared with that of WT BcII. In contrast, the rates of benzylpenicillin, imipenem, and nitrocefin hydrolysis were not significantly compromised. To have a more detailed substrate characterization, other cephalosporin substrates were tested (Table 1). M5 did not show any significant improvement in the hydrolysis of cefotaxime and cephaloglycin, but there were substantial enhancements in the β-lactamase activity toward cephadroxyl, cefaclor, cephradine, cephaloridine, and ceftazidime.
Table 1. Kinetic parameters for the hydrolysis of different β-lactam substrates by WT BcII and mutant M5.
| BcII
|
M5
|
M5/BcII ratio
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrate | kcat, s-1 | KM, μM | kcat/KM, M-1·s-1 | kcat, s-1 | KM, μM | kcat/KM, M-1·s-1 | kcat | KM | kcat/KM |
| Imipenem | 279 | 687 | 4.06 × 105 | 129 | 721 | 1.79 × 105 | 0.462 | 1.05 | 0.440 |
| Benzylpenicillin | 1020 | 662 | 1.54 × 106 | 570 | 574 | 9.93 × 105 | 0.559 | 0.867 | 0.644 |
| Nitrocefin | 30.1 | 9.82 | 3.07 × 106 | 81.3 | 27.8 | 2.92 × 106 | 2.70 | 2.83 | 0.954 |
| Cefotaxime | 80.5 | 43.3 | 1.86 × 106 | 44.9 | 30.3 | 1.49 × 106 | 0.557 | 0.699 | 0.796 |
| Cephaloglycin | ND | ND | 1.89 × 106 | ND | ND | 1.80 × 106 | ND | ND | 0.952 |
| Cephalexin | 3.32 | 125 | 2.65 × 104 | 41.2 | 209 | 1.97 × 105 | 12.4 | 1.67 | 7.42 |
| Cefaclor | 73.5 | 328 | 2.24 × 105 | 410 | 422 | 9.71 × 105 | 5.58 | 1.28 | 4.35 |
| Cephadroxyl | 19.2 | 789 | 2.43 × 104 | 75.2 | 200 | 3.76 × 105 | 3.91 | 0.253 | 15.4 |
| Cephradine | ND | ND | 6.81 × 104 | ND | ND | 3.51 × 105 | ND | ND | 5.15 |
| Ceftazidime | 13.1 | 426 | 3.07 × 104 | 24.4 | 262 | 9.31 × 104 | 1.86 | 0.615 | 3.03 |
| Cephaloridine | 57.9 | 1250 | 4.63 × 104 | 68.1 | 204 | 3.34 × 105 | 1.17 | 0.163 | 7.17 |
Conditions: 10 mM Hepes, 0.2 M NaCl, 50 μg/ml BSA, and 20 μM Zn (pH 7.5) at 30°C. Reported values are the average of at least three independent determinations. Standard deviation values were <5%. ND, not determined
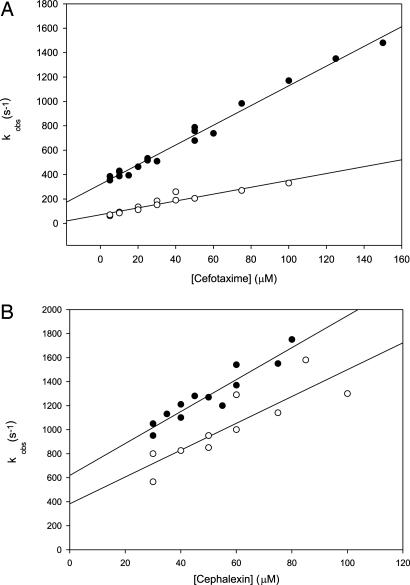
We studied substrate binding to BcII during turnover independently of the reaction progress by following changes in the enzyme Trp fluorescence (36, 37). Cephalexin binding at room temperature was too fast to be characterized in detail and could be better followed at 6°C. Trp fluorescence quenching by binding of cephalexin and cefotaxime was monitored under pseudofirst-order conditions, and the experimental curves could be fitted to single exponential functions. The linear increase of the observed pseudofirst-order rate constants with increasing substrate concentration (Fig. 2) suggests a simple one-step binding process. The data show that the affinity toward cephalexin is not improved in the selected mutant, whereas cefotaxime binding to M5 is favored compared with WT BcII.
Fig. 2.
Measured pseudo-first order rate constants of cefotaxime (A) and cephalexin (B) binding to WT BcII (filled circles) and M5 (open circles). Data fits are shown as solid lines. Enzyme concentrations were 1 μM (A) and 10 μM (B). The reactions were carried out in 10 mM Hepes (pH 7.5), 0.2 M NaCl, and 20 μM added Zn(II) at 6°C. The calculated Ks values were 37.5 and 4.3 μM (cefotaxime hydrolysis by BcII and M5, respectively) and 37 and 44 μM (cephalexin hydrolysis by BcII and M5, respectively). Errors were within ±10%.
In an attempt to correlate these measurements with the in vivo effects of the evolved enzyme, we evaluated the resistance to different cephalosporins conferred by WT BcII and M5 to E. coli cells by disk diffusion assays. In all cases, the outcome of these determinations paralleled the activity and MIC values.
Metal Site Structure of the M5 Mutant. Co(II)-substitution has been largely exploited as a fruitful strategy to obtain structural information about the active sites of zinc enzymes by employing the spectroscopic features of Co(II) (38). This characterization has been also applied to MBLs (16, 19, 39, 40). Despite the fact that the four point mutations found in M5 did not occur in the active site, we prepared the di-Co(II) derivative of this mutant to check whether these mutations could have some effect on the metal site structure. As shown in Fig. 3, the four ligand field bands in the visible region observed in di-Co(II) WT BcII are unaltered in di-Co(II) M5. Instead, a charge transfer band attributed to a Cys → Co(II) interaction is shifted from 347 to 333 nm.
Fig. 3.
Electronic spectra of Co(II)-substituted BcII (black line) and Co(II)-substituted M5 (gray line) in 10 mM Hepes (pH 7.5) and 0.2 M NaCl. The enzyme concentrations were 200 μM, and 3 equivalents of Co(II) were added.
Discussion
We have applied evolutionary pressure to mutant libraries of the MBL from B. cereus (BcII) by using cephalexin (a poorly hydrolyzed substrate) as the driving force for developing further resistance to this antibiotic. This procedure resulted in the selection of several mutant clones able to confer on E. coli, a 64-fold enhanced resistance to cephalexin. Four point mutations with a high frequency of occurrence were identified in these clones. We selected one mutant (m5) harboring exclusively these mutations (N70S, V112A, L250S, and G262S).
The mutant lactamase (M5) displayed an enhanced performance toward cephalexin (the antibiotic used in the selection process), as well as against other cephalosporins poorly hydrolyzed by BcII (see Table 1). The hydrolytic capabilities of M5 toward benzylpenicillin were unaltered, whereas the imipenemase activity was reduced only by a factor of 2 compared with WT BcII. The enhanced cephalosporinase activity in M5 did not compromise the catalytic profile against noncephalosporin substrates.
The hydrolysis of cephalosporins with small substituents at C-3 (cephradine and cephadroxyl, R1 = CH3; and cefaclor, R1 = Cl; see Fig. 1) was enhanced in M5. In contrast, the activity against other cephalosporins with medium-sized or large, extended substituents (cefotaxime, cephaloglycin, and nitrocefin) was mostly unaffected. Molecular dynamics (41) and pre-steady-state studies (36) have suggested that C-3 substituents can be accommodated in a hydrophobic groove in the enzyme active site. These data reveal that the directed evolution experiment was successful in shaping the active site of BcII to better accommodate smaller C-3 substituents, possibly altering this groove.
The activity enhancements toward cephalosporins with small C-3 substituents are mostly due to increases in kcat values, instead of KM. Because KM values can be affected by changes in kcat, we estimated the substrate binding constants (KS) by an independent method. These experiments show that cephalexin binding is not enhanced in M5 (Fig. 2), suggesting that the improved catalytic performance induced by the directed evolution experiment does not result from optimizing substrate binding but from the stabilization of the enzyme-transition state complex by 1.5 kcal/mol.
M5 also was able to hydrolyze cephalosporins with methylene pyridinium substituents at C-3 (ceftazidime and cephaloridine) more efficiently than WT BcII. Docking of these substrates in the enzyme active site (data not shown) shows that the aromatic ring cannot be accommodated into the groove, so that the pyridinium moiety protrudes from the active site, and is not a key structural element of recognition by the hydrophobic groove. The improvements in cephaloridine and ceftazidime hydrolysis are not due to changes in a single kinetic parameter and cannot be clearly rationalized. This result confirms that the effects of the evolution experiment are selective for cephalosporins with small C-3 substituents.
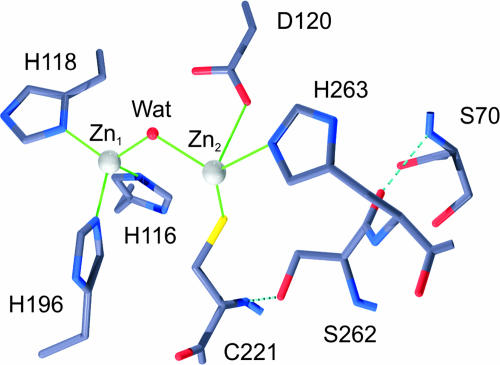
WT BcII is fully active as a dinuclear enzyme. One Zn(II) ion is bound to three His residues and a hydroxide, while the protein ligands of the second Zn(II) are residues Asp-120, Cys-221, and His-263 (Fig. 4) (17). These residues are conserved in all B1 lactamases. None of the four mutations in M5 is located in the active site. Nonetheless, two of them (Asn-70 → Ser and Gly-262 → Ser) correspond to residues that lie below the active site floor. The Val-112 → Ala substitution occurs in a hydrophobic core defining the α/β interface, and the Leu-250 → Ser replacement occurs in the exposed face of an α-helix. Modeling the Asn-70 → Ser and Gly-262 → Ser mutations suggests that the side-chain hydroxyl group of Ser-262 would form a hydrogen bond with the backbone nitrogen of Cys-221 (Fig. 4), i.e., acting as a second-shell ligand (42).
Fig. 4.
Model of the BcII active site with the Gly-262 → Ser and Asn-70 → Ser mutations in the second coordination sphere of Zn(II).
The Gly-262 → Ser mutation, the only amino acid differentiating IMP-1 and IMP-6 (plasmid-encoded enzymes from pathogenic species), is responsible for expanding the substrate profile between these MBLs (32, 43). IMP-3 and IMP-6 (bearing a Gly residue on position 262) have been postulated as precursors of the widespread IMP-1, which has a broader β-lactam spectrum. An elegant codon randomization experiment on IMP-1 has recently shown that Ser-262 alters the substrate specificity of the enzyme (44). All these observations concur with the effect of the mutations induced by directed evolution on BcII.
Oelschlaeger et al. (32, 33) suggested that the Gly-262 → Ser mutation in IMP-1 would reduce the mobility of His-263 through a hydrogen bond network, leaving room to better accommodate cephalosporin with positively charged R1 substituents. Given that M5 is able to better hydrolyze cephalosporins with small, uncharged R1 substituents, other effects should be operative here. The crystal structures of free and inhibited IMP-1 show that Ser-262 indeed forms a hydrogen bond with the amide backbone of Cys-221, which is preserved upon inhibitor binding (14, 45). The absorption spectrum of Co(II)-substituted M5 reveals that the Co(II)–Cys interaction has been perturbed compared with the native enzyme, whereas the tetrahedral coordination of the other metal site is largely maintained. This result allows us to propose that Ser-262 acts as a second-shell ligand of one of the Zn(II) ions, affecting the metal–ligand interactions, and, finally, the substrate orientation of different cephalosporins in the active site. These evidences strongly suggest that Zn2 plays a key role in substrate recognition in MBLs.
The finding that mutations far removed from the active site (V112A and L250S) seem to be involved in the enzymatic activity enhancement suggest new working hypotheses for the structural determinants of reactivity in MBLs, in agreement with recent results that highlight the effect of remote mutations (46–48). This finding could be due to the existence of molecular motions in the active site coupled to remote positions in the protein structure that are able to exert relevant influences in the enzyme reactivity.
Our results allow us to speculate on the effect of evolutionary pressure in Ser-β-lactamases and MBLs. Directed evolution experiments in the extended spectrum β-lactamase TEM-1 also have paralleled results observed in the clinical setting. However, the most relevant mutation resulting from directed evolution on the Ser-β-lactamase TEM-1 does not confer a higher catalytic performance on the enzyme but counteracts the destabilization effects caused by other mutations that enhance the enzymatic activity at the cost of impairing the enzyme stability (29, 49, 50). In contrast, it seems that (at least for cephalosporin hydrolysis), MBLs are still able to evolve further resistance through improvements in the catalytic efficiency.
The increase in MIC in E. coli cells secreting the mutant lactamase M5 parallels an increase in the catalytic efficiency of the enzyme, together with an expansion of the substrate profile. From an evolutionary point of view, this observation may be interpreted as a sign that evolution of more efficient MBLs can occur without sacrificing a broad substrate profile or compromising the enzyme stability. From a clinical point of view, further evolution of MBLs may be expected, which is intrinsically threatening. However, the fact that in vitro evolution experiments are able to mimic natural evolution may be exploited to predict future enzyme mutations and even to design more general inhibitory strategies.
Acknowledgments
We thank Areli Morán for help in some of the DNA sequencing. We thank the reviewers for their useful comments and suggestions. R.M.R. and P.E.T. are recipients of doctoral fellowships from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina. A.J.V. is a Staff Member from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina and an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica and the Howard Hughes Medical Institute (to A.J.V.); Consejo Nacional de Ciencia y Technología Grant G0030-N9607 and Dirección General de Asuntos del Personal Académico Universidad Nacional Autónoma de México Grant IN215201 (to L.S.).
Author contributions: A.J.V. designed research; P.E.T. and R.M.R. performed research; L.S. contributed new reagents/analytic tools; P.E.T., R.M.R., L.S., and A.J.V. analyzed data; and P.E.T. and A.J.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MBL, metallo-β-lactamase; MIC, minimum inhibitory concentration.
References
- 1.Poole, K. (2004) Cell Mol. Life Sci. 61, 2200–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frere, J. M. (1995) Mol. Microbiol. 16, 385–395. [DOI] [PubMed] [Google Scholar]
- 3.Fisher, J. F., Meroueh, S. O. & Mobashery, S. (2005) Chem. Rev. 105, 395–424. [DOI] [PubMed] [Google Scholar]
- 4.Cricco, J. A., Rasia, R. M., Orellano, E. G., Ceccarelli, E. A. & Vila, A. J. (1999) Coord. Chem. Rev. 190–192, 519–535. [Google Scholar]
- 5.Wang, Z., Fast, W., Valentine, A. M. & Benkovic, S. J. (1999) Curr. Opin. Chem. Biol. 3, 614–622. [DOI] [PubMed] [Google Scholar]
- 6.Galleni, M., Lamotte-Brasseur, J., Rossolini, G. M., Spencer, J., Dideberg, O. & Frere, J. M. (2001) Antimicrob. Agents Chemother. 45, 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz, U. & Adolph, H. W. (2004) Cell Mol. Life Sci. 61, 2827–2839. [DOI] [PubMed] [Google Scholar]
- 8.Carfi, A., Pares, S., Duee, E., Galleni, M., Duez, C., Frère, J. M. & Dideberg, O. (1995) EMBO J. 14, 4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Concha, N., Rasmussen, B. A., Bush, K. & Herzberg, O. (1996) Structure (London) 4, 823–836. [DOI] [PubMed] [Google Scholar]
- 10.Ullah, J. H., Walsh, T. R., Taylor, I. A., Emery, D. C., Verma, C. S., Gamblin, S. J. & Spencer, J. (1998) J. Mol. Biol. 284, 125–136. [DOI] [PubMed] [Google Scholar]
- 11.Garau, G., Bebrone, C., Anne, C., Galleni, M., Frere, J. M. & Dideberg, O. (2005) J. Mol. Biol. 345, 785–795. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Saez, I., Hopkins, J., Papamicael, C., Franceschini, N., Amicosante, G., Rossolini, G. M., Galleni, M., Frere, J. M. & Dideberg, O. (2003) J. Biol. Chem. 278, 23868–23873. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Saez, I., Mercuri, P. S., Papamicael, C., Kahn, R., Frere, J. M., Galleni, M., Rossolini, G. M. & Dideberg, O. (2003) J. Mol. Biol. 325, 651–660. [DOI] [PubMed] [Google Scholar]
- 14.Concha, N. O., Janson, C. A., Rowling, P., Pearson, S., Cheever, C. A., Clarke, B. P., Lewis, C., Galleni, M., Frere, J. M., Payne, D. J., et al. (2000) Biochemistry 39, 4288–4298. [DOI] [PubMed] [Google Scholar]
- 15.Bounaga, S., Laws, A. P., Galleni, M. & Page, M. I. (1998) Biochem. J. 31, 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orellano, E. G., Girardini, J. E., Cricco, J. A., Ceccarelli, E. A. & Vila, A. J. (1998) Biochemistry 37, 10173–10180. [DOI] [PubMed] [Google Scholar]
- 17.Fabiane, S. M., Sohi, M. K., Wan, T., Payne, D. J., Bateson, J. H., Mitchell, T. & Sutton, B. J. (1998) Biochemistry 37, 12404–12411. [DOI] [PubMed] [Google Scholar]
- 18.Paul-Soto, R., Bauer, R., Frere, J. M., Galleni, M., Meyer-Klaucke, W., Nolting, H., Rossolini, G. M., de Seny, D., Hernandez-Valladares, M., Zeppezauer, M., et al. (1999) J. Biol. Chem. 274, 13242–13249. [DOI] [PubMed] [Google Scholar]
- 19.de Seny, D., Heinz, U., Wommer, S., Kiefer, M., Meyer-Klaucke, W., Galleni, M., Frere, J. M., Bauer, R. & Adolph, H. W. (2001) J. Biol. Chem. 276, 45065–45078. [DOI] [PubMed] [Google Scholar]
- 20.Rasia, R. M. & Vila, A. J. (2002) Biochemistry 41, 1853–1860. [DOI] [PubMed] [Google Scholar]
- 21.Dal Peraro, M., Vila, A. J. & Carloni, P. (2002) J. Biol. Inorg. Chem. 7, 704–712. [DOI] [PubMed] [Google Scholar]
- 22.Seny, D., Prosperi-Meys, C., Bebrone, C., Rossolini, G. M., Page, M. I., Noel, P., Frere, J. M. & Galleni, M. (2002) Biochem. J. 363, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies, A. M., Rasia, R. M., Vila, A. J., Sutton, B. J. & Fabiane, S. M. (2005) Biochemistry 44, 4841–4849. [DOI] [PubMed] [Google Scholar]
- 24.Dal Peraro, M., Llarrull, L. I., Rothlisberger, U., Vila, A. J. & Carloni, P. (2004) J. Am. Chem. Soc. 126, 12661–12668. [DOI] [PubMed] [Google Scholar]
- 25.Materon, I. C., Queenan, A. M., Koehler, T. M., Bush, K. & Palzkill, T. (2003) Antimicrob. Agents Chemother. 47, 2040–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stemmer, W. P. (1994) Proc. Natl. Acad. Sci. USA 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemmer, W. P. (1994) Nature 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 28.Crameri, A., Raillard, S. A., Bermudez, E. & Stemmer, W. P. (1998) Nature 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 29.Orencia, M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. (2001) Nat. Struct. Biol. 8, 238–242. [DOI] [PubMed] [Google Scholar]
- 30.Vakulenko, S. & Golemi, D. (2002) Antimicrob. Agents Chemother. 46, 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peimbert, M. & Segovia, L. (2003) Protein Eng. 16, 27–35. [DOI] [PubMed] [Google Scholar]
- 32.Oelschlaeger, P., Schmid, R. D. & Pleiss, J. (2003) Biochemistry 42, 8945–8956. [DOI] [PubMed] [Google Scholar]
- 33.Oelschlaeger, P., Mayo, S. L. & Pleiss, J. (2005) Protein Sci. 14, 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., Fitsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 35.Hunt, J. B., Neece, S. H. & Ginsburg, A. (1985) Anal. Biochem. 146, 150–157. [DOI] [PubMed] [Google Scholar]
- 36.Rasia, R. M. & Vila, A. J. (2004) J. Biol. Chem. 279, 26046–26051. [DOI] [PubMed] [Google Scholar]
- 37.Spencer, J., Clarke, A. R. & Walsh, T. R. (2001) J. Biol. Chem. 276, 33638–33644. [DOI] [PubMed] [Google Scholar]
- 38.Bertini, I. & Luchinat, C. (1985) Adv. Inorg. Biochem. 6, 71–111. [PubMed] [Google Scholar]
- 39.Crowder, M. W., Yang, K. W., Carenbauer, A. L., Periyannan, G., Seifert, M. E., Rude, N. E. & Walsh, T. R. (2001) J. Biol. Inorg. Chem. 6, 91–99. [DOI] [PubMed] [Google Scholar]
- 40.Garrity, J. D., Bennett, B. & Crowder, M. W. (2005) Biochemistry 44, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 41.Dal Peraro, M., Vila, A. J. & Carloni, P. (2004) Proteins 54, 412–423. [DOI] [PubMed] [Google Scholar]
- 42.El Yazal, J., Roe, R. R. & Pang, Y. P. (2000) J. Phys. Chem. B 104, 6662–6664. [Google Scholar]
- 43.Iyobe, S., Kusadokoro, H., Ozaki, J., Matsumura, N., Minami, S., Haruta, S., Sawai, T. & O'Hara, K. (2000) Antimicrob. Agents Chemother. 44, 2023–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Materon, I. C., Beharry, Z., Huang, W., Perez, C. & Palzkill, T. (2004) J. Mol. Biol. 344, 653–663. [DOI] [PubMed] [Google Scholar]
- 45.Toney, J. H., Hammond, G. G., Fitzgerald, P. M., Sharma, N., Balkovec, J. M., Rouen, G. P., Olson, S. H., Hammond, M. L., Greenlee, M. L. & Gao, Y. D. (2001) J. Biol. Chem. 276, 31913–31918. [DOI] [PubMed] [Google Scholar]
- 46.Benkovic, S. J. & Hammes-Schiffer, S. (2003) Science 301, 1196–1202. [DOI] [PubMed] [Google Scholar]
- 47.Piana, S., Carloni, P. & Rothlisberger, U. (2002) Protein Sci. 11, 2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piana, S., Carloni, P. & Parrinello, M. (2002) J. Mol. Biol. 319, 567–583. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., Minasov, G. & Shoichet, B. K. (2002) J. Mol. Biol. 320, 85–95. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., Minasov, G. & Shoichet, B. K. (2002) J. Biol. Chem. 277, 32149–32156. [DOI] [PubMed] [Google Scholar]