Abstract
Proteins can form reversible mixed disulfides with glutathione (GSH). It has been hypothesized that protein glutathionylation may represent a mechanism of redox regulation, in a fashion similar to that mediated by protein phosphorylation. We investigated whether GSH has a signaling role in the response of HL60 cells to hydrogen peroxide (H2O2), in addition to its obvious antioxidant role. We identified early changes in gene expression induced at different times by H2O2 treatment, under conditions that increase protein glutathionylation and minimal toxicity. We then investigated the effect of prior GSH depletion by buthionine sulfoximine and diethylmaleate on this response. The analysis revealed 2,016 genes regulated by H2O2. Of these, 215 genes showed GSH-dependent expression changes, classifiable into four clusters displaying down- or up-regulation by H2O2, either potentiated or inhibited by GSH depletion. The modulation of 20 selected genes was validated by real-time RT-PCR. The biological process categories overrepresented in the largest cluster (genes whose up-regulation was inhibited by GSH depletion) were NF-κB activation, transcription, and DNA methylation. This cluster also included several cytokine and chemokine ligands and receptors, the redox regulator thioredoxin interacting protein, and the histone deacetylase sirtuin. The cluster of genes whose up-regulation was potentiated by GSH depletion included two HSPs (HSP40 and HSP70) and the AP-1 transcription factor components Fos and FosB. This work demonstrates that GSH, in addition to its antioxidant and protective function against oxidative stress, has a specific signaling role in redox regulation.
Keywords: chemokines, cytokines, hydrogen peroxide, oxidative stress
Glutathione (GSH) protects the cell from damage induced by high levels of reactive oxygen species (ROS), defined as oxidative stress (1). Its main antioxidant activity consists of the detoxification of peroxides, partly with the aid of various GSH peroxidases. Its role as a major thiol antioxidant is demonstrated by the fact that various conditions of oxidative stress are exacerbated by GSH depletion, for instance by inhibitors of its synthesis, and ameliorated by the addition of GSH or its precursors, including N-acetylcysteine (NAC). Experimentally, thiol antioxidants, including GSH and NAC, have been used as tools to investigate the role of ROS in biological systems.
In addition to scavenging ROS, GSH can form mixed disulfides with proteins, a phenomenon known as protein glutathionylation (2–4). This can occur by various reactions, either by thiol/disulfide exchange between protein cysteines and oxidized GSH (GSH disulfide, GSSG), by direct oxidation, or through the NO-mediated formation of S-nitrosothiols (3, 4).
The effect of protein glutathionylation is generally considered deleterious in the framework of oxidative stress, because it is one of the many forms of thiol oxidation induced by ROS. However, according to the more recent concept of redox regulation, several protein cysteines can be defined as “redox sensitive.” Although disulfide bonds are virtually absent in cytosolic proteins, due to the highly reducing intracellular environment, proteomic studies have shown that many of them can undergo reversible oxidations to form disulfides and glutathionylated proteins (2–5). The reversibility of protein glutathionylation, catalyzed by glutaredoxin and, to a lesser extent, thioredoxin (6), makes this posttranslational modification a likely molecular mechanism by which GSH could act as a redox-dependent signaling molecule, in analogy with protein phosphorylation. However, despite the identification of several proteins undergoing glutathionylation (5, 7), in our opinion a signaling role of glutathionylation is far from being established.
Because glutathionylation is a redox-dependent modification, we reasoned that it might participate in signaling the exposure of the cell to oxidants and have undertaken a study to establish whether GSH has a signaling role in the response to oxidants. To this purpose, we performed studies of gene expression profiling in human promyelocytic leukemia HL-60 cells exposed to hydrogen peroxide (H2O2). Several genes were regulated at different time points in response to H2O2, under conditions that resulted in increased protein glutathionylation. Using this model, we studied the effect of prior GSH depletion by buthionine sulfoximine (BSO) (8) and the GSH depletor diethylmaleate (DEM) (9), to investigate, using two-way ANOVA on microarray data, whether there are patterns of gene regulation that require GSH, because these would be the changes where protein glutathionylation might have a role.
Methods
HL60 Cells and GSH Depletion and Treatments. HL60 cells were cultured in RPMI medium 1640 with 2 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin sulfate (all from Sigma)/10% heat-inactivated FCS. Cells were maintained at a concentration between 0.3 × 106 per ml and 1.5 × 106 per ml. For GSH depletion, they were incubated at the density of 0.75 × 106 per ml in complete medium with 200 μM BSO for 23 h, then with 1 mM DEM for 1 additional hour. Twenty-four hours after starting the depletion treatment, cell density was 0.75 × 106 as compared to 1.0 × 106 per ml in parallel controls and viability (as determined by Trypan blue exclusion) was >98% in both conditions. Control or depleted cells were then incubated with 50 μM H2O2 for 0, 0.5, 2, or 24 h, always in complete medium.
GSH Determinations. Cells (30 × 106) were suspended in 0.3 ml of 5% sulfosalycylic acid. GSH was measured both in the supernatant and in the fraction released from the washed pellet by alkali treatment (protein-bound GSH) (10) using an enzymatic method (11).
RNA Isolation and Microarray Hybridization. All experiments were performed in triplicate. Each sample (20 × 106 cells) was lysed in 3 ml of TRIzol Reagent (Life Technologies, Gaithersburg, MD) and RNA extracted according to the manufacturer's protocol. The obtained RNA was further purified by using the RNeasy (Qiagen, Valencia, CA) system and protocol. Only the samples with the highest purity and integrity (as judged by A260/280 ratio, on 1% agarose/formaldehyde gels and on a Bioanalyzer) (Agilent Technologies, Palo Alto, CA) were pooled and further processed. Biotin labeling of total RNA, hybridization, washing, and scanning of the Affymetrix (Santa Clara, CA) GeneChip Human Genome Hg-U133A were carried out as recommended by the manufacturer. Because there was <2% variability between replicate gene chips carried out with these procedures, technical replicates were not performed. Biological triplicates (samples from three completely independent experiments) were analyzed.
Microarray Data Analysis. Image quantification, background subtraction, and scaling were made with microarray suite (mas) 5.0 software (Affymetrix) with the default parameters for the statistical algorithm. No further background correction of the data was applied. For signal-dependent normalization, the Qspline method (12) was applied with default settings from the affy-package from Bioconductor (13), using the R language and environment. For statistical analysis of differentially expressed genes, we applied ANOVA in the R environment (14), either one way for the extended time course in control cells (0, 0.5, 2, and 24 h of H2O2 treatment) or two-way for the comparable parts of the experiment (control and depleted cells, 0, 0.5, and 2 h of H2O2 treatment), giving values for the significance of differences between the groups (depletion effect), in the groups (H2O2 effect), or with interaction. This gave us a table of four P values for each gene. Because the distribution of the P values for the interaction in two-way ANOVA was homogeneous, we did not take into account this parameter. Two sets of genes (affected by H2O2 treatment and not by GSH depletion and affected by both conditions) were selected and classified according to the similarity in their expression profile, by complete linkage hierarchical clustering using the uncentered Pearson correlation coefficient as the distance measure using the genesis 1.3 software (15). Statistically significant overrepresentation in Gene Ontology categories was checked by using the Gene Ontology Tree Machine (http://genereg.ornl.gov/gotm) (16).
Microarray Data Validation. Quantitative real-time RT-PCR validation was made for 20 genes in four independent experiments (including experiments not tested in microarray) and on-target sequences chosen independently from the target sequences of the arrays. Reverse transcription was carried out at 42°C for 50 min in 50 μl of reverse transcription mixture containing 500 ng of total RNA, 400 units of Reverse Transcriptase (SuperScript II, Invitrogen), 20 units RNase inhibitor (RNase out, Invitrogen), 0.8 mM each dNTPs (Amersham Pharmacia Biosciences), and 1 μg of random primers (Promega). To measure TNF-α mRNA, PCR was performed by using a SyBr green PCR master mix (Applied Biosystems) and the following primers: 5′-GCTTTGATCCCTGACATCTGG-3′ and 5′-AAGTCCTGCAGCATTCTGGC-3′, designed with the primer express software (Applied Biosystems). β-Actin was used as endogenous control (17). All other genes were measured on microfluidic cards with the selected TaqMan Gene Expression Assays pre-loaded into each reaction well (TaqMan low-density arrays, Applied Biosystems), using β-glucuronidase as the endogenous control. All procedures, including data analysis, were performed on the Applied Biosystems PRISM 7900 Sequence Detector (Applied Biosystems).
Results
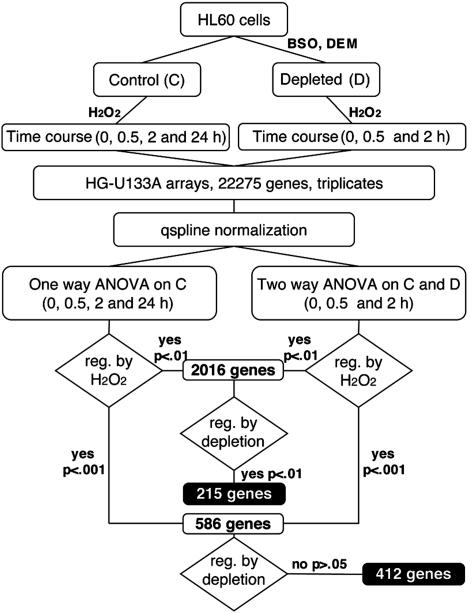
Experimental Design. HL60 cells were depleted of GSH by over-night treatment with 200 μM BSO followed by 1-h incubation with 1 mM DEM. This led to a reduction of 80% in total cellular GSH, without affecting cell viability. Cells were then treated with 50 μMH2O2, which was chosen as the highest concentration that would not decrease cell viability in control cells in the first 2 h. At the time of H2O2 treatment, GSH-depleted cells viability was only slightly decreased (by ≈30%), whereas at 24 h, toxicity in GSH-depleted cells was 90% vs. 30% in control cells. No further toxicity was observed in the next 24 h in control cells, although cell growth was completely inhibited. Gene expression was therefore analyzed up to 24 h in control cells and up to 2 h in depleted cells in three independent experiments (see Fig. 1 for a scheme of the experimental design). Exposure of cells to 50 μM H2O2 for 30 min did not significantly change total GSH levels (control, 185 ± 14; H2O2, 195 ± 10 μg/106 cells), whereas it induced a marked increase in overall protein glutathionylation (protein-bound GSH: control, 17.8 ± 7.8; H2O2; 28.9 ± 2.2 μg/106 cells).
Fig. 1.
Experimental design and data analysis diagram
Microarray Data Analysis. Expression data from mas were normalized by using the signal-dependent Qspline method (12). Statistical analysis was then performed by ANOVA (modified from ref. 13), either two way for the comparable samples in control and depleted cells (up to 2 h of H2O2 treatment) or one way for the 24-h time course in control cells. This gave us a table of four P values for each gene. We then selected two sets of genes: those whose expression was affected by H2O2 treatment (P < 0.001 in either of the two analyses) but not by GSH depletion and those affected by both conditions, as outlined in Fig. 1. The number of genes affected by GSH depletion was markedly lower than that of the unaffected. For this reason, for the second set, we used less stringent statistical conditions (P < 0.01). Genes that were significantly affected by H2O2 treatment either in the one-way or in the two-way analysis at P < 0.001 were 586, of which 412 were not affected by depletion (P > 0.05). Genes affected by H2O2 at P < 0.01 were 2,016, of which 215 were also affected by depletion (P < 0.01).
As a further comparison, we looked for the genes changed by depletion at a high significance (P < 0.001, 139 genes) but not by H2O2 (P > 0.05) and found only 27 genes that, compared with the 412 of the reverse pattern, confirm the absence of gross toxicity effects of depletion per se, and demonstrate that the main changes brought about by GSH depletion reside on the cell response to oxidant stimuli.
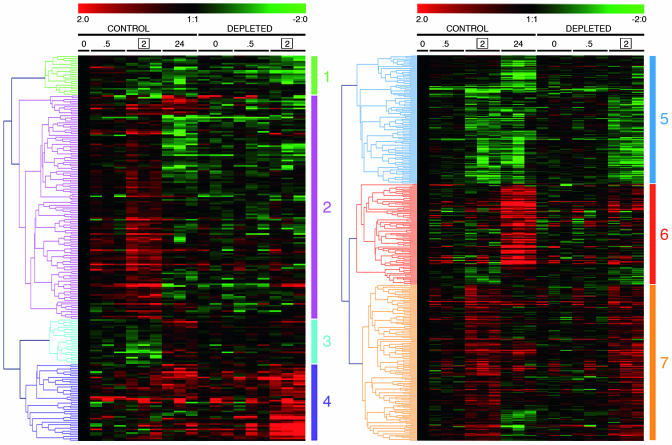
Genes were then classified, according to the similarity in their expression profile, by hierarchical clustering. The set of genes changed only by H2O2 is shown on Fig. 2 Right and has been classified into three clusters, corresponding to down-regulated (cluster 5), late-induced (cluster 6), or early-induced (cluster 7) genes.
Fig. 2.
Hierarchical clustering of GSH-dependent and -independent H2O2-regulated genes. Time points were 0, 0.5, 2, and 24 h for control cells and 0, 0.5, and 2 h for GSH-depleted cells. Data represent log2 ratios vs. respective C0 (no H2O2; no GSH depletion) of triplicate experiments
The genes changed by both conditions, i.e., those whose H2O2 regulation was significantly altered by GSH depletion, are shown in Fig. 2 Left and were classified into four clusters: down-regulation by H2O2 potentiated by BSO/DEM (1), up-regulation by H2O2 inhibited by BSO/DEM (2), down-regulation by H2O2 inhibited by BSO/DEM (3), and up-regulation by H2O2 potentiated by BSO/DEM (4). Clusters 1 and 4 may include genes whose response is amplified due to the higher oxidant stimulus provided by H2O2 in the absence of the antioxidant GSH, or genes whose regulation is inhibited by GSH. Clusters 2 and 3 contain genes whose regulation positively depends on the presence of GSH, i.e., that need GSH for their H2O2-induced regulation. The names of the genes in these four clusters and their probeset identifications (www.affymetrix.com/analysis/netaffx/index.affx) are reported in Figs. 4–7, which are published as supporting information on the PNAS web site.
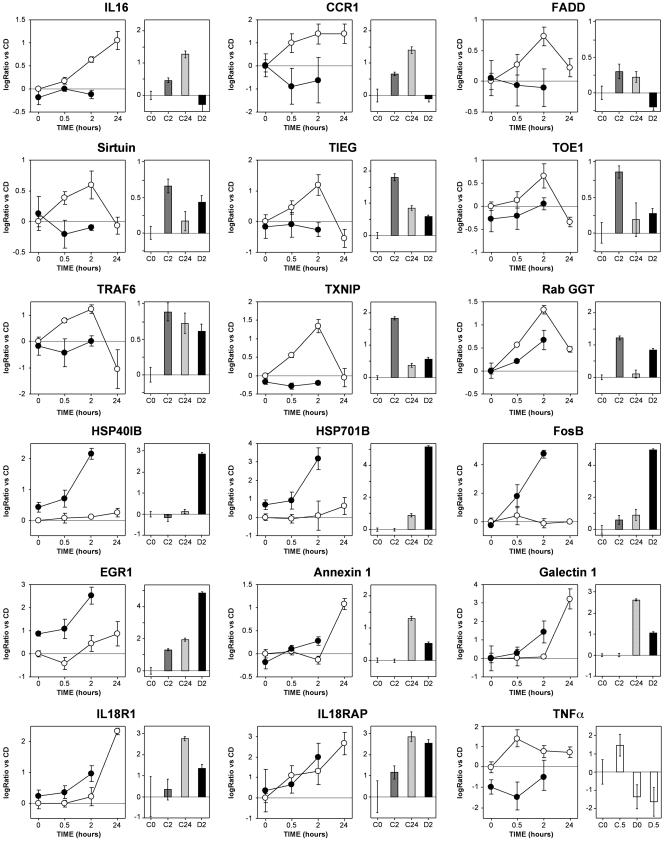
PCR Validation. We selected 20 genes for validation in real-time PCR with target sequences chosen independently from the target sequences of the arrays. Ten genes were taken from cluster 2. For eight of them [IL16 and CC chemokine receptor 1 (CCR1), Fas-associated via death domain protein (FADD), sirtuin, TGFβ inducible early growth response gene (TIEG), target of early growth response receptor 1 (TOE1), thioredoxin inhibitory protein (TXNIP), and type II geranylgeranyl transferase (RabGGT)], the H2O2-induced up-regulation and its dependence on GSH were confirmed by PCR (Fig. 3, at the top). Only TNF receptor-associated factor 6 (TRAF6, Fig. 3 Upper) and v-jun (not shown) had a different expression pattern from that observed in the arrays. In both genes, PCR revealed a stable induction (9-fold for v-jun) from 2 to 24 h, unchanged by depletion. On the contrary, the arrays revealed a transient induction, inhibited by GSH depletion, with a peak at 2 h and a return to lower than basal levels at 24 h. This discrepancy, which probably depends on the different target sequences interrogated by PCR (at the 5′ end) and arrays (at the 3′ end), suggests a peculiar mRNA processing that deserves further investigation.
Fig. 3.
PCR validation of the array data. Results for 18 genes are shown, comparing expression data from microarrays (graphs on the left; open circles, no GSH depletion; closed circles, GSH depleted) and respective results from PCR analysis (bar graphs). Data are expressed as log2 ratio vs. C0. Error bars for the microarray data represent the SEM from triplicate experiments. Error bars for PCR data represent the 95% confidence interval of four independent experiments, each assayed in duplicate. All PCR data were obtained by using TaqMan microfluidic cards, except for TNF-α, which was analyzed by real-time PCR using SyBr green
Five genes were chosen from cluster 4 [two HSPs (HSP40 and HSP70), FosB, early growth response protein 1 (EGR1), and phosphatidylserine receptor]. All of them were confirmed (Fig. 3 and phosphatidylserine receptor, not shown), showing a dramatic effect of GSH depletion on their induction by H2O2. Five additional genes were selected on the basis of their interest [annexin 1, galectin 1, IL-18 receptor type I (IL18R1), IL-18 receptor accessory protein (IL18RAP), and TNF-α). The first four were induced by H2O2 and showed a trend to a higher induction in GSH-depleted cells that became statistically significant when gene expression was analyzed by PCR. TNF-α was induced early upon H2O2 stimulation, an effect that was statistically significant in the PCR validation, although not in the microarray analysis. Not only did GSH depletion inhibit H2O2 induction of this gene, but it also decreased its baseline expression (statistically significant in both assays).
Among the genes whose induction by H2O2 was inhibited by GSH depletion, three major biological process categories were significantly overrepresented, including genes implicated in NF-κB activation, transcription, and DNA methylation (Table 1).
Table 1. Biological process categories according to the Gene Ontology (GO) database over represented among the genes showing GSH-dependent H2O2 induction.
| Go category | Observed | Expected | P value | Gene symbols |
|---|---|---|---|---|
| Positive regulation of NF-κB | 4 | 0.58 | 0.0003 | FADD, TRAF6, EDG2, TNFA |
| Transcription | 34* | 11.67 | 8.E-10 | JUN, KLF10, KIAA0863, ZBTB17, JUND, TOE1, CEBPG, MNT, BRPF1, ZNF140, ZNF175, ZNF35, ZNF305, ZNF571, ZNF45, ZNF304, GATA2, IRX5, ZNF238, ZNF307, ZNF20, HLX1, MCM5, ZNF79, RRN3, FOSL2, ATF7IP, ZNF408, ZNF350, RBM15, T RIM68, ZNF435, FLJ23506, ZNF557 |
| DNA methylation | 2 | 0.14 | 0.009 | ATF7IP, THUMPD2 |
Reported are: the number of regulated genes belonging to the indicated category (observed), the number expected on the basis of the frequency of that class in the array gene list (expected), the significance of the enrichment (P value) (15), and the list of the gene symbols.
Three of those were represented twice.
Discussion
The present work shows that some patterns of gene regulation induced by exposure to H2O2 are GSH-dependent. Interestingly, several of the genes we report to be regulated in a GSH-dependent way are also shown here to be induced by a redox-dependent mechanism. That GSH amplifies the effect of H2O2 on the regulation of some genes, as in clusters 1 and 4, was expected, because GSH depletion, by removing an H2O2-degrading system, obviously increases the extent of oxidation by H2O2. However, these clusters might also include genes whose regulation is inhibited by GSH, such as, for instance, those proteins where glutathionylation is a negative regulator. In particular, cluster 4 included two HSPs and two components of activator protein-1 (AP-1) (Fos and FosB) that are not induced by H2O2 alone and are strongly induced [4- and 8-fold (HSPs) and 25-fold (the two Fos)] in depleted cells, indicating that GSH is a potent inhibitor of their induction. Of note, however, the induction by H2O2 of v-jun, another component of the AP-1 transcription factor, is not potentiated by GSH depletion, suggesting a complex regulation of AP-1 activity by GSH, through a differential regulation of the heterodimer composition. A number of the genes in this cluster are indicative of a higher cell stress and reflect the higher H2O2 toxicity in depleted cells. Some growth arrest-related genes were induced by H2O2 (not shown), in accordance with the observed inhibition of cell growth, but were not GSH-dependent.
The largest cluster (cluster 2) shows an inhibitory effect of GSH depletion on the induction of several genes by H2O2, supporting the initial hypothesis of a role of GSH as a signaling molecule in redox regulation. Among these genes whose induction by H2O2 was GSH-dependent, we found four positive regulators of NF-κB (TRAF6, FADD, EDG2, and TNF-α, with an average and homogeneous 2.2-fold induction), and three of these are also implicated in death receptor pathways. This suggests that H2O2-dependent NF-κB activation could be lower in conditions of decreased GSH. Interestingly, although GSH and NAC inhibit NF-κB activation, GSH is essential for Fas-mediated apoptosis of the hepatocyte (18), and NF-κB activation requires GSSG (19) or GSH biosynthesis (20). Furthermore, there is an optimal GSH-GSSG ratio for NF-κB activation and DNA binding, and this varies for the different promoters (21). It is also important to note that, although glutathionylation involves formation of protein–GSSGs and is obviously inhibited by GSH/GSSG depletion, addition of GSH and its precursor NAC do not augment protein glutathionylation. Indeed, augmenting the levels of GSH will result in a shift of the GSH/GSSG ratio toward a more reducing condition, thus favoring glutaredoxin-mediated protein deglutathionylation (6, 10), an effect that was also observed with other thiol reductants, including NAC (10).
Finally, the 2.5-fold induction of the thioredoxin inhibitor TXNIP (22) by H2O2 was also GSH-dependent, as well as that of the histone deacetylase, also deacetylating other proteins, including p53 and sirtuin (23), which promotes cell survival in mammalian cells and whose yeast homologue extends lifespan (24, 25).
In the present study, we directly addressed the issue of the role of GSH in the signaling events leading to changes in gene expression induced by H2O2, known to augment protein glutathionylation (5, 7). However, it is important to note that protein glutathionylation not only occurs after exposure to oxidants but is also present in basal conditions, and in fact in the present work, we detected a significant amount of glutathionylated protein in untreated cells, as we previously did in human T cell blasts (10). These observations are in agreement with earlier work by Brigelius et al. (26, 27) showing that, in normal liver, ≈1% of total GSH is present as mixed disulfides with proteins (in the range of 30 nmol per gram of tissue). Depending on the cellular redox state, the amount of mixed disulfides may rise up to 20–50% of the total GSH content (28). Thus, it will be important to investigate whether GSH also plays a role in signaling induced by growth factors, cytokines, or bacterial products. In fact, ROS have been documented in recent years as downstream mediators of several distinct signaling pathways, activated by different receptor classes. ROS are produced upon stimulation with growth factors such as platelet-derived growth factor (29), EGF (30), VEGF (31), TGF-β 1 (32), and insulin (33); inflammatory cytokines such as, IL-1β (reviewed in ref. 34) and TNF-α (reviewed in ref. 35); neurotransmitters and peptides, such as serotonin (36), bradikinin (37), and angiotensin II (38); and other stimuli such as T cell receptor-dependent mitogen-activated protein kinase activation (39) and integrin-mediated changes in cell shape (40). Moreover, ROS are generated during the normal cell cycle and govern its progression (41, 42). ROS and NO production is also increased by endotoxin and IFN-γ (43, 44).
There is much evidence that thiols like NAC can regulate the activation of transcription factors and production of cytokines and ultimately affect immune system functions. It has long been known that GSH is essential for several immune functions, such as IL-2 production, IL-2 responses, and cytotoxic T cell activity (45–48), and regulates the Th1-Th2 balance (49). Lower GSH levels in patients with AIDS have been correlated with immune deficiency (47), and alcohol intoxication, a condition that is associated with increased susceptibility to infections (50), also results in GSH depletion (51). Finally, in vivo, GSH depletion worsens survival and antibacterial activity in a model of experimental sepsis (52). Of note, among the GSH-dependent H2O2-regulated genes, we found several immune system-related genes, including cytokines (IL16 and TNF-α) and cytokine receptors (CCR1, IL18R1, and IL18RAP), as well as regulators of NF-κB, a key transcription factor in innate immunity (53). In this context, the definition of the gene expression patterns and transcriptional regulators affected by GSH depletion, along with techniques of redox proteomics to identify glutathionylated proteins (5, 7), may disclose new mechanisms of regulation of immunity.
Supplementary Material
Acknowledgments
This work was supported by grants from Fondazione Cariplo, Milan, Italy; Ministero dell'Istruzione, dell'Università e della Ricerca–Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (MIUR-PRIN) 2003; and MIUR–Fondo per gli Investimenti della Ricerca di Base (MIUR-FIRB) RBAU01Z8C3.
Author contributions: M.F. and P.G. designed research; M.F., L.O.G., U.A.Ø., S.L., R.T., and M.M. performed research; M.F., L.O.G., U.A.Ø., M.M., and P.G. analyzed data; and M.F. and P.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BSO, buthionine sulfoximine; DEM, diethylmaleate; GSH, glutathione; GSSG, GSH disulfide; H2O2, hydrogen peroxide; NAC, N-acetylcysteine; ROS, reactive oxygen species; HSP, heat-shock protein.
References
- 1.Meister, A. & Anderson, M. E. (1983) Annu. Rev. Biochem. 52, 711-760. [DOI] [PubMed] [Google Scholar]
- 2.Fratelli, M., Gianazza, E. & Ghezzi, P. (2004) Exp. Rev. Proteom. 1, 89-100. [DOI] [PubMed] [Google Scholar]
- 3.Ghezzi, P. (2005) Free Radical Res. 39, 573-580. [DOI] [PubMed] [Google Scholar]
- 4.Shelton, M. D., Chock, P. B. & Mieyal, J. J. (2005) Antioxid. Redox Signal. 7, 348-366. [DOI] [PubMed] [Google Scholar]
- 5.Fratelli, M., Demol, H., Puype, M., Casagrande, S., Eberini, I., Salmona, M., Bonetto, V., Mengozzi, M., Duffieux, F., Miclet, E., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 3505-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mannervik, B., Axelsson, K., Sundewall, A. C. & Holmgren, A. (1983) Biochem. J. 213, 519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fratelli, M., Demol, H., Puype, M., Casagrande, S., Villa, P., Eberini, I., Vandekerckhove, J., Gianazza, E. & Ghezzi, P. (2003) Proteomics 3, 1154-1161. [DOI] [PubMed] [Google Scholar]
- 8.Drew, R. & Miners, J. O. (1984) Biochem. Pharmacol. 33, 2989-2994. [DOI] [PubMed] [Google Scholar]
- 9.Costa, L. G. & Murphy, S. D. (1986) Biochem. Pharmacol. 35, 3383-3388. [DOI] [PubMed] [Google Scholar]
- 10.Ghezzi, P., Romines, B., Fratelli, M., Eberini, I., Gianazza, E., Casagrande, S., Laragione, T., Mengozzi, M. & Herzenberg, L. A. (2002) Mol. Immunol. 38, 773-780. [DOI] [PubMed] [Google Scholar]
- 11.Tietze, F. (1969) Anal. Biochem. 27, 502-522. [DOI] [PubMed] [Google Scholar]
- 12.Workman, C., Jensen, L. J., Jarmer, H., Berka, R., Gautier, L., Nielser, H. B., Saxild, H. H., Nielsen, C., Brunak, S. & Knudsen, S. (2002) Genome Biol. 3, research0048. [DOI] [PMC free article] [PubMed]
- 13.Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., et al. (2004) Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlidis, P. (2003) Methods 31, 282-289. [DOI] [PubMed] [Google Scholar]
- 15.Sturn, A., Quackenbush, J. & Trajanoski, Z. (2002) Bioinformatics 18, 207-208. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, B., Schmoyer, D., Kirov, S. & Snoddy, J. (2004) BMC Bioinformatics 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medhurst, A. D., Harrison, D. C., Read, S. J., Campbell, C. A., Robbins, M. J. & Pangalos, M. N. (2000) J. Neurosci. Methods 98, 9-20. [DOI] [PubMed] [Google Scholar]
- 18.Hentze, H., Kunstle, G., Volbracht, C., Ertel, W. & Wendel, A. (1999) Hepatology 30, 177-185. [DOI] [PubMed] [Google Scholar]
- 19.Droge, W., Schulze-Osthoff, K., Mihm, S., Galter, D., Schenk, H., Eck, H. P., Roth, S. & Gmunder, H. (1994) FASEB J. 8, 1131-1138. [PubMed] [Google Scholar]
- 20.Haddad, J. J. & Land, S. C. (2000) Am. J. Physiol. Lung Cell Mol. Physiol. 278, L492-L503. [DOI] [PubMed] [Google Scholar]
- 21.Faruqi, R. M., Poptic, E. J., Faruqi, T. R., De La Motte, C. & DiCorleto, P. E. (1997) Am. J. Physiol. 273, H817-H826. [DOI] [PubMed] [Google Scholar]
- 22.Nishiyama, A., Matsui, M., Iwata, S., Hirota, K., Masutani, H., Nakamura, H., Takagi, Y., Sono, H., Gon, Y. & Yodoi, J. (1999) J. Biol. Chem. 274, 21645-21650. [DOI] [PubMed] [Google Scholar]
- 23.Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149-159. [DOI] [PubMed] [Google Scholar]
- 24.Anderson, R. M., Latorre-Esteves, M., Neves, A. R., Lavu, S., Medvedik, O., Taylor, C., Howitz, K. T., Santos, H. & Sinclair, D. A. (2003) Science 302, 2124-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., de Cabo, R. & Sinclair, D. A. (2004) Science 305, 390-392. [DOI] [PubMed] [Google Scholar]
- 26.Brigelius, R., Muckel, C., Akerboom, T. P. & Sies, H. (1983) Biochem. Pharmacol. 32, 2529-2534. [DOI] [PubMed] [Google Scholar]
- 27.Brigelius, R., Lenzen, R. & Sies, H. (1982) Biochem. Pharmacol. 31, 1637-1641. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, H. F. (1984) Methods Enzymol. 107, 330-351. [DOI] [PubMed] [Google Scholar]
- 29.Sundaresan, M., Yu, Z. X., Ferrans, V. J., Irani, K. & Finkel, T. (1995) Science 270, 296-299. [DOI] [PubMed] [Google Scholar]
- 30.Bae, Y. S., Kang, S. W., Seo, M. S., Baines, I. C., Tekle, E., Chock, P. B. & Rhee, S. G. (1997) J. Biol. Chem. 272, 217-221. [PubMed] [Google Scholar]
- 31.Colavitti, R., Pani, G., Bedogni, B., Anzevino, R., Borrello, S., Waltenberger, J. & Galeotti, T. (2002) J. Biol. Chem. 277, 3101-3108. [DOI] [PubMed] [Google Scholar]
- 32.Ohba, M., Shibanuma, M., Kuroki, T. & Nose, K. (1994) J. Cell Biol. 126, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahadev, K., Wu, X., Zilbering, A., Zhu, L., Lawrence, J. T. & Goldstein, B. J. (2001) J. Biol. Chem. 276, 48662-48669. [DOI] [PubMed] [Google Scholar]
- 34.Brigelius-Flohe, R., Banning, A., Kny, M. & Bol, G. F. (2004) Arch. Biochem. Biophys. 423, 66-73. [DOI] [PubMed] [Google Scholar]
- 35.Garg, A. K. & Aggarwal, B. B. (2002) Mol. Immunol. 39, 509-517. [DOI] [PubMed] [Google Scholar]
- 36.Greene, E. L., Houghton, O., Collinsworth, G., Garnovskaya, M. N., Nagai, T., Sajjad, T., Bheemanathini, V., Grewal, J. S., Paul, R. V. & Raymond, J. R. (2000) Am. J. Physiol. Renal Physiol. 278, F650-F658. [DOI] [PubMed] [Google Scholar]
- 37.Greene, E. L., Velarde, V. & Jaffa, A. A. (2000) Hypertension 35, 942-947. [DOI] [PubMed] [Google Scholar]
- 38.Ushio-Fukai, M., Alexander, R. W., Akers, M., Yin, Q., Fujio, Y., Walsh, K. & Griendling, K. K. (1999) J. Biol. Chem. 274, 22699-22704. [DOI] [PubMed] [Google Scholar]
- 39.Devadas, S., Zaritskaya, L., Rhee, S. G., Oberley, L. & Williams, M. S. (2002) J. Exp. Med. 195, 59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner, E. & Werb, Z. (2002) J. Cell Biol. 158, 357-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi, Y., Ogra, Y. & Suzuki, K. T. (2004) Life Sci. 75, 301-311. [DOI] [PubMed] [Google Scholar]
- 42.Conour, J. E., Graham, W. V. & Gaskins, H. R. (2004) Physiol. Genomics 18, 196-205. [DOI] [PubMed] [Google Scholar]
- 43.Moncada, S. & Higgs, A. (1993) N. Engl. J. Med. 329, 2002-2012. [DOI] [PubMed] [Google Scholar]
- 44.Nathan, C. (2003) J. Clin. Invest. 111, 769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Droge, W., Eck, H. P. & Mihm, S. (1992) Immunol. Today 13, 211-214. [DOI] [PubMed] [Google Scholar]
- 46.Hargrove, M. E., Wang, J. & Ting, C. C. (1993) Cell Immunol. 149, 433-443. [DOI] [PubMed] [Google Scholar]
- 47.Staal, F. J., Ela, S. W., Roederer, M., Anderson, M. T. & Herzenberg, L. A. (1992) Lancet 339, 909-912. [DOI] [PubMed] [Google Scholar]
- 48.Yim, C. Y., Hibbs, J. B., Jr., McGregor, J. R., Galinsky, R. E. & Samlowski, W. E. (1994) J. Immunol. 152, 5796-5805. [PubMed] [Google Scholar]
- 49.Peterson, J. D., Herzenberg, L. A., Vasquez, K. & Waltenbaugh, C. (1998) Proc. Natl. Acad. Sci. USA 95, 3071-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jerrells, T. R., Slukvin, I., Sibley, D. & Fuseler, J. (1994) Alcohol Alcohol Suppl. 2, 425-430. [PubMed] [Google Scholar]
- 51.Altomare, E., Vendemiale, G. & Albano, O. (1988) Life Sci. 43, 991-998. [DOI] [PubMed] [Google Scholar]
- 52.Villa, P., Saccani, A., Sica, A. & Ghezzi, P. (2002) J. Infect. Dis. 185, 1115-1120. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, G. & Ghosh, S. (2001) J. Clin. Invest. 107, 13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.