Abstract
Keratin polypeptides 8 and 18 (K8/18) are the major intermediate filament proteins of simple-type epithelia. K18 Ser-33 phosphorylation regulates its binding to 14-3-3 proteins during mitosis. We studied the significance of keratin binding to 14-3-3 in transgenic mice that overexpress wild-type or Ser-33→Ala (S33A) K18. In S33A but not wild-type K18-overexpressing mice, pancreatic acinar cell keratin filaments retracted from the basal nuclear region and became apically concentrated. In contrast, K18 S33A had a minimal effect on hepatocyte keratin filament organization. Partial hepatectomy of K18-S33A-overexpressing mice did not affect liver regeneration but caused limited mitotic arrest, accumulation of abnormal mitotic figures, dramatic fragmentation of hepatocyte keratin filaments, with retention of a speckled 14-3-3ζ mitotic cell nuclear-staining pattern that usually becomes diffuse during mitosis. Hence, K18 Ser-33 phosphorylation regulates keratin filament organization in simple-type epithelia in vivo. Keratin binding to 14-3-3 may partially modulate hepatocyte mitotic progression, in association with nuclear redistribution of 14-3-3 proteins during mitosis.
Keywords: mitosis|phosphorylation|pancreas|liver
Keratins (K) are the intermediate filament (IF) proteins that are expressed in epithelial cells (1). They consist of a large family of proteins (>20 members, K1–K20) that are divided into type I (K9–K20) and type II (K1–K8) keratins. Simple-type glandular epithelia express K8 (as the major type II keratin) and variable levels of type I keratins (K18, K19, or K20) depending on the cell type, as found in several digestive organs including the liver, pancreas, and intestine (2, 3). Type I and II keratins form obligate noncovalent heteropolymers (e.g., K8/18) that manifest an extended filamentous array, spanning the cytoplasm from the nucleus to the plasma membrane as visualized by immunofluorescence staining. The only well documented function of keratins is to provide mechanical integrity and cytoprotection to epithelial cells in a variety of tissues including skin, liver, eyes, and the oral cavity (4). This function is based on the phenotype of diseases that are found in these tissues as a consequence of keratin mutations (5–7), and the phenotype of several transgenic animal models (5, 8–10).
Given the limited understanding of keratin function, we focused our studies on keratin regulation to gain a handle on this understanding. Two potential important regulatory mechanisms for any protein are posttranslational modifications and interaction with associated proteins. Phosphorylation is predictably important in regulating keratins and other IF proteins because it occurs within their N-terminal “head” and C-terminal “tail” domains, which are the most heterogeneous and, hence, the most likely to impart tissue-specific functions (11–13). Two major phosphorylation sites have been identified for K18, namely Ser-52 and -33. Ser-52 accounts for most of K18 phosphorylation during interphase and increases during mitosis and a variety of stresses (13). Although cell transfection studies suggested that Ser-52 phosphorylation is important for filament organization during mitotic arrest and upon exposure to phosphatase inhibitors (14), no significant impact on filament organization occurred when K18 Ser-52→Ala (S52A) was expressed in transgenic mice (15). However, transgenic mice that expressed K18 S52A were significantly more predisposed to hepatotoxic injury than mice that expressed wild-type (WT) K18 (15). These findings provided direct evidence that keratin hyperphosphorylation, which occurs during a variety of cell stresses, is important in providing a protective mechanism from drug-induced stress in hepatocytes.
Another K18 physiologic phosphorylation site is Ser-33, which is basally phosphorylated at significantly lower levels than Ser-52, but becomes hyperphosphorylated during mitosis (16, 17). K18 S33 phosphorylation during mitosis regulates binding of the 14-3-3 protein family to K18, and mutation of this site in transfected cells abolishes K18/14-3-3 binding and generates collapsed filaments near the nucleus (16). 14-3-3 proteins bind, in a phosphorylation-dependent fashion, to several important cytoplasmic and nuclear proteins including cdc25 and Raf-1 and function, at least in part, to sequester proteins in particular states or subcellular compartments (18–20). Given the informative nature of the transgenic mice that expressed K18 S52A, we hypothesized that transgenic mice that express a K18 Ser-33→Ala mutant (S33A mice) should provide important in vivo information regarding the function of K18 Ser-33 phosphorylation and the significance of its regulation of K18/14-3-3 binding. With S33A mice, we demonstrate that K18 S33 phosphorylation plays an important role in filament organization in the liver and pancreas. This demonstration is the first in vivo example of filament regulation by phosphorylation for any IF protein. In addition, K18 S33 phosphorylation regulates nuclear 14-3-3 redistribution during mitosis, and in doing so, may play a modulating role in hepatocyte mitotic progression after partial hepatectomy.
Materials and Methods
Antibodies and Reagents.
The Ab used (15) were: L2A1 mouse mAb, which recognizes human (h) K18 without crossreaction with mouse (m) keratins; Troma I rat mAb, which recognizes mK8; rabbit Ab 8592, which was raised against hK8/18; rabbit Ab 3055, which recognizes hK18 pS52 and does not crossreact with the corresponding phosphorylation site in mK18; rabbit Ab 8250, which recognizes hK18 pS33 and does crossreact with the equivalent mouse phosphorylation site; mAb LJ4, which recognizes hK8 pS73 and the corresponding mK8 pS79 site; and anti-mK18 Ab (kindly provided by R. Oshima, The Burnham Institute, San Diego). Microcystin-LR (MLR) was from Alexis Corp. (San Diego, CA), and the Transformer mutagenesis kit was from CLONTECH. Anti-14-3-3ζ and σ antibodies were from Santa Cruz Biotechnology.
Transgene Construct and Generation of Transgenic Lines.
The 10-kb hK18 genomic DNA in pGEM (kindly provided by R. Oshima) was mutated at Ser-33 codon (AGC to GCC to generate Ala-33) by using the Transformer kit as recommended by the manufacturer. Both strands of the mutated region were sequenced to confirm the mutation. The K18 S33A genomic DNA was injected into pronuclei of fertilized FVB/N mouse eggs. Progeny mice carrying the hK18 transgene were then chosen, after PCR screening, followed by breeding to select for germ-line transmission by using standard methods. Two mouse lines, S33A1 and S33A2, that express K18 Ser-33→Ala were expanded and used for subsequent studies. The control mice that were used overexpressed a WT 10-kb genomic K18 (TG2 mice) or K18 Ser-52→Ala (S52A mice) as described (15, 21). Transgene copy number and PCR screening of mouse tail genomic DNA for the presence of hK18 was performed as described (15).
Characterization of the Transgenic Mice and Partial Hepatectomy.
For all transgenic mouse hepatectomy experiments, sex- and age-matched mice (>6 weeks old) were weighed just before use. Livers were collected after euthanizing the mice by using CO2 inhalation. Partial hepatectomy was performed by sequentially removing the lateral, left and right median lobes (15) from TG2, S33A1, and S33A2 mice (40–44 mice per transgenic line). Mice were then euthanized 1, 2, 3, 7, 10, 13, 36, and 125 days afterward, and the livers were processed as below. Sham-hepatectomized mice (i.e., mice that had anesthesia, abdominal wall and peritoneal incision, liver exposure, and then closure of the incision) from each transgenic line served as controls. Resected livers were cut into several pieces depending on the experiment and used for: (i) fixation with 10% formalin followed by paraffin embedding, sectioning, then hematoxylin/eosin staining (done by Histo-tec Laboratory, Hayward, CA); (ii) snap freezing in optimum cutting-temperature compound (Miles), sectioning, then fixing briefly in cold acetone for subsequent immunofluorescence staining; and (iii) snap freezing in liquid nitrogen for subsequent biochemical analysis. MLR was injected i.p (30 μg/kg), and then the livers were processed. Student's t test and nonparametric Wilcoxon method were used to calculate the statistical significance between the means. Statistical analysis was performed with JMP version 3.1 (SAS Institute, Cary, NC).
Keratin Isolation, Immunoprecipitation, and Other Methods.
Liver pieces were used to isolate keratins by immunoprecipitation or high-salt extraction. For immunoprecipitation, liver pieces were solubilized in 1% Empigen or 1% Nonidet P-40 in PBS containing a protease/phosphatase inhibitor mixture (22). SDS/PAGE was performed with 10% acrylamide gels (23). Immunoblotting (24) was performed by transferring immunoprecipitates or total liver homogenates (solubilized in 2% SDS in 5% glycerol-containing Laemmli sample buffer) to polyvinylidene difluoride membranes, followed by blotting with anti-keratin antibodies, and then visualization by using enhanced chemiluminescence. Immunofluorescence staining (16) and transmission electron microscopy (25) were performed as described.
Results
Characterization of Transgenic Mice That Overexpress Human K18 Ser-33→Ala.
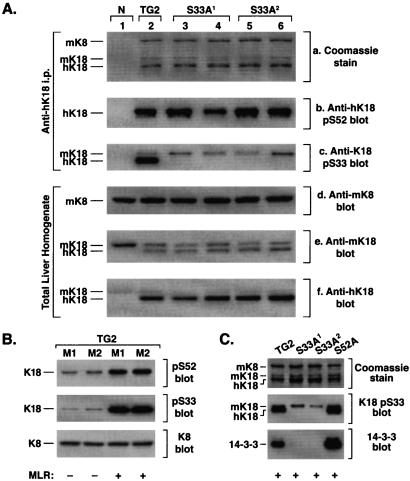
We used a well described 10-kb genomic hK18 clone (15, 21) to design a construct with the only change being a point mutation that generated K18 Ser-33→Ala (S33A). Initially, in transfected cells, we verified that the K18 S33A genomic construct expresses the normally expected K18 product (not shown). This construct was then used to generate several transgenic mouse lines that overexpress K18 S33A, from which two lines, S33A1 and S33A2, with germ-line transmission were selected for subsequent study. The transgene copy numbers in these two lines were 8 and 12, respectively, which is similar to two previously described control mouse lines. The control lines we used (15) overexpress WT hK18 (termed TG2 mice) and hK18 Ser-52→Ala (termed S52A mice) and have 10 and 9 copies, respectively, when tested in the same experiment with the S33A lines. Overexpression was demonstrated biochemically by the ability of mAb L2A1 (which recognizes human but not mK18) to immunoprecipitate the human transgene product from the TG2 and S33A lines but not from normal (N) nontransgenic mice (Fig. 1Aa). The human K18 S33A transgene is not phosphorylated on Ser-33, as expected, when tested by blotting with a phospho(p)-specific anti-K18 pS33 antibody, while K18 S52 phosphorylation remained intact (Fig. 1 Ab and Ac). The anti-hK18 pS52 Ab recognizes only hK18 (because the context of the corresponding mouse site is slightly different than its human counterpart), whereas the anti-hK18 pS33 Ab recognizes human and endogenous mK18 (because that site is highly conserved) (16). Overexpression of hK18 does not alter K8 levels but results in down-regulation of endogenous mK18 expression as determined by blotting of total liver homogenates with mouse and human selective anti-K18 Abs (Fig. 1 Ad–Af) and as observed (15).
Figure 1.
Characterization and expression of K18 S33A and its binding to 14-3-3ζ in transgenic mice. (A) Liver fragments were isolated from the following mouse lines: nontransgenic (N) (one mouse), TG2 (one mouse), S33A1 and S33A2 (two mice each). Fragments were homogenized in 1% Empigen followed by centrifugation. The clarified lysate was used to immunoprecipitate keratins with mAb L2A1 (which recognizes hK18). Immunoprecipitates were analyzed by Coomassie staining (a) and by immunoblotting with anti-hK18 Abs that recognize K18 pS52 (b), or K18 pS33 (c). (d–f) Alternatively, overall keratin expression level was examined by blotting of total liver protein homogenates with anti-mK8, anti-mK18 (which also crossreacts with hK18), and anti-hK18 (which partially crossreacts with mK18). (B) TG2 mice were injected i.p. with MLR in saline, or saline alone, followed by removal of the livers and then homogenization in Laemmli sample buffer. Equal amounts (10 μg) of the total liver lysates were separated by SDS/PAGE followed by blotting with anti-K18 pS52, anti-K18 pS33, or anti-K8 Ab. A blot of nearly equal amounts of K8/18 precipitates (determined by Coomassie staining) gave a similar blotting pattern (not shown). M1 and M2 correspond to separate mice. (C) TG2, S33A1, S33A2, and S52A mice were injected with MLR followed by removal of the livers, solubilization in 1% Nonidet P-40, then keratin immunoprecipitation with mAb L2A1. Precipitates were analyzed by blotting with antibodies to K18 pS33 and 14-3-3ζ. Note that in S33A livers, only the endogenous mK18 remains phosphorylated on S33, whereas 14-3-3ζ coimmunoprecipitates primarily with keratins that are isolated from TG2 and S52A mice.
We tested the effect of the K18 S33A mutation on keratin/14-3-3 binding. Induction of K18 hyperphosphorylation at Ser-33 and Ser-52 (Fig. 1B), with the phosphatase inhibitor MLR, results in significant binding of 14-3-3ζ to K8/18 immunoprecipitates that were obtained from TG2 and S52A but not from S33A mouse livers (Fig. 1C). Longer exposure of the membrane shown in Fig. 1C to film also demonstrates a low level of 14-3-3 binding to endogenous mK18 in the S33A lines as expected (not shown). Therefore, analysis of 14-3-3 binding to K18 in S33A, S52A, and TG2 mouse livers (Fig. 1 B and C) recapitulates what has been described in transfected cells (16) and indicates that these transgenic mice will be useful for studying K18 Ser-33 phosphorylation function in vivo.
Effect of the K18 S33A Mutation on Keratin Filament Organization in Tissues.
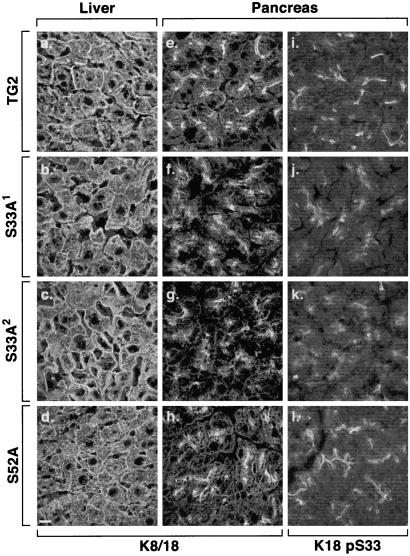
Transfection of K18 S33A into NIH 3T3 cells results in perinuclear keratin filament collapse only when transfected stably but not in transient transfection assays (16), which suggested an important role for K18 S33 phosphorylation and/or 14-3-3 binding in maintaining normal keratin filament organization (16), and prompted us to examine keratin filament organization in a variety of K18 S33A mouse tissues. Keratin filaments seem normal in hepatocytes (Fig. 2 a–d), except for a few scattered cells with short filaments or dots (not shown), and also seemed normal in enterocytes of the small and large intestine (not shown). In contrast, pancreatic acinar cell keratin filaments retracted from the basal nuclear region toward the apical pole and became apically concentrated, which collectively seemed to reduce the extended cytoplasmic staining and disperse the usually fine line of apicolateral staining typically seen in pancreata of WT or S52A K18-overexpressing mice (Fig. 2 e–h). The acinar cell filament reorganization phenotype (shown at the higher magnification of an independent experiment in Fig. 3 a and b) was also confirmed at the ultrastructural level by the prominent appearance of thick filament bundles (compare Fig. 3 c and d) that are not as prominently seen in TG2 pancreata (or in nontransgenic mice pancreata, not shown). The S33 (Fig. 2 i–l) and S52 (not shown) phosphorylated K18 species are polarized in that they are restricted to the apicolateral domain, but their staining pattern becomes diffuse with formation of dots in the S33A transgenic lines (because of endogenous phospho-mK18 staining) as compared with the TG2 and S52A lines. These findings suggest that other “factors” such as keratin posttranslational modifications or associated proteins are present in hepatocytes, but not in acinar cells, which help stabilize the hepatocyte keratin filament network (see Discussion). The differences in acinar cell and hepatocyte keratin filament organization are not due to pancreatic cell damage, because S33A mouse liver and pancreatic histology (as determined by hematoxylin/eosin staining) and serologic tests (amylase and lipase) are normal (not shown).
Figure 2.
Immunofluorescence staining of keratins in transgenic mouse tissues. Liver and pancreas were isolated from the indicated transgenic lines. Tissues were freshly frozen, sectioned, and then fixed in cold acetone. Staining was performed with mAb L2A1, which recognizes the hK18 transgene product (a–h) and anti-K18 pS33 (i–l). Note that the “tight” apicolateral filament distribution, particularly noted in i and l (pancreata of TG2 and S52A mice, respectively), are more dispersed in S33A mice pancreata (j and k; similar findings were noted with anti-hK18 pS52 Ab, not shown). (Bar = 100 μm.)
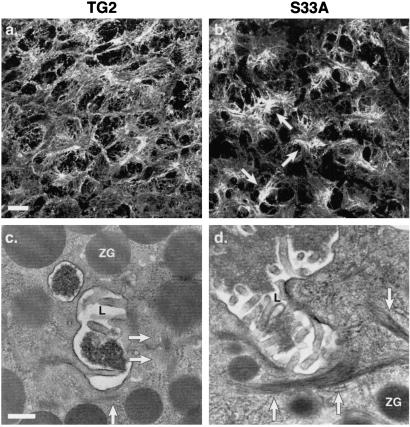
Figure 3.
Immunofluorescence staining and transmission electron microscopy of TG2 and S33A mouse pancreata. (a and b) Immunofluorescence staining was conducted on fresh-frozen tissues obtained from age- and sex-matched TG2 and S33A2 mouse pancreata by using mAb L2A1 as described for Fig. 2. Arrows in b highlight the collapsed apicolateral keratin filament staining that is a consequence of the K18 S33A mutation. (Bar = 100 μm.) (c and d) Transmission electron microscopy was performed on the same pancreata used in a and b. Arrows indicate keratin filament bundles. Similarly appearing bundles were confirmed to be keratins by immune electron microscopy (25, 27). L, lumen; ZG, zymogen granule. (Bar = 0.33 μm.)
Effect of the K18 S33A Mutation on Hepatocyte Mitotic Progression, and 14-3-3 Nuclear Distribution.
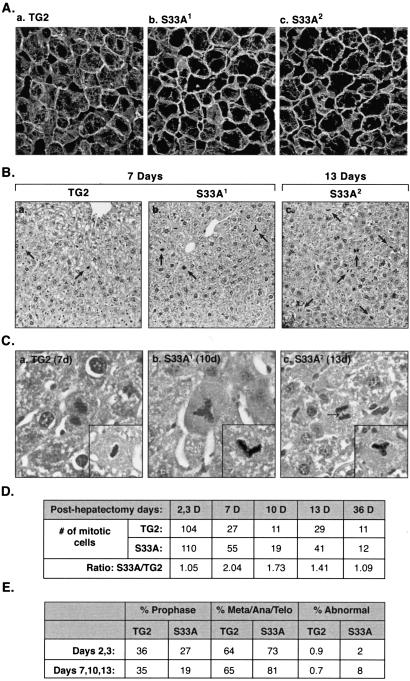
We compared the effect of partial hepatectomy on liver regeneration and histology, and on survival of S33A and TG2 mice. Within 1–3 days, partial hepatectomy resulted in a generalized collapse of the hepatocyte keratin filament network in S33A mice (Fig. 4 Ab and Ac), which was not seen in TG2 (Fig. 4Aa) or S52A (15) mice. The collapsed network returned to normal-appearing filaments within 7–10 days after partial hepatectomy (not shown). Histologic staining of the livers after partial hepatectomy revealed a higher number of mitotic figures in S33A mice than in TG2 mice (Fig. 4B, cells highlighted by arrows). In addition, the S33A posthepatectomy livers had several abnormal mitotic figures, including tripolar and angulated mitotic bodies and bridge formation, which were rarely seen in TG2 livers (Fig. 4C). Quantification of the number of mitotic figures (Fig. 4D) indicated that early on after partial hepatectomy (i.e., 2–3 days) the number of mitotic bodies peak but are similar when comparing TG2 with S33A mice. These mitotic bodies gradually decrease to baseline levels 36 days later, but are found at 1.4- to 2-fold higher numbers in S33A livers 7–13 days after partial hepatectomy (Fig. 4D; P = 0.03). The increased number of mitotic figures in S33A livers during days 7–13 posthepatectomy, as compared with TG2 livers, is likely related to mitotic arrest. This conclusion is supported by the: (i) relative lower percent of prophase cells (P = 0.02); (ii) relative higher percent of metaphase/anaphase/telophase cells (P = 0.02); and (iii) significant increase in abnormal mitotic figures (P = 0.007) (Fig. 4E). No tumors were noted when livers were examined 4 months after partial hepatectomy (not shown).
Figure 4.
Comparison of keratin filament organization and the nature of mitotic progression in TG2 and S33A mouse livers after partial hepatectomy. (A) Livers from 3-day posthepatectomy mice were frozen, sectioned, and then stained with anti-hK18 Ab. Note the dramatic loss of cytoplasmic filaments in the S33A livers. (B) TG2 and S33A mice were subjected to partial hepatectomy. At the indicated days, livers were retrieved, fixed, sectioned, and then stained with hematoxylin/eosin. Note the significantly higher numbers of mitotic bodies in the S33A livers after hepatectomy (indicated by arrows). (C) High magnification images of posthepatectomy livers (after hematoxylin/eosin staining) from TG2 and S33A mice. (Insets) Additional examples of mitotic figures from the indicated mouse line livers. Arrow in histogram c (anaphase cell) highlights an aberrant bridge that is not seen in TG2 mouse livers after hepatectomy (e.g., the late-anaphase cell from TG2 liver in histogram a). (D) At the indicated days (D) after hepatectomy, the total number of mitotic figures was counted (from 20 microscopic ×20 fields), then tabulated by assessing hematoxylin/eosin-stained liver sections of TG2 and S33A mice. (E) The percent of total mitotic figures during prophase, or metaphase (Meta) + anaphase (Ana) + telophase (Telo) for TG2 and S33A mouse livers are indicated 2 and 3 days, or 7/10/13 days after partial hepatectomy. Also, the percent of abnormal mitotic figures (e.g., tripolar, 2-arm angulated, or those with bridges) is shown.
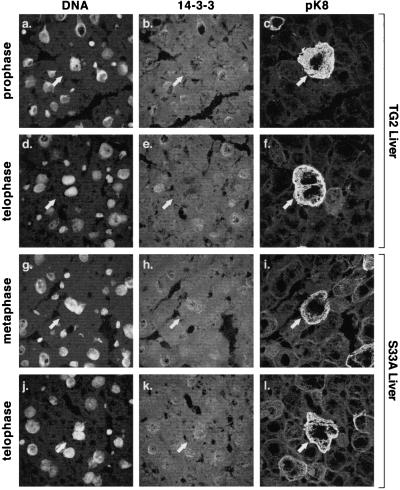
We also examined and compared 14-3-3ζ distributions in TG2 and S33A mouse livers after partial hepatectomy. Assignment of hepatocytes as mitotic was based on the two criteria of the DNA-staining profile and phosphorylation of mouse K8 S79 as detected by staining with the anti-pK8 monoclonal antibody termed LJ4 (26). As shown in Fig. 5 a–f, 14-3-3ζ staining in nonmitotic TG2 mouse hepatocytes manifests a diffuse cytoplasmic pattern (consistent with the majority of 14-3-3ζ distribution being cytosolic; ref 16.) and a punctate nuclear pattern. In contrast, TG2 mouse mitotic hepatocytes lose their 14-3-3ζ nuclear punctate staining (Fig. 5 a–f), whereas mitotic S33A mouse hepatocytes maintain their punctate nuclear 14-3-3ζ staining pattern (Fig. 5 g–l). This dramatic change of nuclear 14-3-3ζ redistribution during mitosis was not seen in cultured tumor cell lines (not shown), which suggests that it is a finding of normal cell growth. Given that the numbers of mitotic hepatocytes at any one time are very limited, it was not possible to detect keratin and 14-3-3 association by co-immunoprecipitation as demonstrated after MLR treatment (Fig. 1C).
Figure 5.
Distribution of 14-3-3ζ during mitosis in TG2 and S33A mice. Mitosis was induced in TG2 and S33A mouse livers by partial hepatectomy. Livers were removed 2 or 3 days after hepatectomy, fixed, and then triple-stained with Toto-1 (DNA stain), anti-14-3-3ζ, and anti-mK8 pS79 (pK8) antibody LJ4. DNA staining was informative, in terms of the mitosis stage, only when clearly delineated mitotic figures were noted and confirmed by staining with the LJ4 antibody that recognizes mitotic hepatocytes (26). Arrows point to the identical triply stained cell for each mitotic stage shown. Note that in contrast to mitotic TG2 hepatocytes, 14-3-3ζ manifests a punctate nuclear staining pattern in mitotic S33A hepatocytes. Similar findings were noted for the 14-3-3σ isoform, although the staining was fainter than for the 14-3-3ζ isoform (not shown).
Discussion
Function of K18 Ser-33 Phosphorylation in Keratin Filament Organization.
The dramatic effect of mutating a single phosphorylation site (K18 Ser-33) on keratin filament organization, in a tissue- and context-dependent fashion, is an important finding for any IF protein. The S33A mutation alters acinar cell keratin filament organization significantly under basal conditions, whereas hepatocyte and enterocyte keratin filaments are minimally affected. The unaltered enterocyte keratin filament organization is not surprising given the substoichiometric expression of K18 relative to K19, with resultant K19 compensation for the K18 mutation. In contrast, the normal-appearing hepatocyte filaments are surprising in light of the acinar cell keratin filament phenotype, particularly because hepatocytes and pancreatic acinar cells express K18 as their major type I keratin. [Acinar cells also express small levels of apicolateral membrane-associated noncytoplasmic K19 (27).] The difference in hepatocyte and acinar cell keratin filament organization is not related to differences in basal K18 S33 phosphorylation levels in the two cell types (not shown), but may be related to the distribution of K18 phospho-Ser-33, other posttranslational modifications, or unknown filament-stabilizing proteins. For example, the K18 pS33 species in hepatocytes distribute as cytoplasmic and membrane-proximal filaments (16) but are essentially absent from the cytoplasm and are found exclusively in proximity to the apicolateral membrane in acinar cells (Fig. 2). Alternatively, the abnormal keratin filament organization in acinar cells under basal conditions may be related to the repeated keratin filament reorganization that occurs upon stimulated secretion (28).
The role of K18 Ser-33 in keratin filament organization may also be independent of K18 binding to 14-3-3 since under basal conditions we do not detect significant keratin/14-3-3 binding. This binding is associated with keratin solubilization and is observed primarily within the soluble K8/18 fraction, which is consistent with 14-3-3 distribution primarily within the cytosolic compartment (17). In addition, it is unlikely that the altered keratin filament organization in acinar cells under basal conditions is caused by mutation-related structural alteration in the K18 protein backbone, because K18 S33A and K18 S33D mutants assemble normally in transiently transfected cells (16), and also because K18 S33A assembles normally in hepatocytes under basal conditions (Fig. 2). These findings indicate that K18 Ser-33 phosphorylation plays an important role in filament reorganization (e.g., after partial hepatectomy) or in basal filament organization (e.g., acinar cells) depending on the tissue context. To that end, several in vitro studies demonstrate the importance of the ordered assembly of filaments (29), and of intrinsic determinants within IF proteins, for their organization (4, 30). Among these determinants are interactions of the head-and-rod domains, which can be modulated by phosphorylation as demonstrated by phosphorylation-mediated inhibition of purified vimentin head-domain interaction with helix 2B of the rod domain (31), supporting the hypothesis that keratin (and likely other IF proteins) filament organization and reorganization are hierarchical and involve multiple phosphorylation/dephosphorylation events that may be coupled to associations/dissociations with other IF-binding proteins.
Our approach of studying the function of keratin phosphorylation in vivo by using transgenic animals has now been validated by using models that express K18 Ser-52→Ala (15) or K18 Ser-33→Ala (this study). This approach is likely to be useful for studying the function of the posttranslational modifications (e.g., phosphorylation and glycosylation) of other IF proteins and has also been used recently in studying the function of non-IF protein phosphorylation (32–34). The approach may entail overexpression of the mutant or WT keratin (as done herein to generate a dominant-negative phenotype), expression of mutant or WT proteins in null backgrounds, or gene replacement. One advantage of simple transgene overexpression is that it mimics what may be seen functionally in natural mutations (e.g., disease situations), although a possible disadvantage is that the phenotype may be underestimated because of endogenous keratin expression. However, overexpression of human K18 results in down-regulation of the endogenous mouse K18 because of tight regulation of type I keratin protein levels by fixed type II keratin levels (Fig. 1A; refs. 15, 22), which allows for adequate representation of the transgene mutant or WT product.
Functional Significance of Keratin/14-3-3 Binding and K18 Ser-33 Phosphorylation during Mitosis.
The effect of the K18 Ser-33 mutation on mitotic progression in dividing hepatocytes in vivo, albeit modest, provides support for the modulatory importance of keratin interaction with 14-3-3 proteins during mitosis. The unique nuclear redistribution of 14-3-3 in dividing hepatocytes during mitosis (Fig. 5), as noted in primary tissues in vivo but not in cultured cell lines, underscores the importance of in vivo studies. Analogous findings were noted during formation of adherens junctions in primary keratinocytes, which differed significantly from what was observed in multiply passaged keratinocytes or cell lines (35). The 14-3-3ζ nuclear redistribution is likely related to the already established K18 S33 mitotic phosphorylation and subsequent K18/14-3-3 binding (16, 17), because it did not occur in S33A mouse livers after partial hepatectomy. Because K18/14-3-3 association occurs in the soluble compartment, the fate of nuclear 14-3-3 puncta, in terms of sequestration by K18, cannot be verified by double-immunofluorescence staining. Our data suggest that 14-3-3 nuclear redistribution is mediated by binding to K18, which likely accounts for the partial mitotic arrest that we observed in S33A mice (Fig. 4). This observation is supported by the established role that 14-3-3 plays in the G2/M checkpoint and mitotic progression (19, 36, 37). The molecule(s) that are affected by inhibition of nuclear 14-3-3 redistribution during mitosis, because of the K18 S33A mutation, remain to be identified. Candidates include the cdc25 phosphatases A, B, and C (37–39), but the hierarchy and cooperative effects of multiple genes (e.g., ref. 40) in mitotic control may explain the somewhat limited effect that we observed on mitotic progression in the S33A mice.
Keratin/14-3-3 binding likely functions as an adapter to bring molecules in close proximity (because of the oligomeric nature of keratins and 14-3-3) or as a displacement mechanism to free other 14-3-3-binding proteins, as shown for vimentin binding with 14-3-3 proteins (41). Support for the displacement mechanism is the lack of any detectable Raf kinase, or other known 14-3-3 binding partners we tested, in association with the keratin/14-3-3 complex (not shown). Our working model includes the following sequence of events: (i) during interphase most of 14-3-3 is cytoplasmic with a small nuclear fraction; (ii) as hepatocytes enter mitosis, K18 S33 phosphorylation levels increase significantly with consequent sequestration of cytoplasmic and nuclear 14-3-3 by keratins. However, 14-3-3 redistribution and sequestration are not possible if K18 S33 is mutated, and (iii) although most cells with the K18 S33A mutation are able to proceed through mitosis, some cells undergo mitotic arrest as a consequence of the S33A mutation.
Acknowledgments
We are very grateful to Dr. Robert Oshima for the use of TG2 mice and for generous gifts of anti-mouse K18 Ab and the genomic K18 construct, to Kris Morrow for preparing the figures, to Dr. Roy Soetikno for assistance with statistical analysis, and to Steve Avolicino (Histo-tec Laboratory, Hayward, CA) for histology staining. This work was supported by National Institutes of Health Grant DK52951, a Veterans Administraion Career Development Award (to M.B.O.), and National Institutes of Health Digestive Disease Center Grant DK56339.
Abbreviations
- h
human
- IF
intermediate filament(s)
- K
keratin
- m
mouse
- MLR
microcystin-LR
- p
phospho
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fuchs E, Weber K. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 2.Moll R, Franke W W, Schiller D L, Geiger B, Krepler R. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 3.Ku N O, Zhou X, Toivola D M, Omary M B. Am J Physiol. 1999;277:G1108–G1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- 4.Coulombe P A, Omary M B. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E, Cleveland D W. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- 6.Irvine A D, McLean W H. Br J Dermatol. 1999;140:815–828. doi: 10.1046/j.1365-2133.1999.02810.x. [DOI] [PubMed] [Google Scholar]
- 7.Ku N O, Gish R, Wright T L, Omary M B. N Engl J Med. 2001;344:1580–1587. doi: 10.1056/NEJM200105243442103. [DOI] [PubMed] [Google Scholar]
- 8.Magin T M, Hesse M, Schroder R. Protoplasma. 2000;211:140–150. [Google Scholar]
- 9.Omary M B, Ku N O, Toivola D M. Hepatology. 2002;35:251–257. doi: 10.1053/jhep.2002.31165. [DOI] [PubMed] [Google Scholar]
- 10.Wong P, Colucci-Guyon E, Takahashi K, Gu C, Babinet C, Coulombe P A. J Cell Biol. 2000;150:921–928. doi: 10.1083/jcb.150.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pant H C, Veeranna, Grant P. Curr Top Cell Regul. 2000;36:133–150. doi: 10.1016/s0070-2137(01)80006-6. [DOI] [PubMed] [Google Scholar]
- 12.Nagata K, Izawa I, Inagaki M. Genes Cells. 2001;6:653–664. doi: 10.1046/j.1365-2443.2001.00454.x. [DOI] [PubMed] [Google Scholar]
- 13.Omary M B, Ku N O, Liao J, Price D. Subcell Biochem. 1998;31:105–140. [PubMed] [Google Scholar]
- 14.Ku N O, Omary M B. J Cell Biol. 1994;127:161–171. doi: 10.1083/jcb.127.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku N O, Michie S A, Soetikno R M, Resurreccion E Z, Broome R L, Omary M B. J Cell Biol. 1998;143:2023–2032. doi: 10.1083/jcb.143.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ku N O, Liao J, Omary M B. EMBO J. 1998;17:1892–1906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao J, Omary M B. J Cell Biol. 1996;133:345–357. doi: 10.1083/jcb.133.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aitken A. Trends Biochem Sci. 1995;20:95–97. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- 19.Fu H, Subramanian R R, Masters S C. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 20.Rosenquist M, Sehnke P, Ferl R J, Sommarin M, Larsson C. J Mol Evol. 2000;51:446–458. doi: 10.1007/s002390010107. [DOI] [PubMed] [Google Scholar]
- 21.Abe M, Oshima R G. J Cell Biol. 1990;111:1197–1206. doi: 10.1083/jcb.111.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku N O, Michie S, Oshima R G, Omary M B. J Cell Biol. 1995;131:1303–1314. doi: 10.1083/jcb.131.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toivola D M, Ku N O, Ghori N, Lowe A W, Michie S A, Omary M B. Exp Cell Res. 2000;255:156–170. doi: 10.1006/excr.1999.4787. [DOI] [PubMed] [Google Scholar]
- 26.Liao J, Ku N O, Omary M B. J Biol Chem. 1997;272:17565–17573. doi: 10.1074/jbc.272.28.17565. [DOI] [PubMed] [Google Scholar]
- 27.Toivola D M, Baribault H, Magin T, Michie S A, Omary M B. Am J Physiol. 2000;279:G1343–G1354. doi: 10.1152/ajpgi.2000.279.6.G1343. [DOI] [PubMed] [Google Scholar]
- 28.O'Konski M S, Pandol S J. J Clin Invest. 1990;86:1649–1657. doi: 10.1172/JCI114887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann H, Aebi U. Curr Opin Struct Biol. 1998;8:177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 30.Coulombe P A, Bousquet O, Ma L, Yamada S, Wirtz D. Trends Cell Biol. 2000;10:420–428. doi: 10.1016/s0962-8924(00)01828-6. [DOI] [PubMed] [Google Scholar]
- 31.Gohara R, Tang D, Inada H, Inagaki M, Takasaki Y, Ando S. FEBS Lett. 2001;489:182–186. doi: 10.1016/s0014-5793(01)02108-1. [DOI] [PubMed] [Google Scholar]
- 32.Chu G, Lester J W, Young K B, Luo W, Zhai J, Kranias E G. J Biol Chem. 2000;275:38938–38943. doi: 10.1074/jbc.M004079200. [DOI] [PubMed] [Google Scholar]
- 33.MacGowan G A, Du C, Cowan D B, Stamm C, McGowan F X, Solaro R J, Koretsky A P, Del Nido P J. Am J Physiol. 2001;280:H835–H843. doi: 10.1152/ajpheart.2001.280.2.H835. [DOI] [PubMed] [Google Scholar]
- 34.Sanbe A, Fewell J G, Gulick J, Osinska H, Lorenz J, Hall D G, Murray L A, Kimball T R, Witt S A, Robbins J. J Biol Chem. 1999;274:21085–21094. doi: 10.1074/jbc.274.30.21085. [DOI] [PubMed] [Google Scholar]
- 35.Vasioukhin V, Bauer C, Yin M, Fuchs E. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 36.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 37.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 38.Conklin D S, Galaktionov K, Beach D. Proc Natl Acad Sci USA. 1995;92:7892–7896. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mils V, Baldin V, Goubin F, Pinta I, Papin C, Waye M, Eychene A, Ducommun B. Oncogene. 2000;19:1257–1265. doi: 10.1038/sj.onc.1203419. [DOI] [PubMed] [Google Scholar]
- 40.Chan T A, Hwang P M, Hermeking H, Kinzler K W, Vogelstein B. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 41.Tzivion G, Luo Z J, Avruch J. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]