Abstract
The x-ray crystal structure of the pyridoxal-5′-phosphate (PLP), S-adenosyl-l-methionine (SAM), and [4Fe–4S]-dependent lysine-2,3-aminomutase (LAM) of Clostridium subterminale has been solved to 2.1-Å resolution by single-wavelength anomalous dispersion methods on a l-selenomethionine-substituted complex of LAM with [4Fe–4S]2+, PLP, SAM, and l-α-lysine, a very close analog of the active Michaelis complex. The unit cell contains a dimer of hydrogen-bonded, domain-swapped dimers, the subunits of which adopt a fold that contains all three cofactors in a central channel defined by six β/α structural units. Zinc coordination links the domain-swapped dimers. In each subunit, the solvent face of the channel is occluded by an N-terminal helical domain, with the opposite end of the channel packed against the domain-swapped subunit. Hydrogen-bonded ionic contacts hold the external aldimine of PLP and l-α-lysine in position for abstraction of the 3-pro-R hydrogen of lysine by C5′ of SAM. The structure of the SAM/[4Fe–4S] complex confirms and extends conclusions from spectroscopic studies of LAM and shows selenium in Se-adenosyl-l-selenomethionine poised to ligate the unique iron in the [4Fe–4S] cluster upon electron transfer and radical formation. The chain fold in the central domain is in part analogous to other radical–SAM enzymes.
Keywords: pyridoxal phosphate, radical mechanism, S-adenosylmethionine
Lysine-2,3-aminomutase (LAM) was first purified and characterized from Clostridium subterminale SB4 in connection with studies of lysine metabolism in Clostridia (1). LAM catalyzes the interconversion of l-α-lysine and l-β-lysine, which proceeds by migration of the amino group from C2 to C3 concomitant with cross-migration of the 3-pro-R hydrogen of l-α-lysine to the 2-pro-R position of l-β-lysine (2). Hydrogen transfer takes place without exchange with solvent protons. Enzymatic isomerization reactions of this type typically require adenosylcobalamin as a coenzyme, and the reactions proceed by radical mechanisms initiated by homolytic cleavage of the Co–C5′ bond to generate the 5′-deoxyadenosyl radical. However, LAM does not require a vitamin B12 coenzyme but instead requires a [4Fe–4S] cluster, S-adenosyl-l-methionine (SAM), and pyridoxal-5′-phosphate (PLP) as coenzymes (3, 4).
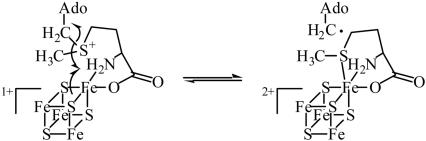
A mechanistic feature in the action of LAM, shared in common with adenosylcobalamin-dependent enzymes, is the participation of the 5′-deoxyadenosyl radical, which initiates free radical formation. However, in the reaction of LAM, this radical initiator arises from a reversible chemical reaction between SAM and the [4Fe–4S]1+ cluster, leading to the homolytic scission of the C5′–S bond in SAM (4, 5). Spectroscopic results indicate that homolysis takes place by reductive cleavage of SAM through electron transfer from the [4Fe–4S]1+ cluster, leading to the [4Fe–4S]2+ cluster and the 5′-deoxyadenosyl radical (6, 7). LAM is a member of the radical–SAM superfamily, which is characterized in part by the cysteine motif CxxxCxxC in the amino acid sequences of family members (8). The three cysteine residues in the motif donate three sulfhydryl ligands to three iron atoms in the [4Fe–4S] cluster, leaving a unique iron that accepts ligands from SAM. The actions of these enzymes entail the transient formation of the 5′-deoxyadenosyl radical as an enzyme-bound intermediate through the reaction of SAM with the iron–sulfur cluster (4).
Spectroscopic studies laid a groundwork for understanding the mechanism by which radical–SAM enzymes catalyze the reductive cleavage of SAM to methionine and the 5′-deoxyadenosyl radical. Selenium x-ray absorption spectroscopy of LAM activated by Se-adenosyl-l-selenomethionine (SeSAM) or in complex with l-selenomethionine revealed a direct ligation of selenium with iron (6). Electron nuclear double resonance experiments demonstrating N/O chelation of the methionyl carboxylate and amino groups of SAM with the unique iron sites of the [4Fe–4S] clusters in pyruvate formate lyase activase and LAM characterized the ligation in the SAM/[4Fe–4S] complex (7, 9). Taken together, these results suggested an inner sphere pathway for electron transfer in the homolytic scission of the C5′–S bond in SAM (7, 9). A mechanism such as that shown in Scheme 1 accounts for the reversible cleavage of SAM at the active site of LAM (7). A different mechanism has been proposed for the irreversible reductive cleavage of SAM in the action of pyruvate formate lyase activase (9).
Scheme 1.
We have solved the x-ray crystal structure of LAM in a complex with the substrate and all of the cofactors to 2.1-Å resolution. The structural model reveals close spatial relationships among SeSAM, the [4Fe–4S]2+ cluster and the external aldimine of l-α-lysine and PLP. This inactive complex differs by a single electron from an active Michaelis complex, which contains the [4Fe–4S]1+ cluster, and the structure is likely to be essentially identical with that of the active complex. The structure supports the mechanism of Scheme 1 for the reversible reductive cleavage of SAM.
Materials and Methods
LAM Expression and Purification. l-Selenomethionine-labeled LAM from C. subterminale SB4 was prepared as a recombinant protein expressed in a methionine auxotroph of an Escherichia coli cell line grown in media containing l-selenomethionine (Aldrich). The gene kamA for LAM (10) was transferred to a pET21a(+) vector (Novagen) containing the lac operator downstream from the T7 promoter, and the resulting expression vector was transformed into competent BL21(DE3) RIL-X cells (Stratagene). A subculture was used to inoculate 16 liters of minimal media (11) containing 0.3 mM l-selenomethionine, 100 μg·ml–1 ampicillin (Sigma), 35 μg·ml–1 chloramphenicol (Sigma), and 0.3% glycerol as the source of carbon. The cells were cultured at pH 7.0 and 37°C in a glass fermentor (VirTis) with vigorous aeration and agitation. After 48 h, the cell density corresponded to an absorbance of ≈1 at 600 nm. Isopropyl β-d-thiogalactoside (1 mM) and additional l-selenomethionine (0.6 mM) were added, and air was replaced with nitrogen gas. After anaerobic culturing for 16 h at 37°C and centrifugation, ≈40 g of cell paste was obtained and stored at –70°C.
The l-selenomethionine-enriched LAM was purified at 22°C in an anaerobic glove chamber (Coy Laboratory Products, Grass Lake, MI) according to a modification of the published procedure (3, 12). Standard buffer (SB) contained 0.03 M K+-N-2-hydroxyethylpiperazine-N-3-propane sulfonic acid at pH 8.0 supplemented with 10 μM PLP, 20 μM FeSO4, 1 mM PMSF, and 1 mM DTT. Cells were suspended in 150 ml of SB and broken by sonication in six 30-s bursts at 4–8°C. After 30 min of centrifugation at 15,000 × g (4°C), the supernatant was treated at 4°C with 3% streptomycin sulfate, followed by centrifugation as above, and then with (NH4)2SO4 at 70% of saturation followed by centrifugation. The precipitate was dissolved in SB containing 0.8 M (NH4)2SO4, applied to a 5- × 15-cm column of Phenyl Sepharose 6 Fast Flow (Amersham Biosciences) equilibrated with SB containing 1.0 M (NH4)2SO4 and eluted with a decreasing linear gradient of (NH4)2SO4 formed from 2 liters of SB and 2 liters of SB with 1.0 M (NH4)2SO4. The yellow-brown protein was collected, precipitated with (NH4)2SO4 at 70% saturation, and centrifuged. The protein was dissolved in 800 ml of SB, applied to a 2.5- × 30-cm column of Q-Sepharose Fast Flow (Amersham Biosciences), and eluted at 3 ml·min–1 with an increasing linear gradient composed from 2 liters of SB with 0.03 M K2SO4 and 2 liters of SB with 0.2 M K2SO4. LAM emerged at the end of the gradient and was precipitated with 70% (NH4)2SO4, redissolved in 5 ml of SB, and applied to a Sephacryl S200 gel filtration column (2.5 × 30 cm) equilibrated and eluted with SB. Peak fractions were concentrated by Centricon 10 centrifugal filters to 20–30 mg·ml–1, divided into aliquots, and frozen in liquid N2.
Purified LAM at 15 mg·ml–1 (0.3 mM subunits) was incubated with 60 mM K-N-2-hydroxyethylpiperazine-N-3-propane sulfonic acid, pH 8.0/0.6 mM FeSO4/0.6 μM PLP/3 mM cysteine for 4 h at 37°C. The enzyme was separated from the reductive incubation components by gel filtration using a 2.5- × 15-cm column of Sephacryl S200 (Amersham Biosciences) preequilibrated and eluted with 30 mM K N-2-hydroxyethylpiperazine-N-3-propane sulfonic acid buffer at pH 8.0 containing 1 mM DTT, 0.25 mM l-lysine, 10 μM PLP, and 20 μM FeSO4. Protein fractions were concentrated by using Centricon 10 spin columns (Amicon YM10, Millipore) to ≈40 mg·ml–1. The concentrated fractions were divided into aliquots and frozen and stored in liquid N2.
Crystallization. Crystals of LAM were grown with sitting drop geometry with drop sizes of 2–10 μl with various ratios of protein-:precipitant. The precipitant consisted of 0.03 M Na-N-2-hydroxyethylpiperazine-N-3-propane sulfonic acid (pH 8.0), 0.2–0.6 M l-lysine·HCl (pH 8.0), 0.2–0.6 M sodium malonate (pH 8.0), 5–15% polyethylene glycol 200, 10–20% polyethylene glycol 8000, 1 mM S-adenosyl-l-homocysteine, and 5 mM DTT. All crystallization reagents were purchased from Sigma or Hampton (San Diego). The concentration of LAM was 20–40 mg·ml–1 (500–1,000 μM subunits) in the buffer in which it was purified and frozen.
Data Collection, Refinement, and Model Building. X-ray diffraction data were collected from two separate crystals of identical source and growth conditions to achieve selenium substructure phases and a protein model refined at 2.1 Å (Table 1; see also Table 2, which is published as supporting information on the PNAS web site). Data set for selenium substructure and initial model. A crystal of l-selenomethionine–LAM was cryoprotected by immersion in liquid nitrogen. Data were collected at 100 K at beamline X-25 of the National Synchrotron Light Source of Brookhaven National Laboratory (Upton, NY) at the selenium absorption maximum as determined by a fluorescence counter measurement. A 120° sweep was carried out in 0.5° increments with exposure times of 10 s per increment. The crystal was inverted by 180° every 10° to minimize the effects of radiation damage to Friedel pairs of reflections. Diffraction data were indexed, integrated, and scaled with the hkl suite of programs including hkl2000 (13).
Table 1. Refinement.
| Parameter | Data set for phasing | Data set for final model |
|---|---|---|
| Wavelength, Å (eV) | 0.9795 (12,659) | 0.9792 (12,662) |
| Temperature, K | 100 | 100 |
| Resolution, Å | 50.0-3.0 | 50.0-2.1 |
| Overall I/σI (highest bin)/σ cutoff | 18.7 (7.1)/0 | 7.4 (3.0)/0 |
| Overall Rmerge (highest bin) | 0.087 (0.263) | 0.088 (0.299)* |
| Overall completeness (highest bin), % | 99.9 (99.5) | 88.3 (62.3) |
| No. of total/unique reflections | 2,147,382/38,399 | 1,347,088/159,068 |
| Spacegroup, unit cell constants | C2, a = 1118.93 Å, b = 81.93 Å, c = 177.96 Å; β = 96.783° | C2, a = 118.90 Å, b = 92.93 Å, c = 177.74 Å; β = 96.738° |
| Vm, Å3·Da-1 (% solvent) | 2.67 (53.88) | 2.67 (53.88) |
| Luzzati error, Å (Rfree-based DPI) | NA | 0.33 (0.21) |
| Rfree(Rcryst), % | NA | 22.5/18.7 |
Rmerge = Σhkl|Ihkl — 〈Ihkl〉|/Σhkl|Ihkl. Rmeas was calculated as in Diederichs and Karplus (12). Rfree is equivalent to Rcryst but consists of 8.1% reflections (7,437) selected at random (19, 42). Working and test sets were combined in Rcryst. Data statistics were calculated with hkl2000 (13), and final model statistics were calculated by refmac (23). DPI, diffraction precision index; NA, not applicable.
Data are for Rmeas.
Selenium atoms (30 of a total of 44 expected) were identified in the 3.0-Å data set (Table 1) by use of the program solve (14), giving a figure of merit of 0.29 to 3.2 Å. Statistical density modification with the program resolve (15) allowed identification of three noncrystallographic symmetry (NCS) operators and improved the figure of merit to 0.69 (overall) and allowed extension of the phases to 3.0 Å. Eighteen selenium sites did not obey the NCS operators and, therefore, were not included in the refinement of the NCS operators. Refinement of all 30 selenium atoms in the program sharp (16, 17) improved the selenium atom positions significantly enough that inclusion of these atoms in resolve density modification allowed identification of a fourth NCS operator. Electron density maps calculated by using phases from resolve were clear enough to visually recognize protein backbone connectivity at a 1σ contour level and to recognize the four [4Fe–4S] clusters at a 10σ contour level. Another 14 selenium atoms were identified and included in a final resolve calculation, which gave a figure of merit of 0.76, with a total of 44 sites obeying the 4-fold NCS operators. Four of these selenium atoms are located within 4.0 Å of each [4Fe–4S] cluster and correspond to SeSAM (see below).
A model comprising ≈40% of the expected 13,216 atoms of the protein was built into the entire asymmetric unit as a polyalanine chain with the automated density tracing feature of resolve. The segments with the best connectivity were collected into a single chain and manually built further by using xfit (18) and cns (19) with strict 4-fold NCS to a correlation coefficient of ≈80% per chain, allowing 75% of the protein and cofactor atoms to be built into the electron density maps calculated with combined solvent-flattened and model phases. Map format interconversion was carried out with the program cns2fsfour and with utilities from the Uppsala Software Factory and ccp4 project (20, 21). Data set for refinement. A single crystal of l-selenomethionine–LAM was cryoprotected as described above. Data were collected at 100 K in beamline 14-ID of the Industrial Macromolecular Crystallography Association Collaborative Access Team of the Advanced Photon Source of Argonne National Laboratory at the selenium absorption maximum as determined by a fluorescence counter measurement. Diffraction data were collected as for data set 1 (see above) and were processed as with data set 1 (Table 1).
Data to 2.1 Å and phases calculated from the previous data set slightly improved the value of Rfree, improved the NCS operator correlation coefficient to 90.0%, and improved the real-space and anomalous Fourier electron density maps to the extent that individual iron atoms of the cluster were resolved in the residual electron density and anomalous difference Fourier maps. Four strong peaks in the anomalous difference Fourier map indicated the position of an atom that was not accounted for as methionine in the amino acid sequence or as SeSAM. The fluorescence scan of a crystal of l-selenomethionine–LAM (Fig. 7, which is published as supporting information on the PNAS web site) clearly confirms the presence of zinc, which had previously been reported (22). Therefore, zinc was modeled at this site, leading to a decrease in Rfree and improvement in the electron density maps. Refinement was continued with refmac (23) by using 4-fold NCS restraint.
The final model consists of 1,638 of 1,664 expected amino acid residues, four each of SeSAM, [4Fe–4S], lysyl-PLP external aldimine, sulfate. There are 629 fixed water molecules. Analysis with procheck (24) shows that of all amino acid residues other than proline or glycine, 1,414 lie in allowed regions (89.03%), 155 lie in additional allowed regions (10.96%), and no residues lie in either generously allowed or disallowed regions (0.0%). There are no NCS-related outliers in the Ramachandran plot. The amino acid moieties of the lysyl-PLP external aldimine and SeSAM adopt allowed Ramachandran angles. A cis-peptide bond is formed between His-221 and Pro-222. Modeling the tetramer or each subunit as a translation–libration–screw group improved the Rfree factor by 2.5%. The mean anistropy of the anisotropic displacement parameters derived from the translation–libration–screw tensors are not significantly different from the average in the Protein Data Bank (PDB) as calculated by using the parvati (Protein Anisotropic Refinement Validation and Analysis Tool) server (Table 2) (25). Residues in the second two helices of the N-terminal region are disordered because some side chains lack electron density or have Beq values of >50 Å2, so the occupancy of the side-chain carboxylates of Glu residues 33 and 43 in this region were set to zero and likewise for Glu residues 148, 239, and 408. The side chains of Lys residues 10, 42, 46, and 385 exhibited little to no electron density; hence, their occupancies were set to zero. Refinement parameters for SeSAM were based on the Cambridge Structural Database [Cambridge Crystallographic Data Centre (43)] entries CERGEL and BADPEB. External aldimine parameters were based primarily on Cambridge Structural Database entries WAC-NAP and VOKJUA. Iron–sulfur cluster parameters were based on Strop et al. (26). Zinc–sulfur parameters were based on PDB ID code 1BTO (27). Figures were generated with povray and either molscript (44), povscript+ (28), or dino.
Results
Overall Structure. The x-ray crystal structure of LAM, solved to 2.1-Å resolution with Rfree factor of 22.5%, contains l-α-lysine as the external aldimine with PLP. The iron–sulfur cluster [4Fe–4S] contains four iron and four sulfide ions, with SeSAM ligated to the cluster. Four sulfate ions and four zinc ions are coordinated by the protein. LAM in the crystal is poised to become an active Michaelis complex upon one-electron reduction of the [4Fe–4S]2+ to [4Fe–4S]1+.
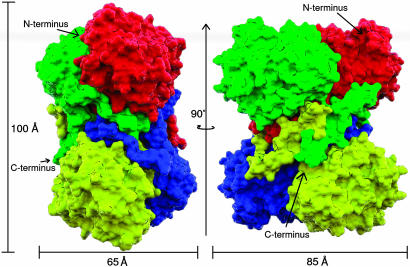
LAM crystallizes as a homotetramer in the asymmetric unit. Cross-linking studies in solution indicate the presence of dimers, tetramers, and hexamers (29), and electron photomicrographs show images in various states of aggregation (T. G. Frey and P.A.F., unpublished observations). The homotetramer consists of a dimer of tightly associated dimers (Fig. 1). In the crystal, the subunits are packed with 222 symmetry in the form of two domain-swapped (45) dimers: The chain folds of a subunit and domain-swapped dimer are shown in Fig. 2. The subunits of the dimer are linked through a zinc ion that coordinates residues from both subunits (Fig. 3). The interface of the domain-swapped dimer buries 8,734 Å2. The dimer–dimer association buries an additional 8,715 Å2 of surface area derived entirely from residues in each C terminus, whereas 40 water molecules mediate contact between the C-terminal coil and residues on the bottom face of each dimer. In all, the tetramer is a highly compact structure of essentially identical subunits. The root mean square deviation (RMSD) of main-chain atoms between subunits in the tetramer are ≤0.35 Å. The tetrameric model accounts for 1,638 of 1,664 residues. All four chains lack electron density for the N-terminal methionine and isoleucine residues and several C-terminal residues. Chains A and C extend to Lys-411, whereas chains B and D extend to Arg-412.
Fig. 1.
Tetrameric packing of LAM subunits. Two views of the tetramer at 0° and rotated by 90°. Each subunit is represented as a separate surface and color. Thus, one domain-swapped, zinc-coordinated dimer corresponds to the red/green or yellow/blue subunit combinations. Molecular surfaces were calculated with msms (42), modeled with dino, and rendered in povray. The surfaces are intended only to distinguish each subunit from the others.
Fig. 2.
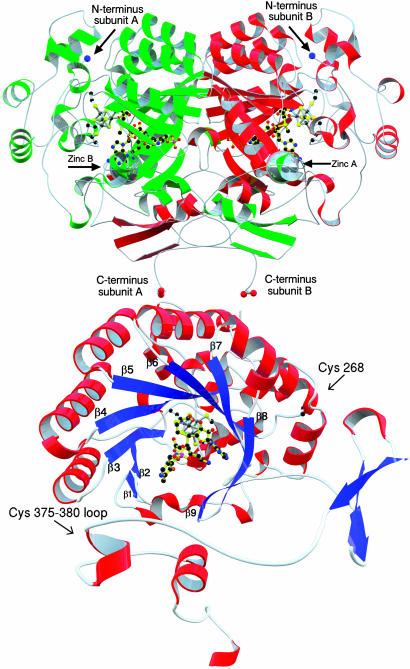
Secondary structure of the LAM subunit from the N terminus of the channel. (Upper) Coloring corresponds to that in Fig. 1 and thus represents the domain-swapped dimer. Zinc is rendered as a silver sphere and is labeled in each subunit. (Lower) LAM monomer. Helices are red, β-strands are blue, and coils are white. Cofactors are rendered with yellow bonds, iron is metallic black, cluster cysteines are rendered with white bonds. The location of the zinc-coordinating cysteines are indicated with arrows (see Fig. 3 for detail).
Fig. 3.
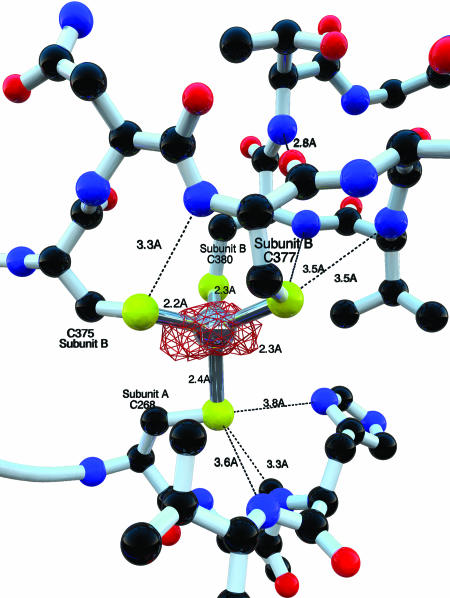
A zinc site in LAM shown with an anomalous difference Fourier map calculated without the contribution of zinc phases and contoured at 6σ. A total of two equivalent zinc sites are required to mediate the covalent attachment of two subunits of LAM to form a dimer.
Each subunit comprises three domains (Fig. 8, which is published as supporting information on the PNAS web site). The central globular domain composed of residues 54–339 is reminiscent of a TIM barrel (β/α)8. However, the barrel lacks two β-strands and forms a (β/α)6-crescent. The β-sheet of the crescent is extended by two β-strands at the C-terminal side, the last of which is antiparallel, and an antiparallel β-strand at the N-terminal side. An N-terminal domain composed of residues 3–53 comprises seven short helices forming a cluster closing off the C terminus of the channel formed by the central crescent of the opposing subunit. The C-terminal domain composed of residues 340–412 contains cysteine residues 375, 370, and 380, which coordinate the zinc ion, and a three-stranded β-sheet that interacts with the last two β-strands of the crescent formed by the opposing subunit containing the fourth zinc ligand (Cys-265). Thus, the quaternary structure involves an assembly of two domain-swapped dimers that are held together by zinc ions in addition to C-terminal coils that converge at the tetramer equatorial region in the formation of an extensively hydrogen-bonded tubular structure. The C-terminal domain accounts entirely for the contacts in the dimer–dimer interface, the remaining contacts being mediated by water molecules.
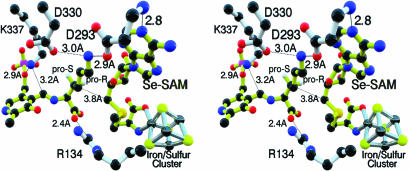
The Active Site. The crescent channel encloses the external aldimine of l-α-lysine and PLP and the complex of SeSAM/[4Fe–4S], which are themselves closely associated in the active site (Figs. 2 and 4). The sequence 125CSMYCRHCTRRR136 binding the iron–sulfur cluster is in the loop emanating from β-strand 2. The GG sequence motif is in the loop connecting β-strand 3 and helix 3 and forms one side of the active site pocket close to the site of the SeSAM methionine moiety ligated to the [4Fe–4S] cluster (Fig. 5).
Fig. 4.
Ball-and-stick, wall-eyed stereodiagram figure of the active site. This wall-eyed stereodiagram emphasizes the stereochemical relationships of the three cofactors, one as lysyl-PLP external aldimine. The C5′ carbon of SeSAM where radical initiation will occur is 3.8 Å from C3 of the lysyl side chain. The conserved Arg-134 denies rotation around the C1–C2 bond of lysyl moiety. Asp-293 interacts with both the ε-amino group of lysine and N1 of the purine ring of SeSAM, and Asp-330 is in hydrogen-bonded contact with the ε-amino group. SeSAM is ligated to the unique iron in the [4Fe–4S] cluster through the selenomethionyl α-amino and carboxylate groups. See Fig. 8 for detailed interactions within the active site. Hydrogen atom positions are provided for reference based on theoretical constraints only and are not determined by the diffraction data.
Fig. 5.
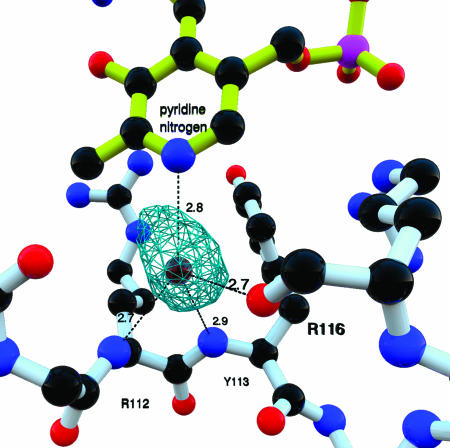
Close-up of the interaction between N1 of PLP and a fixed water molecule. This water molecule is held precisely in place in all four subunits by three main-chain hydrogen bonds from residues in the loop (R112YPDR116). The nature of the hydrogen bonding partners would uniquely position this water such that a water proton is hydrogen bonded with N1 of the pyridine nitrogen, implying that the pyridoxal ring is unprotonated.
The iron–sulfur cluster contains three iron atoms ligated to Cys-125, Cys-129, and Cys-133. The distances and angles for the atoms in this cluster are consistent with prior studies of [4Fe–4S] clusters in proteins (26). The fourth iron atom forms a complex with the carboxylate and α-amino groups of SeSAM. The backbone nitrogen atoms of the cysteine-containing loop come close to two of the sulfur atoms of the [4Fe–4S] cluster, which has been proposed to be significant for modulation of the reduction potential (ref. 26 and references therein).
PLP is located at the N terminus of the channel as the external aldimine with l-α-lysine. The absolutely conserved Gly-321 on the turn between helix 14 and β-strand 8 closes off the N terminus of the channel of the subunit, which is covalently linked by zinc. Although the entire active site is closed to solvent access, there are eight water molecules located in identical positions within the channel in each subunit (Fig. 9, which is published as supporting information on the PNAS web site). The PLP moiety of this adduct is held in place by eight hydrogen bonds to the phosphate, one to the phenolic oxygen and one to the pyridine nitrogen, involving nine residues, four of which are absolutely conserved among LAMs, in addition to water molecules (Fig. 9). Gly-321 of the domain-swapped subunit contributes one of the hydrogen bonds to the phosphate.
A striking feature of the active site is the hydrogen-bonded contact between PLP and a fixed water molecule held in place by backbone hydrogen bonds from the carbonyl of Arg-116, amide hydrogen atoms from Tyr-113 and Arg-112, and N1 of PLP (Fig. 5). The fixed water most likely donates a proton in the hydrogen bond to N1 of PLP, and the pyridine ring is not protonated. This mode of PLP binding is unprecedented among PLP-dependent enzymes.
Lysine is held in place by three hydrogen bonds, two between the ε-amino group of lysine and Asp-330 and Asp-293 and one between the carboxylate group and the conserved Arg-134 (Fig. 4). These interactions uniquely position lysyl-C2 and -C3, between which the amino group and a hydrogen atom are transferred in the mechanism (4, 5). Ser-169 is 2.9 Å from the carboxylate and is likely to be important. In the structure of this complex, the lysyl side chain is positioned close to the adenosyl portion of SeSAM, with the 3-pro-R hydrogen near adenosyl-C5′, which is within 4 Å of C3 in the lysyl side chain. Upon reductive cleavage of SAM, the 5′-deoxyadenosyl radical is in excellent position to abstract the 3-pro-R hydrogen from the lysyl-PLP external aldimine, the first step in the rearrangement of l-α-lysine to l-β-lysine (4, 5). The radical-based mechanism includes four radicals, three of which have been characterized structurally and kinetically by EPR spectroscopy (30–34).
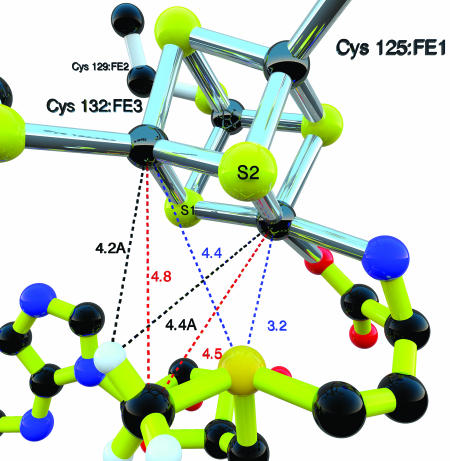
In the structure of SeSAM ligated to the [4Fe–4S] cluster, selenium is fixed within 3.2 Å of the unique iron (Fig. 6), to which the α-amino and carboxylate groups of the methionyl moiety of SeSAM are also ligated (Fig. 4). Thus, selenium is well positioned to ligate iron upon electron transfer and cleavage of the C5′–Se bond in the mechanism of Scheme 1, as observed in the x-ray absorption spectroscopy experiments (6). In the reaction of SAM, sulfur presumably reacts in the same way.
Fig. 6.
Close-up of the SeSAM/[4Fe–4S] cluster region. Shown are the structural relationships and distances between the Se atom in LAM or methyl group of SeSAM and cluster atoms. Distances shown are for subunit A, with an RMSD of 0.1 Å for all subunits. The distances shown can be compared with those determined spectroscopically in experiments on LAM or pyruvate–formate lyase-activating enzyme: x-ray absorption spectroscopy in the postcleavage, radical state of LAM; Se-to-Fe distance, 2.7 Å (6). Electron nuclear double resonance of pyruvate–formate lyase-activating enzyme: methyl-C-Fe, 4–5 Å (9); methyl-H-Fe, 3.0–3.8 Å (9). Hydrogen atom positions are theoretical and are not determined by the diffraction data.
Discussion
The structure of LAM increases to four the current number of three-dimensional structures of radical–SAM enzymes. These enzymes share a (β/α)6 repeat motif in their structural cores. Alignment of this motif for all four structures places the iron–sulfur cluster at the same location relative to the motif. The β-sheet formed by this motif is extended on both sides in MoaA, HemN, and LAM to form a larger crescent channel closed by an additional domain, whereas the sheet is extended by two additional β/α repeats in BioB which close to form a TIM barrel. Despite these structural similarities, these enzymes diverge in ways related to their biochemical actions. MoaA functions in molybdopterin biosynthesis (35), BioB in biotin synthesis (36), and HemN in heme biosynthesis in anaerobes (37). Unlike the others, LAM catalyzes a PLP-dependent reversible reaction. From the standpoint of structural fold, LAM is most similar to HemN, the structure of which is similar to the central domain of the LAM subunit. HemN is monomeric in solution and in the crystal and therefore distinguished from LAM in that the domain that closes the crescent channel resides in the same subunit as the crescent domain. In BioB, the TIM barrel is fully formed and there is an additional [2Fe–2S] cluster. MoaA also binds an additional iron–sulfur cluster. Among these enzymes, only LAM uses PLP as a coenzyme, and it is the only one to date that is tetrameric and formed from domain-swapped dimers held together by coordination to zinc.
Specific interactions among amino acid residues in the active site of LAM appear essential for its function. The 310-helix spanning Cys-129 to Cys-132 in the CxxxCxxC motif is conserved in all structures of radical–SAM enzymes to date. The carbonyl oxygen of Gly-171 is within hydrogen-bonding distance of the amino nitrogen of the selenomethionine moiety of SeSAM. The role of the GG motif in the SAM binding site is not clear. Based on proximity, it could be involved in ligation of SAM to the [4Fe–4S] cluster, but it might also be important for assembly of the cluster, being flexible enough to allow insertion and subsequent trapping of the [4Fe–4S] cluster during biosynthesis. The nitrogen of this same residue is within hydrogen-bonding distance of the backbone carbonyl of Met-124, the residue preceding the cysteine motif binding the [4Fe–4S] cluster. This latter interaction is found in all four radical–SAM enzyme structures to date. The GG loop conformation is significant in that it breaks the β-sheet hydrogen bonding pattern to the next strand by elimination of the hydrogen bond between Gly-171 and Ile-121. This results in a buckling conformation of GG in toward the β channel.
The presence of SeSAM as the ligand of the unique iron in the [4Fe–4S] cluster was a surprise. This ligand was expected to be S-adenosyl-l-homocysteine (SAH) from the crystallization mixture. However, SAH was ruled out as follows: Electron density appeared in a position consistent with a SAM methyl group and not with SAH. Moreover, the anomalous difference Fourier map included a peak at the position corresponding to the sulfur of SAM and at a magnitude incommensurate with sulfur and on the same order of magnitude as other selenium nuclei in the structure. SeSAM built into the structure fitted the electron density perfectly, with distances and angles at Se consistent with small-molecule crystal structures for thioethers and selenoethers. Apparently SeSAM bound to LAM in the bacterial culture survived the purification procedure and remained tightly bound to the [4Fe–4S] cluster.
Notwithstanding analogies in the roles of the 5′-deoxyadenosyl radical in the actions of the SAM-dependent LAM and adenosylcobalamin-dependent enzymes, the structure of LAM differs in a mechanistically significant way from the coenzyme B12-dependent enzymes. As shown in Fig. 4, C5′ of SAM lies within 3.8 Å of C3 in the lysyl side chain, and this distance should become shorter upon reductive cleavage of the C5′–S bond. In contrast, the substrate binding sites of adenosylcobalamin-dependent enzymes are much further from the Co–C5′ bond of coenzyme B12, typically 6–9 Å (38–41). These distances inspire the advancement of propositions that the 5-deoxyadenosyl radical undergoes conformational changes allowing approach of adenosyl-C5′ to the substrate in the actions of adenosylcobalamin-dependent enzymes. No such conformational change in the 5-deoxyadenosyl radical is required to account for hydrogen transfer in the action of LAM.
Supplementary Material
Acknowledgments
We thank Drs. Michael Becker (National Synchrotron Light Source) and Lisa Keefe and Carin Stamper (Industrial Macromolecular Crystallography Association Collaborative Access Team) for beamline time, Dr. Keith Brister and the staff at BioCARS for help and support, Dr. Gary Navrotski at BioCARS for assistance with fluorescence detection, and Gregory A. Petsko and Mark A. Wilson for helpful discussions. Use of BioCARS Sector 14 was supported by National Center for Research Resources (National Institutes of Health) Grant RR07707. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources of the National Institutes of Health. Use of the Industrial Macromolecular Crystallography Association Collaborative Access Team beamline 17-ID was supported through a contract with the Illinois Institute of Technology. Use of the Advanced Photon Source was supported by Contract W-31-109-Eng-38 from the U.S. Department of Energy Office of Basic Energy Sciences. This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK28607 (to P.A.F.) and National Science Foundation Grant DBI 9874458 (to D.R.).
Author contributions: P.A.F. and D.R. designed research; B.W.L. and F.J.R. performed research; F.J.R. contributed new reagents/analytic tools; B.W.L. and D.R. analyzed data; and B.W.L., P.A.F., and D.R. wrote the paper.
Abbreviations: LAM, lysine-2,3-aminomutase; NCS, noncrystallographic symmetry; PLP, pyridoxal-5′-phosphate; SAM, S-adenosyl-l-methionine; SeSAM, Se-adenosyl-l-selenomethionine.
Data deposition: Coordinates and diffraction data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2A5H).
References
- 1.Chirpich, T. P., Zappia, V., Costilow, R .N. & Barker, H. A. (1970) J. Biol. Chem. 245, 1778–1789. [PubMed] [Google Scholar]
- 2.Aberhart, D. J., Gould, S. J., Lin, H. J., Thiruvengadam, T. K. & Weiller, B. H. (1983) J. Am. Chem. Soc. 105, 5461–5470. [Google Scholar]
- 3.Petrovich, R. M., Ruzicka, F. J., Reed, G.H. & Frey, P. A. (1992) Biochemistry 31, 10774–10781. [DOI] [PubMed] [Google Scholar]
- 4.Frey, P. A. (2001) Annu. Rev. Biochem. 70, 121–148. [DOI] [PubMed] [Google Scholar]
- 5.Moss, M. & Frey, P. A. (1987) J. Biol. Chem. 262, 14859–14862. [PubMed] [Google Scholar]
- 6.Cosper, N. J., Booker, S. J., Frey, P. A. & Scott, R. A. (2000) Biochemistry 39, 15668–15673. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., Walsby, C. J., Hoffman, B. M. & Frey, P. A. (2003) J. Am. Chem. Soc. 125, 11788–11789. [DOI] [PubMed] [Google Scholar]
- 8.Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. (2001) Nucl. Acids Res. 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsby, C. J., Hong, W., Broderick, W. E., Cheek, J., Ortillo, D., Broderick, J. B. & Hoffman, B. M. (2002) J. Am. Chem. Soc. 124, 3143–3151. [DOI] [PubMed] [Google Scholar]
- 10.Ruzicka, F. J., Lieder, K. & Frey, P. A. (2000) J. Bacteriol. 182, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steipe, B., DeMange, P., Eckerskorn, C., Kellermann, J., Budisa, N. & Huber, R. (1995) Eur. J. Biochem. 230, 788–796. [DOI] [PubMed] [Google Scholar]
- 12.Diederichs, K. & Karplus, P.A. (1997) Nat. Struct. Biol. 4, 269–275. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger, T. C. & Berendzen, J. (2000) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terwilliger, T. C. (2000) Acta Crystallogr. D 55, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003) Acta Crystallogr. D 59, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 17.Brodersen, D. E., de La Fortelle, E., Vonrhein, C., Bricogne, G., Nyborg, J. & Kjeldgaard, M. (2000) Acta Crystallogr. D 56, 431–441. [DOI] [PubMed] [Google Scholar]
- 18.McRee, D. (1992) J. Mol. Graphics. 10, 44–46. [Google Scholar]
- 19.Brunger, A. T. Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P. Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 20.Kleywegt, G. J., Zou, J. Y., Kjelgaard, M. & Jones, T. A. (2001) in International Tables for Crystallography, eds. Rossmann, M. G. & Arnold, E. (Kluwer, Dordecht, The Netherlands), Vol. F, pp. 353–356, 366–367. [Google Scholar]
- 21.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 22.Petrovich, R. M., Ruzicka, F. J., Reed, G. H. & Frey, P. A. (1991) J. Biol. Chem. 266, 7656–7660. [PubMed] [Google Scholar]
- 23.Murshudov, G. N., Vagin, A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 24.Macarthur, M. W., Moss, D. S., Thornton, J. M. & Laskowski, R. A. (1993) J. Appl. Cryst. 26, 283–291. [Google Scholar]
- 25.Merritt, E.A. (1999) Acta Crystallogr. D 55, 1109–1117. [DOI] [PubMed] [Google Scholar]
- 26.Strop, P., Takahara, P. M., Chiu, H.-J., Angove, H. C., Burgess, B. K. & Rees, D. C. (2000) Biochemistry 40, 651–656. [DOI] [PubMed] [Google Scholar]
- 27.Cho, H., Ramaswamy, S. & Plapp, B. V. (1997) Biochemistry 36, 382–389. [DOI] [PubMed] [Google Scholar]
- 28.Fenn, T. D., Ringe, D. & Petsko, G. A. (2003) J. Appl. Cryst. 36, 944–947. [Google Scholar]
- 29.Song, B. & Frey, P. A. (1991) J. Biol. Chem. 266, 7651–7655. [PubMed] [Google Scholar]
- 30.Ballinger, M. D., Frey, P. A. & Reed, G. H. (1992) Biochemistry 31, 10782–10789. [DOI] [PubMed] [Google Scholar]
- 31.Wu, W., Lieder, K. W., Reed, G. H. & Frey, P. A. (1995) Biochemistry 34, 10532–10537. [DOI] [PubMed] [Google Scholar]
- 32.Wu, W., Booker, S., Lieder, K. W., Bandarian, V., Reed, G. H. & Frey, P. A. (2000) Biochemistry 39, 9561–9570. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J., Bandarian, V., Reed, G. H. & Frey, P. A. (2001) Arch. Biochem. Biophys. 387, 281–288. [DOI] [PubMed] [Google Scholar]
- 34.Magnusson, O. T., Reed, G. H. & Frey, P. A. (2001) Biochemistry 40, 7773–7782. [DOI] [PubMed] [Google Scholar]
- 35.Hanzelmann, P. & Schindelin, H. (2004) Proc. Natl. Acad. Sci. USA 101, 12870–12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. (2004) Science 303, 76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layer, G., Moser, J., Heinz, D. W., Jahn, D. & Schubert, W. D. (2003) EMBO J. 22, 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mancia, F., Smith, G. A. & Evans, P. R. (1999) Biochemistry 38, 7999–8005. [DOI] [PubMed] [Google Scholar]
- 39.Mansoorabadi, S., Padmakumar, N., Fazliddinova, N., Vlasie, M., Banerjee, R. & Reed, G. H. (2005) Biochemistry 44, 3153–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boas, J. F., Hicks, P. R. & Pilbrow, J. R. (1978) J. Chem. Soc. Faraday Trans. 2 74, 417–431. [Google Scholar]
- 41.Toraya, T. (2003) Chem. Rev. 103, 2095–2127. [DOI] [PubMed] [Google Scholar]
- 42.Brunger, A. T. (1992) Nature 355, 472–474. [DOI] [PubMed] [Google Scholar]
- 43.Allen, F. H. (2002) Acta Cryst. B 58, 380–388. [DOI] [PubMed] [Google Scholar]
- 44.Kraulis, P. J. (1999) J. Appl. Cryst. 24, 946–950. [Google Scholar]
- 45.Bennett, M. J., Choe, S. & Eisenberg, D. (1994) Proc. Natl. Acad. Sci. USA 91, 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.