Abstract
Bacteria of Shigella spp. are responsible for shigellosis in humans. They use a type III secretion system to inject effector proteins into host cells and induce their entry into epithelial cells or trigger apoptosis in macrophages. We present evidence that the effector OspG is a protein kinase that binds various ubiquitinylated ubiquitin-conjugating enzymes, including UbcH5, which belongs to the stem cell factor SCFβ-TrCP complex promoting ubiquitination of phosphorylated inhibitor of NF-κB type α (phospho-IκBα). Transfection experiments indicated that OspG can prevent phospho-IκBα degradation and NF-κB activation induced by TNF-α stimulation. Infection of epithelial cells by the S. flexneri wild-type strain, but not an ospG mutant, led to accumulation of phospho-IκBα, consistent with OspG inhibiting SCFβ-TrCP activity. Upon infection of ileal loops in rabbits, the ospG mutant induced a stronger inflammatory response than the wild-type strain. This finding indicates that OspG negatively controls the host innate response induced by S. flexneri upon invasion of the epithelium.
Keywords: IκB, inflammation, invasion, pathogen, ubiquitination
The intestinal barrier is endowed with detection and defense mechanisms to achieve tolerance to commensal microorganisms and protection against invading microorganisms (1). Invasion by extracellular and intracellular pathogens is sensed by various signaling pathways converging to activate NF-κB, a member of the Rel family of transcription factors involved in the activation of a large number of genes in response to pathogens, stress signals and proinflammatory cytokines (2). Under nonstimulating conditions, NF-κB is retained in the cytoplasm through its association with inhibitory proteins (IκBs). A variety of signaling pathways activate IκB kinases to phosphorylate IκBs, leading to ubiquitination of phospho-IκBs and their degradation by the proteasome (3), which allows translocation of NF-κB to the nucleus, activation of NF-κB-regulated genes, and establishment of an inflammatory response.
Ubiquitination, resulting in the covalent attachment of the 76-residue ubiquitin to target proteins, involves three sequential steps performed by one ubiquitin-activating enzyme (E1), a limited number of ubiquitin-conjugating enzymes (E2s; also known as Ubc in enzyme designations), and a large number of ubiquitin-ligating enzymes (E3s), respectively (4). Each E3 recognizes a set of substrates and cooperates with one or a few E2s. The E3 complex SCFβ-TrCP, which promotes ubiquitination of phospho-IκBα, consists of five proteins: the scaffold protein Cullin1, the adaptor protein Skp1, the RING domain protein Roc1, the E2 UbcH5b, and the F box protein β-TrCP, which interacts with phospho-IκBα (5).
Bacteria of Shigella spp. are the agent of shigellosis in humans, a disease characterized by the destruction of the colonic epithelium that is responsible for 1 million deaths per year (6). These bacteria use a type III secretion (TTS) system to enter epithelial cells and trigger apoptosis in macrophages (7). TTS systems comprise (i) a secretion apparatus that spans the bacterial envelope; (ii) translocators that transit through the TTS apparatus and insert into the membrane of the host cell to form a pore; (iii) effectors that transit through the TTS apparatus and the translocator pore to be injected into the cell cytoplasm, where they interfere with a variety of cellular functions; (iv) molecular chaperones; and (v) specific transcription regulators (8). The S. flexneri TTS system is encoded by a 213-kb virulence plasmid (9). The TTS apparatus is activated upon contact of bacteria with epithelial cells (10). Transcription of a set of genes encoding effectors is regulated by the TTS apparatus activity (11) and controlled by MxiE, a transcription activator of the AraC family (12, 13).
The repertoire of S. flexneri effectors includes ≈20 proteins identified as substrates of the TTS apparatus (9). We present the functional analysis of the effector OspG, a 196-residue protein whose production is regulated by secretion activity (9, 14). A two-hybrid screen in yeast and in vitro studies indicated that OspG binds ubiquitinylated E2s, including UbcH5. Transfection experiments were used to investigate the potential role of OspG in interfering with activation of the NF-κB pathway that involves UbcH5. Characterization of the phenotype of an ospG mutant by using in vitro and in vivo models of infection indicated that OspG is involved in the down-regulation of the host innate response induced by invasive bacteria.
Methods
Bacterial Strains. The invasive S. flexneri strain M90T-Sm and the virulence plasmid-cured strain BS176 are described in ref. 15. To construct the ospG mutant DWS14, a PCR-amplified DNA fragment encompassing nucleotides 61-360 of ospG was cloned between the XbaI and EcoRI sites of the suicide plasmid pSW23T, giving raise to pSWOspGTr. This plasmid was transferred by conjugation to the wild-type strain M90T-Sm, and integration of the suicide plasmid into the ospG gene carried by the virulence plasmid was verified by PCR and restriction analysis of the virulence plasmid. A PCR fragment encompassing ospG was cloned between the EcoRI and HindIII sites of pUC18 to construct pUC18-OspG, which was used to complement the ospG mutant.
Materials. Horseradish peroxidase-coupled avidin and anti-UbcH5 and anti-UbcH7 antibodies were from Boston Biochem (Cambridge, MA); MG132, ubiquitin, biotinylated ubiquitin, and ubiquitin-activating enzyme were from Affiniti Research (Mamhead, U.K.); anti-c-myc antibody was from Sigma; anti-IκBα antibody was from Santa Cruz Biotechnology; anti-phospho-IκBα antibody was from Cell Signaling Technology (Beverly, MA); and recombinant human TNF-α wasfromR&D Systems.
Plasmid Constructions. PCR-amplified fragments carrying the ospG coding sequence were cloned between the NcoI and BglII sites of pKJ1 to construct pKJ-OspG (OspG-His), between the BamHI and EcoRI sites of pRK5myc to construct pRK5myc-OspG (myc-OspG), and between the BamHI and EcoRI sites of pGEX4T2 to construct pGEX4T2-OspG (GST-OspG). Site-directed mutagenesis of pGEX4T2-OspG and pRK5myc-OspG was performed to construct pGEX4T2-OspG-K53A and pRK5myc-OspG-K53A. pUbcH7-GFP, pUbcH5a-GFP, pcDNA3-GFP, and pET15-UbcH5b are described in refs. 16 and 17. A PCR fragment encoding UbcH5b was inserted into pcDNA3-GFP to construct pUbcH5b-GFP (UbcH5b-GFP), and PCR fragments encoding UbcH7 and UbcH5 were cloned between the NcoI and BamHI sites and NcoI and BglII sites of pKJ1 to construct pKJUbcH7 (UbcH7-His) and pKJUbcH5b (UbcH5b-His).
Yeast Two-Hybrid Screening. The ospG coding sequence was amplified by PCR and cloned into plasmid pB27 to screen the library constructed in plasmid pP6 by using random-primed cDNA made from human placenta poly(A) RNA, as described in ref. 18. The insert carried by prey plasmids in positive clones was amplified by PCR and sequenced to identify the corresponding gene in the GenBank database by using a fully automated procedure.
In Vitro Assays. His- and GST-tagged proteins were purified by affinity chromatography and stored in 50 mM Tris·HCl, pH 7.6/50 mM NaCl/20% glycerol. HEK-293T cells transfected with pUbcH7-GFP, pUbcH5a-GFP, pUbcH5b-GFP, or pRK5myc-OspG were lysed in radioimmunoprecipitation assay (RIPA) buffer [20 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM MgCl2/10% (vol/vol) glycerol/1% Nonidet P-40] containing a protease inhibitor mixture. Extracts containing UbcH5a-GFP, UbcH5b-GFP, or UbcH7-GFP were mixed with extracts containing myc-OspG and supplemented with anti-myc antibodies and protein G-Sepharose beads. Immunoprecipitated proteins were analyzed by SDS/PAGE and immunoblotting with anti-GFP antibodies. To detect the interaction between OspG and endogenous E2s, 500 μl (≈1 mg of proteins) of HEK-293T cell extract in RIPA buffer was mixed with GST or GST-OspG (100 μg) and glutathione-Sepharose beads for 1 h at 4°C. Beads were washed three times with RIPA buffer, and bound proteins were analyzed by SDS/PAGE and immunoblotting with anti-UbcH5 and anti-UbcH7 antibodies. Ubiquitinylation of UbcH5b-His and UbcH7-His by E1 was performed in a 20-μl reaction mixture containing 50 mM Tris·HCl (pH 7.5), 10 mM ATP, 5 mM MgCl2, 0.2 mM DTT, 2 μg of biotinylated ubiquitin, 0.5 μg of E1, and 2 μg of E2 at 37°C for 1 h in the presence or absence of 5 μM GST or GST-OspG. After ubiquitinylation, 100 μg of purified GST or GST-OspG and glutathione-Sepharose beads was added to the reaction mixture. Phosphorylation assays were performed in a 20-μl reaction mixture containing 50 mM Tris·HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 5 μM GST-OspG, 5 μCi (1 Ci = 37 GBq) [γ-32P]ATP and, when indicated, 5 μg of ubiquitin at 37°C for 1 h.
Cell Transfections. HEK-293T cells were plated at a density of 105 cells per well in 6-well plates and, 24 h later, transfected with 1 μg of pRK5myc-OspG and 0.5 μg of plasmid pRcCMV-mIκB. TNF-α was added to media 40 h after transfection. At the indicated times after TNF-α stimulation (see Fig. 3), cells were harvested and suspended in 2× SDS sample buffer. When indicated, the proteasome inhibitor MG132 was added at 50 μM 4 h before TNF-α stimulus. IκBα and phospho-IκBα were detected by SDS/PAGE analysis and immunoblotting. Activation of NF-κB-dependent promoter was assayed as described in ref. 19. HEK cells were plated as 5 × 104 cells per well in 24-well plates. The following day, 0.1 μg of luciferase reporter plasmid pIgγ-luc (20), 0.1 μg of β-galactosidase reporter plasmid placZ (19), and 0.1, 0.2, or 0.5 μg of pRK5myc-OspG or pRK5myc-OspG-K53A were used to transfect these cells with FuGENE 6. The total DNA amount was adjusted to 0.7 μg with pRK5myc. TNF-α (10 ng/ml) was added to media 40 h after transfection, and cells were lysed 6 h later in luciferase assay buffer [25 mM Tris·HCl, pH 7.9/8 mM MgCl2/1mMDTT/1% (vol/vol) Triton X-100/15% (vol/vol) glycerol]. Luciferase activity was determined and normalized to β-galactosidase activity as described in ref. 19. Each assay was performed in triplicate and repeated three times.
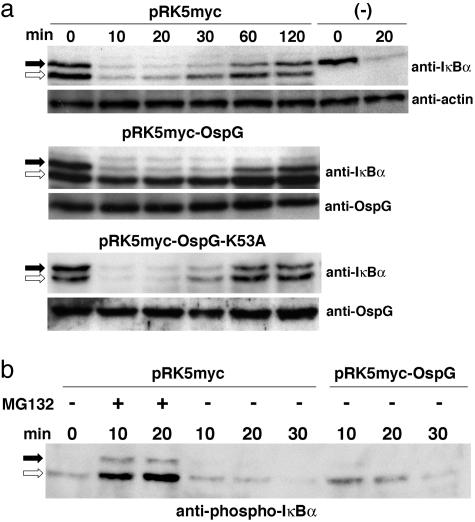
Fig. 3.
OspG blocks degradation but not phosphorylation of IκBα induced by TNF-α.(a) Extracts of HEK-293T cells transfected by pRcCMV-mIκBα encoding mIκBα and either pRK5myc, pRK5myc-OspG, or pKR5myc-OspG-K53A or non-transfected (-) were prepared at the indicated times after TNF-α stimulation and analyzed by SDS/PAGE and immunoblotting with anti-IκBα and anti-actin antibodies. (b) Extracts of cells transfected by pRcCMV-mIκBα and either pRK5myc or pRK5myc-OspG and treated (+) or not (-) with the proteasome inhibitor MG132 were prepared at the indicated times after TNF-α stimulation and analyzed by SDS/PAGE and immunoblotting with anti-phospho-IκBα antibodies. Filled and open arrows indicate the positions of endogenous hIκBα and recombinant mIκBα, respectively
In Vitro and in Vivo Infections. Infection of semiconfluent monolayers of HeLa cells by derivatives of the wild-type, ospG, and ospG/pUC18-OspG strains expressing the AfaE adhesin were performed as described in ref. 21. Bacteria were incubated with epithelial cells for 10 min at 25°C to promote adhesion, and nonadhesive bacteria were removed by aspiration, and cells were transferred at 37°C. At the indicated times after temperature shift (see Fig. 5), cells were lysed and lysates were analyzed by SDS/PAGE and immunoblotting. Rabbit intestinal loop infections were performed as described in ref. 22. Suspensions of 5 × 109 bacteria (0.5 ml) were injected into 5-cm rabbit small intestine loops. Loops were returned to the abdominal cavity, and rabbits were killed after 8 h. Each bacterial strain was tested in three rabbits. Tissue sampling, staining with haematoxylin and eosin or Giemsa, and histopathological analysis were performed as described in ref. 22.
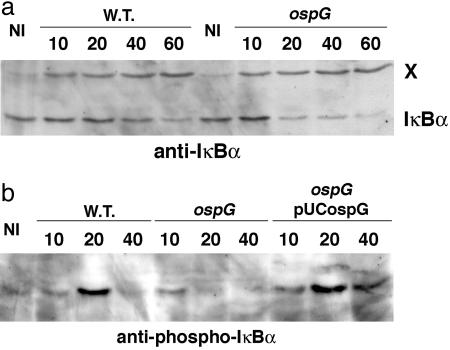
Fig. 5.
Inactivation of ospG induces a more rapid degradation of IκBα in infected epithelial cells. HeLa cells infected by derivatives of wild-type or ospG strains producing the AfaE adhesin were incubated at 25°C for 10 min to allow adhesion of bacteria to cells and then shifted to 37°C. At the indicated time points after temperature shift, cells were lysed and extracts were analyzed by SDS/PAGE and immunoblotting with anti-IκBα (a) and anti-phospho-IκBα (b) antibodies. A protein (indicated by X) recognized by the anti-IκBα serum was used as an internal loading control. NI, noninfected
Results
OspG Binds Ubiquitinylated E2s. To identify partners of interaction of OspG within the eukaryotic cell, an OspG bait was used in a two-hybrid system in yeast to screen a library of preys constructed from human cDNA. The insert carried by 198 prey plasmids selected among ≈5 × 107 clones was identified by sequencing. Among these plasmids, 95 encoded almost the entire sequence (≈150 residues) of proteins of the E2 (Ubc) family, including E2D2 (UbcH5b), E2D3 (UbcH5c), E2E2 (UbcH8), E2E3 (UbcH9), E2L3 (UbcH7), and RIG-B. Two approaches were used to validate the interaction between OspG and E2s.
OspG carrying an N-terminal myc tag and GFP, UbcH5a-GFP, UbcH5b-GFP, and UbcH7-GFP were expressed independently in HEK-293T cells. Mixtures of cell extracts containing myc-OspG and each GFP-tagged E2 were used for immunoprecipitations with anti-myc antibodies, and precipitates were analyzed by immunoblotting with anti-GFP antibodies (Fig. 1a). GFP-tagged E2s, but not GFP, were coimmunoprecipitated with myc-OspG.
GST-OspG purified from Escherichia coli was mixed with an extract of HEK-293T cells and glutathione-Sepharose beads, and proteins bound to beads were analyzed by immunoblotting with anti-UbcH5 (Fig. 1b) and anti-UbcH7 antibodies (data not shown). Approximately 50% of UbcH5 and UbcH7 molecules present in the cell extract were copurified with GST-OspG.
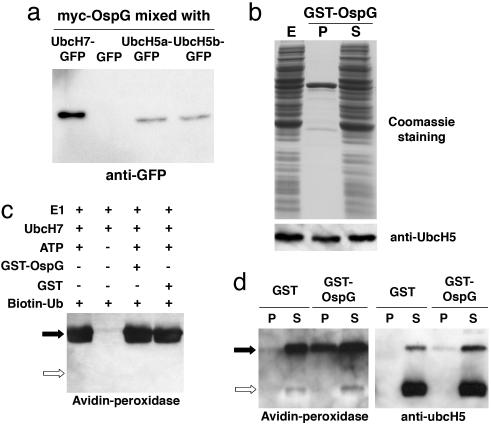
Fig. 1.
OspG binds Ub-E2s. (a) Mixtures of HEK-293T cell extracts containing myc-OspG and GFP, UbcH7-GFP, UbcH5a-GFP, or UbcH5b-GFP were incubated with anti-myc antibodies, and immunoprecipitates were analyzed by SDS/PAGE and immunoblotting with anti-GFP antibodies. (b) An extract of HEK-293T cells (E) was incubated with GST-OspG and glutathione-Sepharose beads. After centrifugation, proteins present in pellets (P) and supernatants (S) were analyzed by SDS/PAGE and Coomassie blue staining or immunoblotting with anti-UbcH5 antibodies. (c) Products of ubiquitinylation reactions carried out in the presence (+) or absence (-) of ubiquitin-activating enzyme E1, UbcH7-His, ATP, GST-OspG, GST, and biotinylated ubiquitin (Biotin-Ub) were analyzed by SDS/PAGE and probed with peroxidase-labeled streptavidin (Avidin-peroxidase) to detect ubiquitinylated UbcH7-His. Filled and open arrows indicate the position of Ub-UbcH7 and UbcH7, respectively. (d) Product of ubiquitinylation reactions containing E1, ATP, UbcH5b-His, and biotinylated ubiquitin were mixed with GST-OspG or GST and glutathione-Sepharose beads. After centrifugation, proteins present in pellets and supernatants were analyzed by SDS/PAGE and detected by blotting with anti-UbcH5 antibodies or peroxidase-labeled streptavidin. Filled and open arrows indicate the position of Ub-UbcH5 and UbcH5, respectively. In a and b, proteins were heated to 100°C in a sample buffer containing DTT, which cleaves the phosphodiester bond between ubiquitin and E2s, whereas, in c and d, DTT was omitted
To test the effect of OspG on ubiquitinylation of E2s by E1, the ubiquitinylation reaction was performed by using purified proteins. The presence of GST-OspG was without effect on ubiquitinylation of UbcH7-His (Fig. 1c) or UbcH5b-His (data not shown) by E1 in vitro. To test whether OspG binds ubiquitinylated E2s (Ub-E2s), ubiquitinylation of UbcH5b-His was performed in the presence of biotinylated ubiquitin. The reaction mixture was then supplemented with purified GST or GST-OspG, and proteins bound to GST or GST-OspG were analyzed by SDS/PAGE and immunoblotting with anti-UbcH5 antibodies or peroxidase-labeled streptavidin (Fig. 1d). Ub-UbcH5b, but not UbcH5b or ubiquitin, was purified with GST-OspG, indicating that OspG binds Ub-UbcH5b. Exclusive binding of OspG to Ub-E2s in vitro and selection of E2-containing preys using an OspG bait in yeast suggest that E2 preys were ubiquitinylated in yeast. Thus, the two-hybrid system in yeast permitted detection of an interaction requiring a posttranslational modification of the prey. The ubiquitin moiety is probably part of the OspG binding site on different Ub-E2s, whose sequences, in the case of UbcH5b and UbcH7, share only 35% identity.
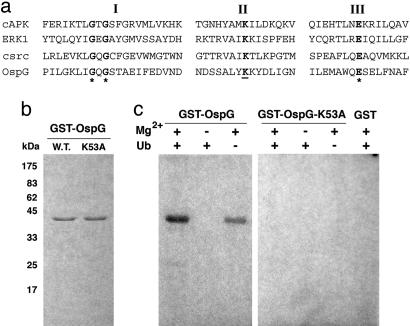
OspG Is Endowed with Autophosphorylation Activity. The OspG sequence exhibits similarities to the catalytic domain of eukaryotic protein kinases (Fig. 2a) and lacks subdomains VIII-XI (23). OspG residue Lys-53 corresponds to PKA Ca residue Lys-72, whose replacement abolishes kinase activity (24, 25). To determine whether OspG is endowed with autophosphorylation activity, as are several other eukaryotic protein kinases, GST, GST-OspG, and GST-OspG-K53A, in which Lys-53 was replaced by Ala, were incubated in the presence of [γ-32P]ATP and Mg2+, and the reaction products were analyzed by SDS/PAGE and autoradiography (Fig. 2b). GST-OspG, but not GST-OspG-K53A, exhibited a Mg2+-dependent autophosphorylation activity. This activity and binding of OspG to Ub-E2s led us to investigate whether OspG phosphorylates Ub-E2s. Addition of ubiquitin, E1, and either UbcH5-His or UbcH7-His to the phosphorylation assay did not reveal any labeling of E2s, Ub-E2s, or ubiquitin (data not shown), suggesting that these proteins are not substrates for phosphorylation by OspG.
Fig. 2.
Autophosphorylation activity of OspG. (a) Amino acid sequence alignment of the first three catalytic motifs (indicated by roman numerals) of human cAMP-dependent protein kinase α subunit (cAPK), rat extracellular signal-regulated kinase 1 (ERK1), human protooncogene kinase src (csrc), and S. flexneri OspG (OspG). Residue K53 of OspG replaced with Ala in OspG-K53A is underlined. (b) Purified GST-OspG (W.T.) and GST-OspG-K53A (K53A) were analyzed by SDS/PAGE and Coomassie blue staining. (c) Products of a phosphorylation assay performed in the presence or absence of GST-OspG (OspG), GST-OspG-K53A (OspG-K53A), GST, Mg2+, ubiquitin (Ub), and [γ-32P]ATP were analyzed by SDS/PAGE and autoradiography
OspG Prevents Degradation of Phospho-IκBα. UbcH5b is a component of the SCFβ-TrCP complex, which promotes phospho-IκBα ubiquitination and its subsequent degradation by the proteasome (3). To investigate whether OspG can interfere with IκBα degradation induced upon stimulation of eukaryotic cells with TNF-α, we performed transfection experiments with pRK5myc-OspG. Because only a fraction (≈20%) of the cell population was effectively transfected, the potential effect of OspG in preventing IκBα degradation could not be analyzed on the whole population. Taking advantage of the difference in migration of human IκBα (hIκBα) and mouse IκBα (mIκBα) in SDS/PAGE analysis (26), we performed cotransfection experiments with plasmids encoding OspG and mIκBα to analyze the fate of IκBα molecules in transfected cells expressing both OspG and mIκBα. Cells cotransfected with the plasmid encoding mIκBα and either the vector pRK5-myc or its derivatives encoding myc-OspG or myc-OspG-K35A were stimulated by TNF-α to induce IκBα phosphorylation, and cell extracts were analyzed by SDS/PAGE and immunoblotting (Fig. 3a). After TNF-α stimulation, endogenous hIκBα and recombinant mIκBα were degraded in cells cotransfected with the vector control, indicating that mIκBα was subjected to the same regulatory pathway as hIκBα in this experimental setup. Degradation of mIκBα was blocked in cells cotransfected with the plasmid encoding myc-OspG but not in cells cotransfected with the plasmid encoding myc-OspG-K35A, indicating that wild-type OspG can block TNF-α-induced IκBα degradation. Because of the low transfection efficiency, most hIκBα molecules were not protected from degradation in the sample corresponding to cells transfected with pRK5myc-OspG. Cell extracts were also analyzed by using anti-phospho-IκBα antibodies (Fig. 3b). In cells transfected by the control plasmid, accumulation of phospho-hIκBα and phospho-mIκBα was observed in the presence of the proteasome inhibitor MG132, consistent with a TNF-α induced phosphorylation of hIκBα and mIκBα and their degradation by the proteasome. In the absence of MG132, increased amounts of phospho-mIκBα were detected by following TNF-α stimulation of cells transfected with the plasmid encoding myc-OspG compared with cells transfected with the control plasmid. This result suggests that OspG does not affect the TNF-α-induced pathway leading to IκBα phosphorylation and that blockage of IκBα degradation occurs at the step of phospho-IκBα ubiquitination.
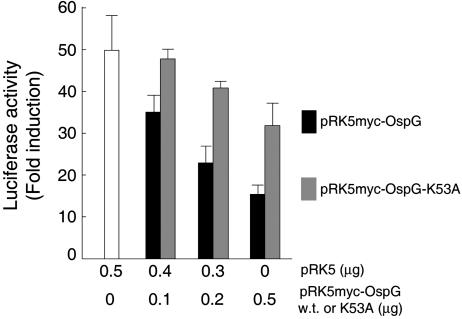
OspG Prevents Activation of an NF-κB Responding Promoter. Accumulation of phospho-mIκBα in cells expressing myc-OspG suggested that translocation of NF-κB to the nucleus and activation of NF-κB-regulated promoters might be impaired in these cells. To test this hypothesis, cells were transfected with a reporter plasmid encoding luciferase under the control of the NF-κB responding promoter pIgγ and a plasmid encoding either myc-OspG or myc-OspG-K35A, and luciferase activity was assayed 6 h after TNF-α stimulation (Fig. 4). Luciferase activity was induced 50-60 times in control cells; this induction was reduced by 70% in cells producing myc-OspG. In contrast, only a minor decrease in luciferase induction was observed in cells producing myc-OspG-K35A, which suggests that OspG is blocking the pathway leading to activation of NF-κB by preventing degradation of endogenous phospho-hIκBα and that the catalytic activity of OspG is involved in this process. The slight decrease in luciferase activity observed in cells producing myc-OspG-K53A suggests that binding of OspG-K53A to UbcH5 might also interfere with the ubiquitin ligase activity of SCFβ-TrCP.
Fig. 4.
OspG inhibits activation of a NF-κB-regulated promoter. Luciferase and β-galactosidase activities were assayed after TNF-α stimulation of cells cotransfected with pIgγ-luc (encoding luciferase under the control of a NF-κB-regulated promoter), placZ (encoding β-galactosidase), pRK5myc, and either pRK5myc-OspG (w.t.) or pRK5myc-OspG-K53A (K53A) at the indicated amounts. Bars indicate fold inductions in luciferase activities normalized to β-galactosidase activities
Inactivation of ospG Increases IκBα Degradation in Infected Epithelial Cells. To determine the contribution of OspG in pathogenesis, we constructed an ospG mutant. This mutant did not exhibit any difference compared with the wild-type strain with respect to entry into epithelial cells and dissemination of intracellular bacteria from cell to cell (data not shown). To compare the kinetics of hIκBα degradation after infection of HeLa cells, we used derivatives of wild-type and ospG strains expressing the AfaE adhesin to synchronize entry. Lysates of HeLa cells infected for 10, 20, 40, and 60 min were analyzed by SDS/PAGE and immunoblotting with anti-IκBα and anti-phospho-IκBα antibodies (Fig. 5). hIκBα degradation occurred 60 min after infection by the wild-type strain but was detectable as soon as 20 min after infection by the ospG mutant. Shortly after infection, accumulation of phospho-hIκBα was detected in cells infected by the wild-type strain and the ospG mutant harboring pUC18-OspG (encoding wild-type OspG) but not by the ospG mutant, indicating that OspG produced by bacteria infecting epithelial cells prevents or at least delays phospho-hIκBα degradation.
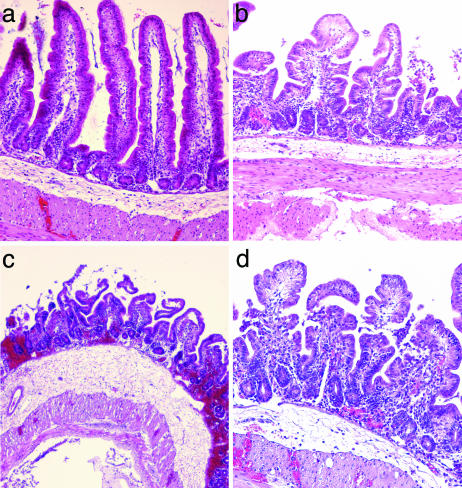
Inactivation of ospG Increases the Inflammatory Response in Vivo. The phenotype of the ospG mutant was characterized in vivo by using the ligated ileal loop model of infection in rabbits. Histological samples of loops infected for 8 h by the wild-type strain, the virulence plasmid-cured noninvasive strain, the ospG mutant, or the ospG mutant harboring pUC18-OspG were stained with hematoxylin and eosin (Fig. 6). Infection by the wild-type strain was characterized by a shortening of the villi, an erosion of the epithelium, and an increase in the number of polymorphonuclear leukocytes (PMN) present in the lamina propria (22). The ospG mutant induced a stronger destruction of the mucosa than the wild-type strain: large areas of the epithelium appeared necrotic, villi were almost completely eliminated, and the lamina propria exhibited extensive hemorrhages and a massive infiltrate of PMN. In addition, the subepithelial region of loops infected by the ospG mutant exhibited a more important edema and an increased inflammatory infiltrate compared with loops infected by the wild-type strain. Complementation of the ospG mutant by pUC18-OspG restored a phenotype similar to that of the wild-type strain. These results indicate that inactivation of ospG leads to the induction of a stronger inflammatory response upon infection in vivo.
Fig. 6.
The ospG mutant induces a stronger inflammatory reaction than the wild-type strain upon infection of ileal loops in rabbits. Ileal loops infected for 8 h by the virulence plasmid-cured strain (a), the wild-type strain (b), the ospG mutant (c), and the ospG mutant harboring plasmid pUC18-OspG (d) were stained with hematoxylin and eosin
Discussion
NF-κB is at the heart of signaling cascades leading to mucosal immune responses (2) and is a target for bacterial strategies aimed at dampening innate immunity (1). Bacteroides tethaiotaomicron enhances binding of the nuclear hormone receptor peroxisome proliferator-activated receptor subtype γ to the NF-κB subunit RelA and promotes export of RelA from the nucleus (27). Upon infection of intestinal epithelial cells in vitro, an avirulent strain of Salmonella has been shown to block SCFβ-TrCP function and prevent ubiquitination of IκBα normally induced by virulent Salmonella or TNF (28). There is also evidence that a number of TTS effectors target innate immunity responses in both animals and plants (29). The cysteine protease YopJ from Yersinia, which cleaves the SUMO group from sumoylated proteins, and the phospho-tyrosine phosphatases YopH from Yersinia and StpP from Salmonella enterica all interfere with several transduction pathways, including those leading to NF-κB activation. Although detailed mechanisms of inhibition have not been elucidated, these effectors may point to potential targets to develop innovative antiinflammatory drugs.
Recruitment of PMN to the epithelium facilitates access of S. flexneri to the basolateral pole of epithelial cells and increases invasion in vitro and in vivo (22, 30). Mechanisms of the mucosal innate immune response causing the inflammatory infiltration of PMN are being characterized. These mechanisms respond to intracellular sensing of muropeptides by the Nod1 pathway (31) and extracellular sensing of endotoxin by the Toll-like receptor 4 pathway (32). A major feature of this response is the induction of proinflammatory genes, including those encoding chemokines, such as IL-8, a potent chemoattractant for PMN (33). Infection by Shigella is likely to promote an innate response that is qualitatively and quantitatively specific to this pathogen because of its repertoire of virulence factors. Indeed, the functional analysis of the TTS effector OspG provides evidence that this protein interferes with activation of the NF-κB pathway. When produced in HEK-293T cells, OspG prevented phospho-IκBα degradation induced by TNF-α stimulation and, when produced by invasive bacteria, OspG delayed phospho-IκBα degradation induced upon entry of bacteria into HeLa cells. The observation that OspG binds to but does not phosphorylate Ub-UbcH5b suggests that OspG acts at the step of phospho-IκBα ubiquitination. Furthermore, in transfection experiments, blockage of degradation of phospho-IκBα and of activation of an NF-κB responding promoter were lost when residue Lys-53 of OspG was replaced by Ala, which abolished OspG autophosphorylation activity. This result suggests that the mechanism of SCFβ-TrCP inhibition by OspG involves phosphorylation of a component of SCFβ-TrCP after association of OspG to Ub-UbcH5b. IκBα degradation occurred after 1 h of infection of HeLa cells by the wild-type strain, probably because the increase in the number of intracellular bacteria overwhelmed the OspG protective effect, because there is no sustained production and delivery of OspG when bacteria are intracellular. Indeed, OspG production is tightly regulated by the TTS apparatus activity and controlled by MxiE (14), and transcription of MxiE-regulated genes is induced upon entry of bacteria into cells but repressed during multiplication of bacteria within epithelial cells (11). A role of OspG in interfering with activation of NF-κB in vivo is supported by the observation that the inflammatory response induced upon infection of ileal loops by the ospG mutant was more severe than with the wild-type strain. A function of OspG during infection of humans might be to secure early stages of Shigella interaction with the intestinal epithelium to facilitate colonization and invasion by an initially low number of luminal bacteria. In addition to its ability to inhibit IκBα degradation, OspG might interfere with ubiquitination of other SCFβ-TrCP substrates, such as the transcription cofactor β-catenin, which acts together with TCF/LEF-1 (T cell factor/lymphocyte enhancer-binding factor 1) to regulate cell proliferation and differentiation (34). Moreover, the ability of OspG to bind to different Ub-E2s suggests that OspG might target and interfere with the function of a large number of E3s.
Colonization and invasion of mucosal surfaces by pathogenic bacteria require down-regulation of host innate responses. Beyond its role in induction of acute-phase antimicrobial defense genes, NF-κB is also a major regulator of the adaptive immune response. Mice lacking individual NF-κB proteins show defects in B- and T-cell proliferation, activation, cytokine production, and isotype switching (35). NF-κB also controls production of factors, including IL-18, IFN-γ, IL-12, and costimulatory molecules, such as CD80/CD86, which are required for the development of an efficient T helper-1 response against invasive pathogens (36, 37). By interfering with NF-κB activation, OspG might also play a role in preventing this switch, leading to a less efficient and short-lasting anti-Shigella T helper-2-type response.
Acknowledgments
We thank A. M. Weissman (National Institutes of Health, Frederick, MD), H. C. Ardley (University of Leeds, Leeds, U.K.), and S. Memet, D. Philpott, and R. Weil (Institut Pasteur) for kindly providing some plasmids and L. Selig, L. Arbibe, and J. W. Rohde for helpful discussions. This study was supported by the Institut Pasteur and the International Vaccine Institute. P.J.S. is a scholar of the Howard Hughes Medical Institute.
Author contributions: D.W.K., P.L., and C.P. designed research; D.W.K., G.L., A.-L.P., and P.J.S. performed research; G.L. contributed new reagents/analytic tools; D.W.K., G.L., A.-L.P., P.J.S., and C.P. analyzed data; and D.W.K., P.J.S., and C.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-ligating enzyme; TTS, type III secretion; IκBα, inhibitor of NF-κB type α; hIκBα, human IκBα; mIκBα, mouse IκBα; PMN, polymorphonuclear leukocyte(s).
References
- 1.Sansonetti, P. J. (2004) Nat. Rev. Immunol. 4, 953-964. [DOI] [PubMed] [Google Scholar]
- 2.Li, Q. T. & Verma, I. M. (2002) Nat. Rev. Immunol. 2, 725-734. [DOI] [PubMed] [Google Scholar]
- 3.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- 4.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- 5.Zheng, N., Schulman, B. A., Song, L. Z., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703-709. [DOI] [PubMed] [Google Scholar]
- 6.Kotloff, K. L., Winickoff, J. P., Ivanoff, B., Clemens, J. D., Swerdlow, D. L., Sansonetti, P. J., Adak, G. K. & Levine, M. M. (1999) Bull. W. H. O. 77, 651-666. [PMC free article] [PubMed] [Google Scholar]
- 7.Cossart, P. & Sansonetti, P. J. (2004) Science 304, 242-248. [DOI] [PubMed] [Google Scholar]
- 8.Hueck, C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., d'Hauteville, H., Kunst, F., Sansonetti, P. & Parsot, C. (2000) Mol. Microbiol. 38, 760-771. [DOI] [PubMed] [Google Scholar]
- 10.Menard, R., Sansonetti, P. & Parsot, C. (1994) EMBO J. 13, 5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demers, B., Sansonetti, P. J. & Parsot, C. (1998) EMBO J. 17, 2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mavris, M., Page, A. L., Tournebize, R., Demers, B., Sansonetti, P. & Parsot, C. (2002) Mol. Microbiol. 43, 1543-1553. [DOI] [PubMed] [Google Scholar]
- 13.Parsot, C., Ageron, E., Penno, C., Mavris, M., Jamoussi, K., d'Hauteville, H., Sansonetti, P. & Demers, B. (2005) Mol. Microbiol. 56, 1627-1635. [DOI] [PubMed] [Google Scholar]
- 14.Le Gall, T., Mavris, M., Martino, M. C., Bernardini, M. L., Denamur, E. & Parsot, C. (2005) Microbiology 151, 951-962. [DOI] [PubMed] [Google Scholar]
- 15.Allaoui, A., Mounier, J., Prevost, M. C., Sansonetti, P. J. & Parsot, C. (1992) Mol. Microbiol. 6, 1605-1616. [DOI] [PubMed] [Google Scholar]
- 16.Ardley, H. C., Tan, N. G. S., Rose, S. A., Markham, A. F. & Robinson, P. A. (2001) J. Biol. Chem. 276, 19640-19647. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, J. P., Bates, P. W., Yang, M., Vierstra, R. D. & Weissman, A. M. (1995) J. Biol. Chem. 270, 30408-30414. [DOI] [PubMed] [Google Scholar]
- 18.Colland, F., Jacq, X., Trouplin, V., Mougin, C., Groizeleau, C., Hamburger, A., Meil, A., Wojcik, J., Legrain, P. & Gauthier, J. M. (2004) Genome Res. 14, 1324-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philpott, D. J., Yamaoka, S., Israel, A. & Sansonetti, P. J. (2000) J. Immunol. 165, 903-914. [DOI] [PubMed] [Google Scholar]
- 20.Munoz, E., Courtois, G., Veschambre, P., Jalinot, P. & Israel, A. (1994) J. Virol. 68, 8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Nhieu, G. T., BenZeev, A. & Sansonetti, P. J. (1997) EMBO J. 16, 2717-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perdomo, O. J. J., Cavaillon, J. M., Huerre, M., Ohayon, H., Gounon, P. & Sansonetti, P. J. (1994) J. Exp. Med. 180, 1307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanks, S. K. & Hunter, T. (1995) FASEB J. 9, 576-596. [PubMed] [Google Scholar]
- 24.Hanks, S. K. & Quinn, A. M. (1991) Methods Enzymol. 200, 38-62. [DOI] [PubMed] [Google Scholar]
- 25.Tyler, J. S. & Friedman, D. I. (2004) J. Bacteriol. 186, 3472-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weil, R., Whiteside, S. T. & Israel, A. (1997) Immunobiology 198, 14-23. [DOI] [PubMed] [Google Scholar]
- 27.Kelly, D., Campbell, J. I., King, T. P., Grant, G., Jansson, E. A., Coutts, A. G. P., Pettersson, S. & Conway, S. (2004) Nat. Immunol. 5, 104-112. [DOI] [PubMed] [Google Scholar]
- 28.Neish, A. S., Gewirtz, A. T., Zeng, H., Young, A. N., Hobert, M. E., Karmali, V., Rao, A. S. & Madara, J. L. (2000) Science 289, 1560-1563. [DOI] [PubMed] [Google Scholar]
- 29.Espinosa, A. & Alfano, J. R. (2004) Cell. Microbiol. 6, 1027-1040. [DOI] [PubMed] [Google Scholar]
- 30.Perdomo, J. J., Gounon, P. & Sansonetti, P. J. (1994) J. Clin. Invest. 93, 633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girardin, S. E., Boneca, I. G., Carneiro, L. A. M., Antignac, A., Jehanno, M., Viala, J., Tedin, K., Taha, M. K., Labigne, A., Zahringer, U., et al. (2003) Science 300, 1584-1587. [DOI] [PubMed] [Google Scholar]
- 32.d'Hauteville, H., Khan, S., Maskell, D. J., Kussak, A., Weintraub, A., Mathison, J., Ulevitch, R. J., Wuscher, N., Parsot, C. & Sansonetti, P. J. (2002) J. Immunol. 168, 5240-5251. [DOI] [PubMed] [Google Scholar]
- 33.Pedron, T., Thibault, C. & Sansonetti, P. J. (2003) J. Biol. Chem. 278, 33878-33886. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T. (1999) Genes Dev. 13, 505-510. [DOI] [PubMed] [Google Scholar]
- 35.Gerondakis, S., Grumont, R., Rourke, I. & Grossmann, M. (1998) Curr. Opin. Immunol. 10, 353-359. [DOI] [PubMed] [Google Scholar]
- 36.Kojima, H., Aizawa, Y., Yanai, Y., Nagaoka, K., Takeuchi, M., Ohta, T., Ikegami, H., Ikeda, M. & Kurimoto, M. (1999) J. Immunol. 162, 5063-5069. [PubMed] [Google Scholar]
- 37.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]