Abstract
Somatic hypermutation of Ig variable region genes is initiated by activation-induced cytidine deaminase; however, the activity of multiple DNA polymerases is required to ultimately introduce mutations. DNA polymerase η (Polη) has been implicated in mutations at A/T, but polymerases involved in C/G mutations have not been identified. We have generated mutant mice expressing DNA polymerase (Polθ) specifically devoid of polymerase activity. Compared with WT mice, Polq-inactive (Polq, the gene encoding Polθ) mice exhibited a reduced level of serum IgM and IgG1. The mutant mice mounted relatively normal primary and secondary immune responses to a T-dependent antigen, but the production of high-affinity specific antibodies was partially impaired. Analysis of the JH4 intronic sequences revealed a slight reduction in the overall mutation frequency in Polq-inactive mice. Remarkably, although mutations at A/T were unaffected, mutations at C/G were significantly decreased, indicating an important, albeit not exclusive, role for Polθ activity. The reduction of C/G mutations was particularly focused on the intrinsic somatic hypermutation hotspots and both transitions and transversions were similarly reduced. These findings, together with the recent observation that Polθ efficiently catalyzes the bypass of abasic sites, lead us to propose that Polθ introduces mutations at C/G by replicating over abasic sites generated via uracil-DNA glycosylase.
Keywords: abasic site, low-fidelity DNA polymerase, activation-induced cytidine deaminase, uracil-DNA glycosylase
Functional Ig genes are assembled in developing B cells by recombination-activating gene-mediated rearrangement of the germline V, D, and J gene segments (1–3). This process generates a primary repertoire of B cells expressing diversified surface immunoglobulins. Upon antigen stimulation and in the presence of T cell help, B cells undergo further diversification of their Ig genes, namely somatic hypermutation (SHM) and class switch recombination (CSR), in the germinal centers (GCs) of secondary lymphoid organs such as spleen, lymph node, and Peyer's patches (4). Both SHM and CSR are initiated by a single enzyme, activation-induced cytidine deaminase, which catalyzes the deamination of C to U on DNA and/or possibly on an as-yet-hypothetical endonuclease mRNA (5–7). Although the mechanism of SHM is still not fully understood, it is thought that mutations are ultimately introduced by error-prone DNA polymerases during the DNA repair process (7, 8).
Approximately 10 new low-fidelity DNA polymerases have been identified in the past several years (9, 10). DNA polymerase θ (Polθ) is a ≈300-kDa family A polymerase with a unique structure, having a helicase domain in its N-terminal portion and a polymerase domain in its C terminus (11–13). Polq, the gene encoding Polθ, has a homolog in Drosophila, the mus308 gene (14). Mutant flies exhibited hypersensitivity to DNA interstrand crosslinking agents such as cisplatinum but not to alkylating agents such as methyl methanesulfonate (15). Seki et al. (13) recently purified human POLQ and examined its catalytic activity. POLQ has extremely low fidelity, making frequent errors on undamaged templates. Moreover, POLQ was shown to be the only enzyme to efficiently catalyze both the insertion and extension steps that allow bypass of abasic sites (16).
The function of Polθ in mammalian cells remains poorly understood. In a phenotype-based mutagenesis screen for chromosome instability mutants, Shima et al. (12) identified a mouse mutation called chaos1 (chromosome aberration occurring spontaneously), which exhibited elevated levels of spontaneous micronuclei in reticulocytes. The chaos1 mutation introduced a T-to-C base substitution in the Polq gene, which caused a serine-to-proline substitution at amino acid residue 1932 in this very large protein. Very recently, these investigators have generated Polθ-deficient mice, which have a phenotype very similar to chaos1, thus confirming the identity of Polq and chaos1 (17). Surprisingly, besides increased micronuclei in reticulocytes, the Polθ-deficient mice did not show any obvious phenotype and displayed the same sensitivity as normal mice to the DNA crosslinking agent mitomycin C and to γ-irradiation.
Unlike other low-fidelity DNA polymerases, which are ubiquitously expressed, human and mouse Polq exhibit a tissue-specific expression pattern. We found preferential expression in lymphoid tissues; most interestingly, abundant Polq transcripts were detected in GC B cells, the target cells for both SHM and CSR (18). The lymphoid tissue-specific expression pattern of Polq in both human and mouse suggested that Polθ might have a specialized role in mammalian lymphocytes.
To investigate the polymerase function of Polθ in SHM, here we have generated mice specifically devoid of Polθ polymerase activity (Polq-inactive), leaving helicase and other potentially important functional domains intact. Compared with WT mice, Polq-inactive mice exhibited a moderate decrease in overall mutation frequency in the JH4 intronic sequence of responding B cells, a genomic region chosen for analysis to avoid antigen-selection bias. Remarkably, however, only mutations at C/G were specifically reduced in Polq-inactive mice, whereas mutations at A/T were not affected. We propose that the polymerase activity of Polθ mediates the generation of C/G mutations by replicating over abasic sites formed by uracil-DNA glycosylase-catalyzed excision of the uracil residues generated by activation-induced cytidine deaminase.
Materials and Methods
Generation of Mice Lacking the Polymerase Core Domain. A targeting vector was constructed to replace exons 25 and 26 with the neomycin gene. R1 ES cells were transfected with the linearized targeting vector and 2 days after transfection cultured in the presence of 600 μg/ml G418 and 2 μM ganciclovir (only for the first 2 days). ES colonies were expanded and subjected to Southern blot analysis to identify homologous recombination. The deletion of exons 25 and 26 was further confirmed by RT-PCR analysis by using primers s6623 (5′-AGCGGGAAAAGCACCTGAAC-3′) and as7597 (5′-CGTCGTGAAGTTGAAGGATG-3′). RT-PCR was performed at 95°C for 2 min followed by 30 cycles of amplification at 95°C for 5 s, 58°C for 10 s, and 72°C for 2 min by using Taq polymerase (Toyobo, Osaka). Chimera mice were bred with C57BL/6N to obtain heterozygous mice. After backcrossing to C57BL/6N for three generations, heterozygous mice were bred with each other to obtain homozygous mice. Mice were kept at specific pathogen-free conditions, and animal experiments were approved by the Animal Facility Committee of the RIKEN Yokohama Institute.
Assay for Polymerase Activity. Polyclonal rabbit antibodies were raised against a peptide (QNDRTGLLPKRKLKG) located near the C terminus of mouse Polθ. For immunoprecipitation, 5 × 107 spleen cells were incubated for 30 min on ice in 0.5 ml of lysis buffer, as described (16) The lysate was centrifuged for 10 min at 15,000 rpm, and the supernatant was precleared four times with 0.5 ml of protein A beads. The precleared lysate was incubated with 10 μl of protein A beads at 4°C for 4 h and washed five times with the lysis buffer. Half of the washed beads were used for the polymerase assay and the remaining half of the beads for Western blot analysis. The polymerase assay was performed essentially as described by using 32P-labeled 16 mer (5′-CACTGACTGTATGATG-3′) annealed to a 30 mer (5′-CTCGTCAGCATCTACATCATACAGTCAGTG-3′) oligonucleotide as a template (16).
Flow Cytometry Analysis and Proliferation Assay. Flow cytometry analysis was performed as described (19). For proliferation assays, B cells were purified by negative selection from spleen of 8- to 12-wk-old mice by using a B cell enrichment set (BD Biosciences, San Jose, CA). Stimulation with anti-IgM antibodies, CD40 ligand, and LPS and 3H-thymidine uptake was essentially as described (20).
Immune Response. Five WT and eight Polq-inactive mice (9–10 wk old) were injected i.p. with 100 μg of 4-hydroxy-3-nitrophenyl-acetyl (NP) coupled to chicken gammaglobulin (NP-CGG) (Biosearch) precipitated with alum and boosted 4 wk later. Mice were bled weekly, and serum titers of NP-specific IgG1 were analyzed by ELISA, by using NP-specific monoclonal high- (clone C6) and low- (clone N1G9) affinity antibodies as a standard (21).
SHM Assay. Five pairs of age- and sex-matched WT and Polq-inactive mice (16 wk old) were immunized with 100 μg of NP-CGG and 2 weeks later, B220+PNA+ GC B cells were sorted. All mice were IgMb/b as determined by FACS analysis of peripheral blood leukocytes by using allotype-specific antibodies (FITC anti-IgMa and PE-anti-IgMb, BD Biosciences). Genomic DNA was isolated from sorted GC B cells and amplified with forward primer J558Fr3 (5′-CAGCCTGACATCTGAGGACTCTGC-3′) and reverse primer JHCHint (5′-CTCCACCAGACCTCTCTAGACAGC-3′), as described (22). PCR was carried out with KOD-Plus polymerase (Toyobo) under the following conditions: 94°C for 5 min and then 94°C for 20 s, 65°C for 30 s, and 68°C for 70 s for 30 cycles. The PCR products were cloned into the pCR2.1 vector for sequencing. Only clones with unique V(D)J junctions were analyzed.
Results
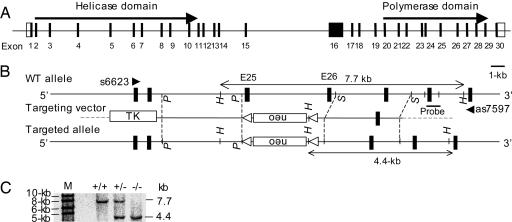
Generation of Mice Specifically Devoid of Polθ Polymerase Activity. To specifically inactivate the DNA polymerase activity of Polθ while leaving the helicase and other potentially important domains intact, we deleted exons 25 (185 base pairs) and 26 (112 base pairs), which encode a conserved aspartic acid residue known to be essential for the polymerase catalytic activity (23, 24) and the tyrosine residue that binds the incoming nucleotide (11, 14), respectively (Fig. 1 A and B). Homologous recombination in ES cells was monitored by Southern blot analysis (Fig. 1C). Because the deletion of these two exons was in-frame and the Polq promoter was intact, the targeted allele could theoretically be transcribed and translated. As predicted, RT-PCR analysis by using primers flanking exons 25 and 26 (s6623 and as7597, Fig. 1 A) gave rise to a 975-bp band in WT and a 678-bp band in mutant cells (Fig. 2A), although the level of mutant Polq was slightly reduced, possibly due to the intronic insertion of the neomycin gene. We further sequenced each of the two bands and confirmed that they were derived from WT mRNA and mRNA lacking exons 25 and 26 (data not shown). Consistent with the expression of the mutant mRNA, Western blot analysis revealed an immunoreactive protein in the mutant cells with a molecular weight similar to the WT Polθ (Fig. 2B). Because the antibodies used in this assay recognize an epitope near the C-terminal region of Polθ, we conclude that mutant cells express a truncated Polθ protein lacking the polymerase core domain at a slightly reduced level compared to WT cells.
Fig. 1.
Disruption of Polθ polymerase core motif. (A) Scheme of the mouse Polq gene. Solid boxes, exons; open boxes, noncoding regions. (B) Targeting strategy. The targeting vector was designed to replace exons 25 and 26 with a neomycin gene. Restriction enzyme sites (P, PvuII; S, SphI; H, HindIII), the probe for Southern blot analysis (solid bar), and PCR primers (s6623 and as7597) are indicated. (C) Southern blot analysis of HindIII-digested ES DNA
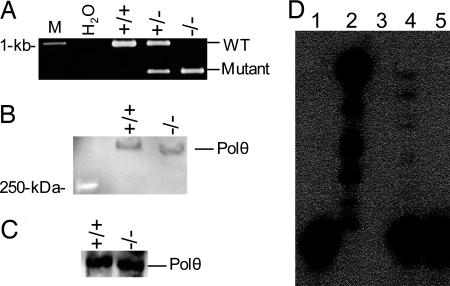
Fig. 2.
Polq-knockout mice express a mutant Polθ devoid of polymerase activity. (A) RT-PCR analysis using primers s6623 and as7597 (Fig. 1B) to detect the truncated mRNA. (B) Western blot analysis of Polθ expression in the splenocytes. (C) WT and mutant Polθ were immunoprecipitated from mouse spleen with rabbit polyclonal antibodies against Polθ and subjected to Western blot with the same antibodies. (D) The immunoprecipitated WT and mutant Polθ were analyzed for DNA polymerase activity as described in Materials and Methods. Lane 1, primer template alone; lane 2, exonuclease-free Klenow fragment; lane 3, empty well; lane 4, WT Polθ; lane 5, mutant Polθ
To confirm that the mutant enzyme was devoid of DNA synthesis activity, we immunoprecipitated Polθ from WT and mutant splenocytes. Western blot analysis revealed that a similar amount of WT and mutant Polθ was immunoprecipitated from both samples (Fig. 2C). As expected, WT (Fig. 2D, lane 4), but not mutant (Fig. 2D, lane 5), Polθ exhibited a template-dependent polymerase activity. These results demonstrate that the mutant mice express a truncated Polθ that has greatly reduced DNA polymerase activity but is likely intact for other potentially important functions.
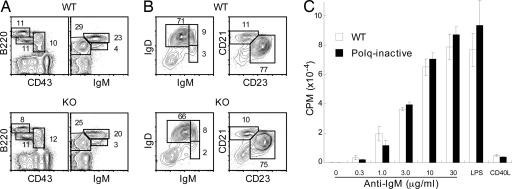
Normal B Cell Development and Maturation in Polq-Inactive Mice. FACS analysis of bone marrow cells revealed no significant differences in the percentages of B220+CD43+ progenitor and B220+CD43-IgM- precursor cells (Fig. 3A). B cell maturation in the spleen was also normal, because there was a similar ratio of IgMhighIgDdull (immature), IgMhighIgDhigh (transitional), and IgMdullIgDhigh (mature) populations in WT and Polq-inactive mice (Fig. 3B). In addition, no apparent differences between WT and Polq-inactive mice were observed in the follicular (CD23highCD21dull) and marginal zone (CD23dullCD21high) B cells (Fig. 3B). In our analysis of six pairs of WT and Polq-inactive mice, we have found no significant differences in B and T lymphocyte development and function (data not shown). In addition to their normal frequency, Polq-inactive B cells were functionally normal, as assessed by proliferation in response to anti-IgM antibodies, LPS, and CD40 ligand (Fig. 3C) and to various combinations of these stimuli (not shown), although the response to low doses of anti-IgM stimulation appeared to be slightly reduced in Polq-inactive B cells. These results demonstrate that Polq-inactive B cells undergo normal differentiation and maturation and respond normally to an array of in vitro stimuli that signal through a variety of receptors, including the B cell antigen receptor, Toll-like receptor 4, and the surrogate for T cell help, CD40.
Fig. 3.
Normal B cell development and in vitro responses in Polq-inactive mice. (A) FACS profiles of bone marrow cells in the lymphoid gate. (B) FACS profiles of B220+ cells in spleen. (C) Proliferative responses of purified spleen B cells. Cells (5 × 105/ml, 100 μl per well in 96 flat-bottom plates) were cultured for 48 h in medium alone or in the presence of different doses of anti-IgM antibodies, 10 μg/ml LPS, or 1/3 dilution of CD40L and pulsed with 3H-thymidine for the last 6 h
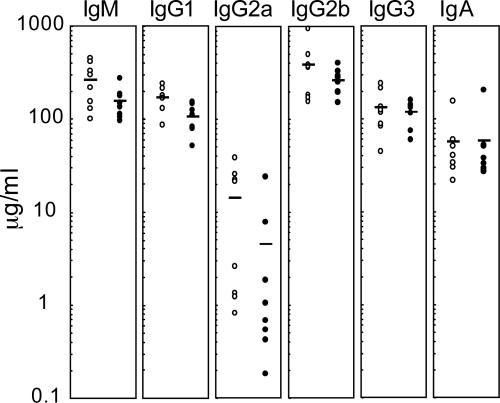
Reduction in Serum Ig Levels in Polq-Inactive Mice. To analyze B cell function in vivo, we first measured the levels of serum Igs in nonimmunized mice. Except for IgA, we found a general reduction in serum Ig levels in Polq-inactive mice (Fig. 4). The average Ig levels ± SD (μg/ml) in WT and Polq-inactive mice were 262 ± 132 and 154 ± 57 for IgM, 169 ± 47 and 107 ± 34 for IgG1, 14.1 ± 14 and 4.52 ± 8.1 for IgG2a, 381 ± 257 and 261 ± 81 for IgG2b, 130 ± 66 and 118 ± 34 for IgG3, and 56.0 ± 42 and 57.2 ± 59 for IgA, respectively. The reduction in the levels of IgM and IgG1 was statistically significant (P = 0.05 and 0.009, respectively), whereas the reduction in the levels of IgG2a, IgG2b, and IgG3 was not.
Fig. 4.
Serum Ig levels are slightly reduced in Polq-inactive mice. Eight pairs of age-matched WT and Polq-inactive mice (9–10 weeks old) were bled, and the serum Ig levels were measured by ELISA. Open circles, WT; solid circles, Polq-inactive mice. Bars, average titer
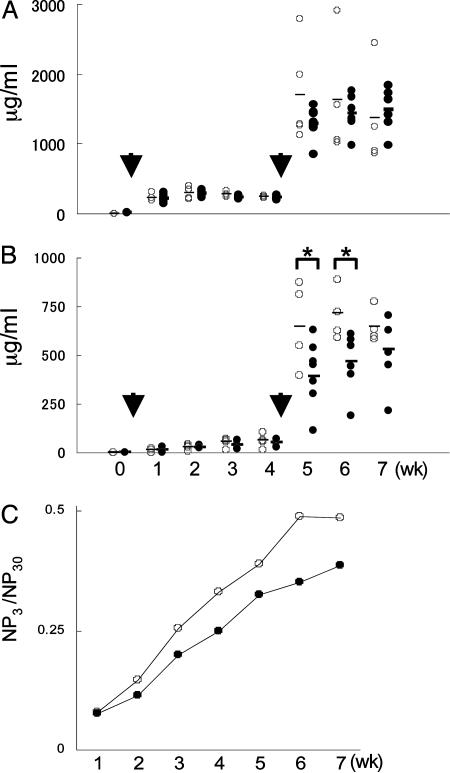
A Partial Impairment in the Production of High-Affinity Antibodies in Polq-Inactive Mice. We then examined the primary and secondary immune responses to the T-dependent antigen NP-CGG. Polq-inactive mice produced slightly reduced amounts of total (low- and high-affinity) anti-NP antibodies in both primary and secondary immune responses, as measured in an ELISA assay with NP30-BSA (Fig. 5A). However, the production of high-affinity antibodies, as measured with NP3-BSA, was more clearly reduced in the Polq-inactive mice (Fig. 5B). In fact, the observed reduction in the titers of total anti-NP antibodies could be explained by the reduction in the titers of high-affinity anti-NP antibodies. The reduced affinity maturation in Polq-inactive mice was further illustrated by comparing the ratio of NP3- and NP30-binding antibody titers (Fig. 5C). We also examined the appearance of the B220+PNA+ GC B cells after NP-CGG immunization. Polq-inactive and WT mice contained a similar frequency of GC B cells in spleen on days 10 and 14 after antigen injection (data not shown). These results may suggest a defect in selection of high-affinity B cells in the GC or that the Polq-inactive B cells have intrinsic defects in the SHM of Ig V genes.
Fig. 5.
Immune responses and affinity maturation in Polq-inactive mice. Mice (five WT and eight Polq-inactive) were immunized with 100 μg of NP-CGG precipitated with alum and boosted 4 wks later. Arrows indicate immunization times. Mice were bled weekly, and NP-specific serum IgG1 antibodies were measured by ELISA. Open and solid circles represent WT and Polq-inactive mice, respectively. (A) Titers of total (high- and low-affinity) NP-specific antibodies. (B) Titers of high-affinity NP-specific antibodies. *, P < 0.1 (unpaired Student's t test). (C) The average ratio of high-/low-affinity antibodies
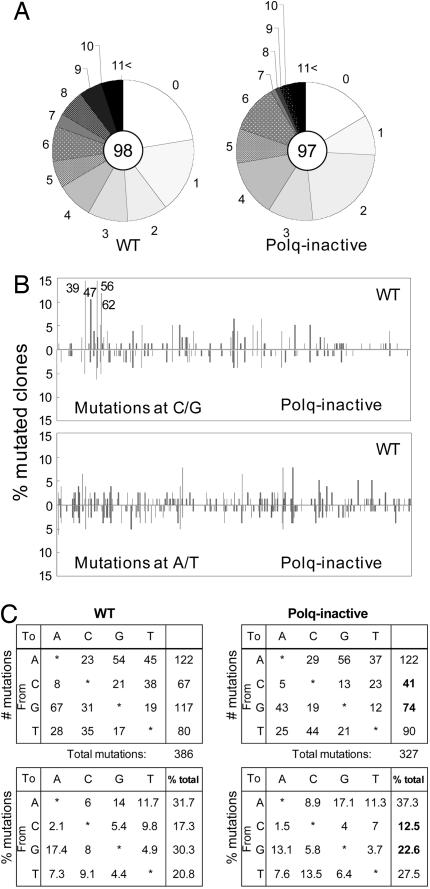
A Selective Reduction of Mutations at C/G in Ig Genes of Polq-Inactive Mice. To gain insight into the mechanisms underlying the reduced levels of high-affinity antibodies, we immunized mice with NP-CGG, isolated the GC B cells, and examined SHM. We restricted our analysis to the intronic sequence downstream of the JH4 region rather than the coding sequence of the VH186.2 gene, which is the dominant V gene segment used in the NP response, because the VH186.2 sequence is under strong antigen-driven selection, hence the unselected mutation pattern would likely be obscured (22, 25, 26). We analyzed 98 and 97 unique clones from WT and Polq-inactive mice, respectively (Fig. 6A; data for sequences are available upon request). Polq-inactive mice exhibited a slight reduction in the total mutation frequency in the JH4 intron (Table 1). However, when the mutation frequency of C/G vs. A/T mutations was calculated, we found that, whereas mutation frequency at A/T was similar between WT and Polq-inactive mice (0.522% and 0.514%, respectively), C/G mutation frequency was reduced by 41% (0.279% vs. 0.476% in WT mice, Table 1; Fig. 6B). In fact, the reduction in the overall mutation frequency in Polq-inactive mice was solely attributable to the reduction in mutations at C/G. The specific reduction of mutations at C/G was further illustrated by calculating the ratio of AT:CG mutations, which was 1.097 in WT and 1.843 in Polq-inactive mice (Table 1). Similarly, the relative representation of mutations at C/G and A/T was skewed (Fig. 6C; P < 0.001, χ2 test). The reduction of mutations at C/G was more focused on the intrinsic SHM hotspots (Fig. 6B). In particular, the C/G mutations at the four hotspots located at nucleotide positions 39, 47, 56, and 62 were all significantly reduced in Polq-inactive mice (Fig. 6B, P < 0.05). Both transition and transversion mutations were similarly affected in the Polq-inactive mice (Fig. 6C). No differences were observed in the ratio of transitions to transversions within C/G and A/T. These results indicate that the polymerase activity of Polθ is important for the generation of mutations at C/G.
Fig. 6.
Specific reduction of C/G mutations in the JH4 intronic sequences in Polq-inactive mice. (A) Pie charts depict the accumulation of mutations in unique sequences. The total number of unique clones analyzed is shown in the center of each circle. (B) Distribution of C/G (Upper) and A/T (Lower) mutations over the JH4 intronic region. The nucleotide number shown in Upper indicates the four hotspots (39, TGTT; 47, AGTT; 56, AGCA and 62, TGCA) (25, 27, 29, 38, 39)
Table 1. Somatic mutations in JH4 intronic sequences (509 base pairs).
| WT | Polq inactive | |
|---|---|---|
| Number of clones | 98 | 97 |
| Mutated clones, % | 76 (77.6) | 81 (83.5%) |
| Total length of mutated sequences | 38,684 | 41,229 |
| Total number of mutations (at AT:CG) | 386 | 327 |
| (202:184) | (212:115) | |
| Total mutation frequency, % | 0.998 | 0.793 |
| Mutation frequency at A/T, % | 0.522 | 0.514 |
| Mutation frequency at C/G, % | 0.476 | 0.279 |
| Ratio of AT: CG mutations | 1.097 | 1.843 |
Discussion
In the present study, we generated and analyzed mice expressing a mutant Polθ specifically devoid of polymerase activity. To our knowledge, this represents a unique mutant mouse line to have reduced C/G but relatively normal A/T mutations. The active site, rather than a gene ablation mutagenesis strategy, was adopted, because we reasoned that in the complete absence of Polθ, its function might be compensated for by other DNA polymerases. At least 10 lesion bypass DNA polymerases have been identified in higher eukaryotes. The presence of the mutant Polθ could potentially prevent irrelevant DNA polymerases from taking part in the process of SHM and allow us to explore the requirement for the DNA synthesis activity of Polθ in SHM. In this regard, it is possible that Polθ-null mice might show a quite different phenotype as compared with our Polq-inactive mice.
B cell development and maturation were normal in Polq-inactive mice. In addition, the Polq-inactive and WT B cells responded similarly to several different in vitro stimuli. Interestingly, we observed a reduction in serum Ig levels in nonimmunized mice, suggesting that Polq-inactive B cells might be partially impaired in CSR. In this regard, it is notable that mice deficient in msh2 or msh6, components of the mismatch repair pathway, exhibit a specific reduction in mutations at A/T and also have defects in CSR (27, 28). Although Polθ contains a helicase-like domain, helicase activity, which is known to play a critical role in DNA replication and recombination, has not been experimentally verified. Because Polq-inactive mice express a mutant protein with intact helicase and other potentially important functional domains, it is unclear how a mutant Polθ devoid of DNA synthesis activity might affect CSR. The reduction in the titer of serum IgM, which is mainly derived from B1 B cells, further suggests that Polq-inactive mice may have defects in the differentiation of specific B cell subpopulations.
Available evidence supports the DNA deamination model in which activation-induced cytidine deaminase deaminates C to U and generates a U/G mismatch (27, 29–33). This U/G lesion is thought to be resolved by three distinct but related pathways (8, 34). The phase 1a pathway generates G to A and C to T transitions by directly replicating over the U/G mismatch where U is recognized as T. The phase 1b pathway generates both transitions and transversions at C/G by replicating over the noninstructive abasic site formed after excision of U via uracil DNA glycosylase. U/G mispair can also be recognized by components of the mismatch repair pathways and trigger a short-patch mutagenic DNA repair, leading to mutations at A/T pairs (phase 2 pathway).
In all these scenarios, mutations are finally introduced by DNA polymerases. Indeed, a number of low-fidelity DNA polymerases have been implicated in SHM of Ig genes. Inhibition of DNA polymerase ζ (Polζ) expression by antisense RNA was shown to reduce overall mutation frequency, but not mutation pattern, in the CL-01 B cell line and in transgenic mice (35, 36). DNA polymerase ι (Polι) has also been suggested to play a role in SHM in a GC-type Burkitt's lymphoma line (37). However, 129 mice, which lack a functional Polι due to a nonsense mutation in the Poli gene, exhibit a normal frequency and pattern in Ig gene mutations (32, 38, 39). Deficiency in DNA polymerase η (Polη) has been shown to correlate with dramatically reduced mutations at A/T pairs in both human and mice (32, 33, 40–42). Polη does not seem to be involved in the mutations at C/G pairs, because neither the frequency nor the spectrum at C/G mutations was altered in the absence of Polη (32, 33). These observations suggest that different DNA polymerases might be involved in different mutagenic pathways. Until now, however, a DNA polymerase(s) specifically involved in C/G mutations has not been identified.
In the present study, we have shown that Polθ, or more precisely its DNA synthesis activity, is important for the generation of mutations at C/G pairs. These findings, together with the observations that human POLQ is the only enzyme that efficiently catalyzes both the insertion and the extension steps for bypass of abasic sites (16), implicate Polθ in the phase 1b mutagenic pathway. Biochemical analysis has demonstrated that POLQ preferentially inserts A opposite an abasic site (6). If POLQ has the same catalytic specificity in vivo, then it would generate primarily G to A and C to T transitions. If so, inactivation of Polθ polymerase activity should have resulted in fewer transitional mutations at C/G. However, both transition and transversion mutations at C/G were similarly reduced in the Polq-inactive mice. This observation suggested that C/G transversions were also affected in Polq-inactive mice. It is conceivable that Polθ may be able to insert nucleotides other than A opposite abasic sites under in vivo conditions where other necessary accessory factors are present. Indeed, recombinant human Polη and DNA polymerase κ (Polκ) exhibited dramatically different catalytic properties, including DNA synthetic activity and ability to bypass an abasic site, when assayed in the presence of accessory factors PCNA, replication factor C and replication protein A (43, 44). It is also possible that Polθ may catalyze the extension step from mismatch termini formed by other low-fidelity polymerases, which may insert C and T, as well as A opposite an abasic site and generate both transition and transversion mutations.
Disruption of Polθ polymerase activity resulted in approximately one-third reduction in both transition and transversion mutations at C/G pairs. Assuming that the transition mutations generated by phase 1a pathway are unaffected in Polq-inactive mice, Polθ polymerase activity should generate most of the transition mutations and a part of the transversion mutations at C/G in the phase 1b pathway. Therefore, a substantial portion of the transversion mutations at C/G must be catalyzed by as-yet-unidentified DNA polymerases. Candidate enzymes include Rev1, which predominantly generates transversions at C/G (45) and interestingly is up-regulated in GC B cells (unpublished results), and Polζ, which is a mismatch extender (46) and has been implicated in both A/T and C/G mutations (35, 36).
Acknowledgments
We thank Chitose Ishihara, Takako Maruyama, Hiromi Ebihara, and Akiko Ukai for excellent technical assistance; Drs. Masahiko Hatano, Akemi Sakamoto, and Peter Burrows for critical comments and helpful discussions; and Satoko Miyauchi for secretarial assistance. This work was supported by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology in Japan.
Author contributions: K.M., T.S., M.T., T.T., and J.O.-W. designed research; K.M., R.O., A.T., H.K., K.K., and J.O.-W. performed research; R.O. and T.A. contributed new reagents/analytic tools; and J.O.-W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NP, 4-hydroxy-3-nitrophenyl-acetyl; NP-CGG, NP coupled to chicken gammaglobulin; SHM, somatic hypermutation; Polθ/ζ/η/ι/κ, DNA polymerase θ/ζ/η/ι/κ; CSR, class switch recombination; GC, germinal center.
References
- 1.Tonegawa, S. (2003) Nature 302, 575-581. [DOI] [PubMed] [Google Scholar]
- 2.Schatz, D. G., Oettinger, M. A. & Baltimore, D. (1989) Cell 59, 1035-1048. [DOI] [PubMed] [Google Scholar]
- 3.Oettinger, M. A., Schatz, D. G., Gorka, C. & Baltimore, D. (1990) Science 248, 1517-1523. [DOI] [PubMed] [Google Scholar]
- 4.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20, 165-196. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 6.Neuberger, S. M., Harris, S. R., Di Noia, J. & Petersen-Mahrt, K. S. (2003) Trends Biochem. Sci. 28, 305-312. [DOI] [PubMed] [Google Scholar]
- 7.Honjo, T., Muramatsu, M. & Fagarasan, S. (2004) Immunity 20, 659-668. [DOI] [PubMed] [Google Scholar]
- 8.Neuberger, M. S., Di Noia, J. M., Beale, R. C., Williams, G. T., Yang, Z. & Rada, C. (2005) Nat. Rev. Immunol. 5, 171-178. [DOI] [PubMed] [Google Scholar]
- 9.Goodman, M. F. (2002) Annu. Rev. Biochem. 71, 17-50. [DOI] [PubMed] [Google Scholar]
- 10.Hubscher, U., Maga, G. & Spadari, S. (2002) Annu. Rev. Biochem. 71, 133-163. [DOI] [PubMed] [Google Scholar]
- 11.Sharief, F. S., Vojata, P. J., Ropp, P. A. & Copeland, W. C. (1999) Genomics 59, 90-96. [DOI] [PubMed] [Google Scholar]
- 12.Shima, N., Hartford, S. A., Duffy, T., Wilson, L. A., Schimenti, K. J. & Schimenti, J. C. (2003) Genetics 163, 1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki, M., Marini, F. & Wood, R. D. (2003) Nucleic Acids Res. 31, 6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, P. V., Mazina, O. M., Leonhardt, E. A., Case, R. B., Boyd, J. B. & Burtis, K. C. (1996) Mol. Cell. Biol. 16, 5764-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd, J. B., Sakaguchi, K. & Harris, P. V. (1990) Genetics 125, 813-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seki, M., Masutani, C., Yang, L. W., Schuffert, A., Iwai, S., Bahar, I. & Wood, R. D. (2004) EMBO J. 23, 4484-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shima, N., Munroe, R. J. & Schimenti, J. C. (2004) Mol. Cell. Biol. 24, 10381-10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura, K., Bahar, R., Seimiya, M., Chiyo, M., Wada, A., Okada, S., Hatano, M., Tokuhisa, T., Kimura, H., Watanabe, S., et al. (2004) Int. J. Cancer 109, 9-16. [DOI] [PubMed] [Google Scholar]
- 19.Seimiya, M., Wada, A., Kawamura, K., Sakamoto, A., Ohkubo, Y., Okada, S., Hatano, M., Tokuhisa, T., Watanabe, T., Saisho, H., et al. (2004) Eur. J. Immunol. 34, 1322-1332. [DOI] [PubMed] [Google Scholar]
- 20.Wang, J., Koizumi, T. & Watanabe, T. (1996) J. Exp. Med. 184, 831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa, K., Akasako-Furukawa, A., Shirai, H., Nakamura, H. & Azuma, T. (1999) Immunity 11, 329-338. [DOI] [PubMed] [Google Scholar]
- 22.Schenten, D., Gerlach, L. V., Guo, C., Velasco-Miguel, S., Hladik, L. C., White, L. C., Friedberg, C. E., Rajewsky, K. & Esposito, G. (2002) Eur. J. Immunol. 32, 3152-3160. [DOI] [PubMed] [Google Scholar]
- 23.Patel, P. H. & Loeb, L. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marini, F., Kim, N., Schuffert, A. & Wood, R. D. (2003) J. Biol. Chem. 278, 32014-32019. [DOI] [PubMed] [Google Scholar]
- 25.Frey, S., Bertocci, B., Delbos, F., Quint, L., Weill, J. C. & Reynaud, C. A. (1998) Immunity 9, 127-134. [DOI] [PubMed] [Google Scholar]
- 26.Jolly, C. J., Klix, N. & Neuberger, M. S. (1997) Nucleic. Acids Res. 25, 1913-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada, C., Ehrenstein, M. R., Neuberger, M. S. & Milstein, C. (1998) Immunity 9, 135-141. [DOI] [PubMed] [Google Scholar]
- 28.Martomo, S. A., William, W. Y. & Gearhart, P. J. (2004) J. Exp. Med. 200, 61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rada, C., Williams, G. T., Nilsen, H., Barnes, D. E., Lindahl, T. & Neuberger, M. S. (2002) Curr. Biol. 12, 1748-1755. [DOI] [PubMed] [Google Scholar]
- 30.Rada, C., Di Noia, J. M. & Neuberger, M. S. (2004) Mol. Cell 161, 63-171. [DOI] [PubMed] [Google Scholar]
- 31.Faili, A., Aoufouchi, S., Weller, S., Vuillier, F., Stary, A., Sarasin, A., Reynaud, C. A. & Weill, J. C. (2004) J. Exp. Med. 199, 265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delbos, F., De Smet, A., Faili, A., Aoufouchi, S., Weill, J. C. & Reynaud, C. A. (2005) J. Exp. Med. 201, 1191-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martomo, S. A., William, W. Y., Wersto, R. P., Ohkumo, T., Kondo, Y., Yokoi, M., Masutani, C., Hanaoka, F. & Gearhart, P. J. (2005) Proc. Natl. Acad. Sci. USA 102, 8656-8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, G. S., Brandt, V. L. & Roth, D. B. (2004) Mol. Cell 16, 505-508. [DOI] [PubMed] [Google Scholar]
- 35.Zan, H., Komori, A., Li, Z., Cerutti, A., Schaffer, A., Flajnik, M. F., Diaz, M. & Casali, P. (2001) Immunity 14, 643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz, M., Verkoczy, L. K., Flajnik, M. F. & Klinman, N. R. (2001) J. Immunol. 167, 327-335. [DOI] [PubMed] [Google Scholar]
- 37.Faili, A., Aoufouchi, S., Flatter, E., Gueranger, Q., Reynaud, C. A. & Weill, J. C. (2002) Nature 419, 944-947. [DOI] [PubMed] [Google Scholar]
- 38.McDonald, J. P., Frank, E. G., Plosky, B. S., Rogozin, I. B., Masutani, C., Hanaoka, F., Woodgate, R. & Gearhart, P. J. (2003) J. Exp. Med. 198, 635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu, T., Azuma, T., Ishiguro, M., Kanjo, N., Yamada, S. & Ohmori, H. (2005) Immunol. Lett. 98, 259-264. [DOI] [PubMed] [Google Scholar]
- 40.Zeng, X., Winter, D. B., Kasmer, C., Kraemer, K. H., Lehmann, A. R., Gearhart, P. J. (2001) Nat. Immunol. 2, 37-541. [DOI] [PubMed] [Google Scholar]
- 41.Rogozin, I. B., Pavlov, Y. I., Bebenek, K., Matsuda, T. & Kunkel, T. A. (2001) Nat. Immunol. 2, 530-536. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, T. M., Vaisman, A., Martomo, S. A., Sullivan, P., Lan, L., Hanaoka, F., Yasui, A., Woodgate, R. & Gearhart, P. J. (2005) J. Exp. Med. 201, 637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haracska, L., Johnson, R. E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L. & Prakash, S. (2001) Mol. Cell. Biol. 21, 7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haracska, L., Unk, I., Johnson, R. E., Phillips, B. B., Hurwitz, J., Prakash, L. & Prakash, S. (2002) Mol. Cell. Biol. 22, 784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson, L. J. & Sale, J. E. (2003) EMBO J. 22, 1654-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S. & Prakash, L. (2000) Nature 406, 1015-1019. [DOI] [PubMed] [Google Scholar]