Abstract
Myosin V is an efficient processive molecular motor. Recent experiments have shown how the structure and kinetics of myosin V are specialized to produce a highly processive motor capable of taking multiple 36-nm steps on an actin filament track. Here, we examine how two identical heads coordinate their activity to produce efficient hand-over-hand stepping. We have used a modified laser-trap microscope to apply a ≈2-pN forward or backward force on a single-headed myosin V molecule, hypothesized to simulate forces experienced by the rear or lead head, respectively. We found that pulling forward produces only a small change in the kinetics, whereas pulling backward induces a large reduction in the cycling of the head. These results support a model in which the coordination of myosin V stepping is mediated by strain-generated inhibition of the lead head.
Keywords: load dependence, molecular motor, processivity, optical trap
Myosin V is a two-headed, actin-based motor (1) used in intracellular trafficking (2). Previous studies demonstrated that myosin V is processive (3–5). That is, it takes many 36-nm steps without dissociating from an actin track. Myosin V normally has two identical heavy chains, consisting of a catalytic domain capable of binding actin and hydrolyzing ATP, six light chain-binding domains per head forming a rigid lever arm (5–7), a coiled-coil dimerization domain, and a cargo interaction domain. Experiments where only one myosin head is labeled show that the two heads alternate position and the molecule walks hand over hand (8). Each head is a fully functional motor, as shown by the observation that single-headed constructs are capable of producing actin movement although these constructs are not processive (5, 6).
The two heads of a processively walking myosin V are asymmetric, with the lead head in predominantly a prestroke configuration and the rear head in predominantly a poststroke configuration. The lead and rear heads straddle the actin filament in a “telemark” stance (9, 10) (Fig. 1). For a forward step to take place the rear head has to release from the actin filament while the lead head remains bound. Once the rear head leaves actin, the bound prestroke-configured lead head strokes its lever arm, pushing the rear head forward toward where it searches by Brownian motion for a new actin-binding site 36 nm from the bound head. Once the new leading head binds to actin, the roles of the heads switch and the molecule must wait for the new rear head to release from actin to take another forward step. If myosin V operates efficiently, ATP binding should occur only in the rear head. ATP-induced dissociation in the lead head would not be productive for forward-stepping.
Fig. 1.
The myosin V catalytic cycle. (A) A prestroke myosin V head (red) is shown binding to an actin filament. Once the trailing head unbinds from actin, it is stroked forward by the bound head that transitions from its prestroke state (red) to a poststroke state (green). Once the head has stroked, it can complete the catalytic cycle by first releasing ADP, and then binding a new ATP molecule. ATP binding causes the head to release from the actin to be advanced to the next actin-binding site. While unbound, the head also hydrolyzes its ATP, cocking the head into a prestroke geometry. A, actin; M, myosin. (B) The prestroke-to-poststroke transition is part of an equilibrium that could be influenced by external force (F) on the molecule.
Previous studies have shown that the rate-limiting step in the myosin V catalytic cycle is ADP release, in both single-headed unloaded molecules (11) and two-headed walking molecules (12). For these two observations to be consistent, the catalytic cycle should be arrested in one of the heads of a two-headed walking molecule. This state would lead to efficient unidirectional movement if the arrested head were always the bound lead head. An important question therefore is how do two heads separated by 36 nm communicate their biochemical state to each other to coordinate the hand-over-hand walking.
Our working hypothesis is that the lead head is strained backward by its connection to the rear head and likewise the rear head is strained forward by the lead head. To observe biochemical changes in a molecule with different geometric constraints we have used a laser-trap microscope to specifically put a large forward force or large backward force on a single myosin V head to mimic the intramolecular strain felt by the lead and rear heads of the two-headed motor (Fig. 1B).
Methods
All optical trap assays were performed in flow cells prepared as described (6, 13). Assay buffer included 25 mM imidazole HCl (pH 7.4), 25 mM KCl, 4.5 μM calmodulin, 1 mM EGTA, 10 mM DTT, 4 mM MgCl2, an oxygen scavenging system to retard photobleaching (25 μg·ml–1 glucose oxidase, 45 μg·ml–1 catalase, 30.8 mM 2-mercaptoethanol, and 0.5% glucose), and an ATP regeneration system (0.1 mg·ml–1 creatine phosphokinase and 1 mM creatine phosphate). The ATP regeneration system was omitted in assays where 1,000 μM ADP was added.
Motors were adsorbed to a nitrocellulose coverslip via a specific surface attachment by using monoclonal anti-GFP antibody (3E6, Q-Biogene, Irvine, CA). After treatment with antibody (0.05 mg/ml), the surface was blocked by additional treatment with 1 mg/ml BSA. Dilutions of motor were then flown into the cell.
The dwell distribution fits to a normalized distribution (k1 k2)/(k1 – k2) × {e–k2t – e–k1t} for two rates (k1 and k2) or k e–kt for a single rate (k). Errors were calculated as the SD of the negative of the inverse of the second derivative of the maximum-likelihood calculation, the SD of fit rates derived from 200 bootstrap trials, or the SD of fit rates derived from 100 Monte Carlo simulations. Using stage displacement leads to an uncertainty in the exact start of the binding of myosin to the actin of ≤10 ms. Events <10 ms were not counted to avoid false positive identification of binding events. For rates <20 s–1, the systematic error in fits caused by this uncertainty is within the range of random error reported. Rates between 20 and 50 s–1 are systematically overestimated by 10–20%. Rates >50 s–1 are subject to large errors in data of this resolution and should be interpreted as “fast.”
Results
Application of Force on Single Molecules. Addition of a feedback-controlled piezo-electric stage to a dual-beam laser-trap microscope creates a system that allows a large forward or backward force on a single myosin bound to a trapped actin dumbbell (Fig. 2A). Experiments were performed by using a dual-beam optical-trap microscope (6). A P-733.2CL NanoPositioning Stage (Physik Instruments, Karlsruhe, Germany) was added to the stage. The piezo stage was positioned and oscillated by using analog inputs under the control of labview software (National Instruments, Austin, TX).
Fig. 2.
Experimental setup and operation. (A) Experimental setup. An actin dumbbell held in a dual-beam laser trap is brought into contact with a myosin molecule adsorbed onto a bead on the surface of the flow cell. The flow cell is mounted on a piezoelectric stage that is oscillated along the axis of the actin dumbbell. Bead position is monitored by quadrant photodiode detection. Binding of the myosin to the actin dumbbell causes a displacement of the beads.A computer monitors the bead position and stops the stage oscillation when binding occurs. (B) Stage and bead position after displacement. The stage (green) rapidly moves 190 nm, with a 500-Hz damped oscillation caused by the mechanical resonance in the piezoelectric stage. Initially, the dumbbell (blue) is also displaced because of hydrodynamic interaction with the surface but it is pulled back to its baseline position in the trap within 5 ms. The blue trace is an average of 100 events to remove the effects of random thermal noise. (C) Example data showing a series of binding events for MV-6IQ-S1 in 10 μM ATP. The bead positions for both beads in the dumbbell are shown. (D) The red traces show example traces of binding events after an upward or downward displacement of the stage.
Biotinylated actin filaments were stretched between two trapped 1-μm diameter polystyrene beads coated with streptavidin to form a dumbbell. Actin dumbbells were brought into contact with myosin adsorbed to 1.5-μm glass beads that were tightly adhered to the surface of the flow cell.
Stepwise input into the piezo stage produces a rapid displacement of the stage platform. The stage travels 190 nm within 2 ms (Fig. 2B, green trace). The 80-nm peak-to-peak, 500-Hz oscillation in the stage after the initial displacement rapidly decays to <10 nm within 30 ms. A trapped dumbbell is also displaced by the stage movement caused by hydrodynamic interactions with the flow cell surface (Fig. 2B, blue trace). A dumbbell when not associated with a surface-bound motor returns to its baseline position within an average of 4 ms. After a 6-ms delay, the feedback computer checks the signal from the quadrant photo diodes, which are tracking the positions of the dumbbell beads (0.5-ms observation at 20,000-Hz sample rate). If the dumbbell has returned to within 50 nm of its baseline position, the stage is stepped in the opposite direction. If the quadrant photodiode signal is above threshold, indicating that the dumbbell is bound to a motor on the surface, the computer suspends oscillation and repeats the observation until the bead signal returns to baseline. Observation and calculation takes 3–4 ms. In the absence of binding events, the total time between steps is ≈10 ms, 6 ms to allow the system to return to equilibrium and 4 ms to determine that the motor has not bound, for a net oscillation frequency of 50 Hz, or 100 step events per s.
Force Induces Asymmetric Changes in Single Myosin Heads. Chicken myosin V cDNA was expressed as a single-headed S1 construct in insect cells, along with calmodulin and chicken myosin V essential light chain (6, 14). The myosin V heavy chain with all six light chain-binding motifs (IQ repeats) intact (MV-6IQ-S1) was truncated at Lys-910. Shortened lever arm versions with only the first IQ repeat (MV-1IQ-S1) or the first four IQ repeats (MV-4IQ-S1) were created by truncation at Arg-791 and Arg-863, respectively.
Myosin binding to the trapped actin filament dumbbell prevents the trapped beads from moving freely with respect to the surface. After displacement, the actin connection to surface-bound myosin holds the dumbbell away from the center of the traps (Fig. 2D, red traces). Within 6 ms, the bound dumbbells are distinguished from unbound events. Binding events range from 50 to 200 nm from neutral position. The net stiffness of the dumbbell is 0.015–0.020 pN·nm–1, so a bound myosin experiences a 2 ± 1-pN force along the actin filament axis. The symmetry of the oscillation yields an even distribution of binding events with a forward force (in the direction of normal myosin travel) and backward force on the myosin head.
When the myosin comes unbound from actin, the dumbbell abruptly returns to a neutral position (Fig. 2C). A dwell time is defined as from the displacement of the stage to the unbinding of the myosin. Dwell time distributions were fit to a simplified kinetic scheme of either a single irreversible rate or two sequential irreversible rates by using a maximum-likelihood estimate (15).
The orientation of the actin filament was determined at the beginning of each experiment by collecting ≈1 min of data with the stage oscillation turned off. During this period, between 3 and 50 binding events were observed. The MV-6IQ-S1 construct yielded binding events with displacements consistent with previously observed 20-nm steps (6).
Once oscillation began, a single myosin could yield binding events for up to 1 h, producing data sets with up to 5,000 discrete binding events. The median number of events was 263 per dumbbell.
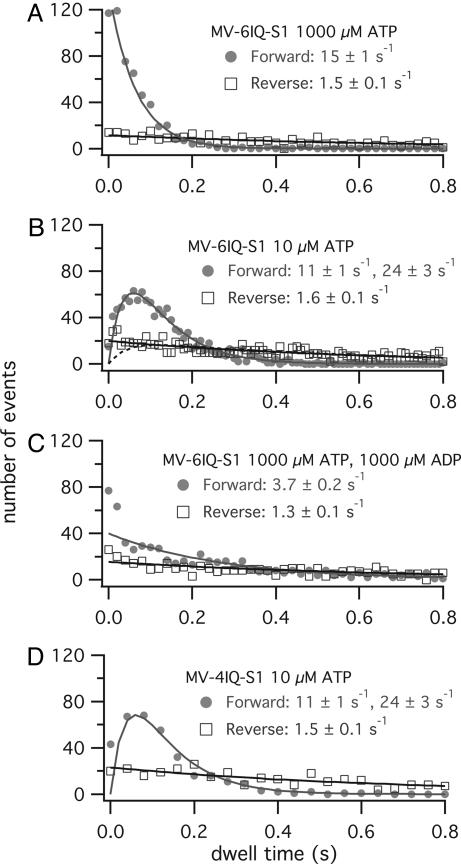
A single-headed construct with a full-length lever arm (MV-6IQ-S1) showed significant differences in dwell time distribution when subjected to forward and backward pulling (Fig. 3). At 1,000 μM ATP, 930 binding events were scored. For 532 events, the motor experienced a forward force. A maximum-likelihood fit of the dwell time distribution yielded a single rate of 15 ± 1 s–1. This rate is close to previously observed values for processive stepping of myosin V at saturating ATP (12); 15 s–1 corresponds to the rate-limiting rate of ADP release for an unloaded molecule (12–16 s–1, ref. 11). The 398 events where the motor experienced a backward force had a different distribution of dwell times, best fit as a single rate of 1.5 ± 0.1 s–1 (Fig. 3A). Thus, a backward force, hypothesized to mimic a strained lead head of a processively walking dimer, greatly inhibits the rate of release of that head.
Fig. 3.
Dwell-time histograms for MV-6IQ-S1. Data are shown for both pulling forward (•, mimicking a trailing head) or pulling backward (□, mimicking a leading head). Data are fit as two sequential rates fk1,k2(t) = {k1 k2/(k1 – k2)} × (e–k2t – e–k1t) or fit as a single exponential rate f(t) = ke–kt. (A) At 1,000 μM ATP, the forward pulling is fit to a single rate of 15 s–1 (n = 532). The backward-pulling distribution fits to a single rate of 1.5 s–1 (n = 398). (B) At 10 μM ATP, the forward pulling is fit to two rates (n = 1,071), and the backward pulling is fit to a single rate (n = 1,222). The dashed line is a fit to two rates, 1.7 and 24 s–1. The deviation shows that any other steps other than the 1.6-s–1 observed off rate must be ≫24 s–1. (C) At 1,000 μM ATP, the forward pulling is fit to a single rate of 3.7 s–1 (n = 662) and the backward pulling is fit to 1.3 s–1 (n = 553). (D) Dwell-time histogram for MV-4IQ-S1 at 10 μM ATP.
The longer dwell times (lower rate constant) for a backward pulled head remained the same even when the ATP concentration was reduced to 10 μM. The rate determined for the dwell time distribution for 1,222 backward-pulling events was 1.6 ± 0.1 s–1. At 10 μM ATP, the dwell time distribution for 1,071 forward-pulling events was best fit as two rates of 11 ± 1 and 24 ± 3 s–1 (Fig. 3B).
Addition of 1,000 μM ADP to 1,000 μM ATP slowed the 662 forward-pulling events to one detectable rate of 3.7 ± 0.2 s–1, as expected because addition of the high concentration of ADP competes for ATP binding after the ADP release. ADP at 1,000 μM had a minimal effect on the dwell time for backward pulling. The dwell time distribution for 553 events fit to a single 1.3 ± 0.1 s–1 rate (Fig. 3C). Thus, this rate is independent of both ATP and ADP concentrations.
Even when the strongly bound state time was made extremely slow by reducing ATP to 10 μM and keeping ADP at 1,000 μM, the 44 backward-pulling dwell times fit as a single 1.4 ± 0.2 s–1 rate, whereas the distribution of forward-pulling events was very long. These very long dwells made it difficult to collect a sufficient amount of events to distinguish two rates from a single rate. The 17 dwells collected can be fit to a net single rate of 0.5 ± 0.2 s–1.
A single-headed construct containing the first four IQ repeats (MV-4IQ-S1) showed similar behavior to the full-length six-IQ construct. At 10 μM ATP, the dwell time distribution fits to two rates (11 ± 1 and 24 ± 3 s–1) for forward pulling (n = 328) and a single rate (1.5 ± 0.1 s–1) for backward pulling (n = 403) (Fig. 3D).
When the lever arm was shortened to a single IQ repeat (MV-1IQ-S1), the difference between dwell times during forward and backward pulling was greatly reduced (Fig. 4). Over the range of 1 to 2,000 μM ATP the dwell time distribution was best fit to two rates. One rate was fairly constant with forward and backward pulling and dependent on ATP concentration. The other rate was constant over the ATP range and was ≈2-fold slower when the molecule was pulled backward.
Fig. 4.
Force and ATP dependence for MV-1IQ-S1. Rates shown are derived from a maximum-likelihood fit of the dwells to two sequential rates. Rates from different ATP concentrations are plotted on a log-log scale. The fits are estimates based on one ATP-independent rate (k1) and one saturating ATP-dependent rate (k2).
The role of intramolecular strain on myosin V kinetics was further explored by using a two-headed myosin construct and examining the dwell times in processive staircases as a function of step size. We hypothesized that the intramolecular strain on the dimer should relate to the size of the last step taken. It might be expected, for example, that a molecule that just took a larger step may experience more intramolecular strain. Alternatively, molecules that take steps in the range of ≈18 nm will experience the most torque, which could result in a larger change in conformational-dependent changes in rate constants. A two-headed myosin V construct with four IQ repeats (MV-4IQ-HMM) was used, as in Purcell et al. (6). During processive stepping, the MV-4IQ-HMM construct had a wide distribution of step sizes. Stepping against 1-pN force in a force-feedback-controlled laser trap, the SD was 10 nm of a 24-nm step. Looking at all of the steps pooled together, the dwell time distribution fit as two rates, 23 and 10 s–1 at 20 μM ATP and 19 and 5 s–1 at 10 μM ATP, suggesting an ADP release step of ≈20 s–1 and ATP-induced release at ≈0.5 μM–1·s–1. To examine the dwell times in processive staircases as a function of step size, data were collected in 4-nm bins for the 10-μM ATP data, and the distributions for the dwell times before and after the steps were fit to two rates (Fig. 5). For the poststep dwells, the apparent ADP release rate showed a strong acceleration after 18-nm steps, consistent with torque on the molecule playing a significant role.
Fig. 5.
Off-axis strain accelerates ADP release. Cartoon representation of MV-4IQ-HMM attached to a segment of actin. The lead head is shown bound at the second, sixth, seventh, and 11th actin subunit, corresponding to a span of 5.5, 16.5, 19.3, and 30.3 nm between the two heads. The intermediate distances require the myosin to twist around the actin helix, as shown by the arrows indicating the orientation of each myosin head. Processive steps from MV-4IQ-HMM at 10 μM ATP were collected into 4-nm bins, and the times for the preceding and following dwells were fit to two rates. Closed symbols show the two rates that best-fit prestep dwells, and open symbols show the rates derived from poststep dwells.
Discussion
The data in this article show that the direction of an applied force creates a clear asymmetry in the bound state time of a single myosin V head bound to actin. The question is how to map the changes observed onto the established kinetic pathway for myosin. At limiting ATP, the myosin V head has two rate-limiting steps, ADP release (12–16 s–1) and ATP binding (0.9–1.6 μM–1·s–1) (11, 16, 17). Forward pulling on the MV-6IQ-S1 construct in 10 μM ATP produces a dwell time distribution with two apparent rates, 11 and 24 s–1 (Fig. 3B). Raising the ATP to 1,000 μM produces a one-rate fit of 15 s–1. At this concentration, the rate of the ATP-induced strong-to-weak transition is probably >100 s–1, faster than the detection limit of the assay. The rates at 11 and 15 s–1 are within error of the established ADP release rate. Data from MV-1IQ-S1 are consistent with a second-order rate constant for ATP-induced actin unbinding of ≈2 μM–1·s–1. In addition, the rates for pulling forward are similar regardless of the length of the lever arm.
Pulling backward on MV-6IQ-S1 reduces the apparent dwell time to a single measurable rate. Under this condition, the dwell time distribution is not affected by the concentration of ADP or ATP. This finding suggests that the unbinding occurs before the ADP release step (Fig. 6). We hypothesize that a backward load is inhibiting the weak-binding to strong-binding transition of myosin V to actin. This load dependence suggests that this transition involves a mechanical displacement. This transition is hypothesized to occur while the actin-bound motor is in its ADP and actin-bound state, changing from its weakly actin-bound (A·M·ADP) to its strongly actin-bound (A·M*·ADP) state. A backward load leads to an increase in the motor's population of weakly bound state, resulting in the motor's dissociation from the actin filament at ≈1.5 s–1. Recent studies on unloaded single-head myosin V molecules provide kinetic evidence for at least two actomyosin ADP states (18, 19). The observed rate of unbinding under backward load is several orders of magnitude faster than the slow off rate of strongly bound ADP myosin (0.04 s–1) (11, 19) or nucleotide-free rigor myosin V (0.007 s–1) (19). Hannemann et al. (19) identified a second actomyosin-ADP state that dissociates from actin on time scales consistent with the dissociation observed here under backward load. This state represents only a fraction of the unloaded molecules (≈10%); however, our results suggest that backward force influences this equilibrium to populate the rapidly dissociating state.
Fig. 6.
A model for how force coordinates the kinetics of the myosin V head. Energy profile for a head bound to actin subjected to an external force. Under unloaded conditions (black curve) the transition between an intermediately bound state (AMD) to a strongly bound state (AM*D) is rapid and strongly biased to formation of the strongly bound state. After the myosin reaches this state, there is a rate-limiting transition where the motor releases ADP. If a motor experiences a forward force (green curve, mimicking a trailing head), the free energy is reduced as a function of the position along the reaction coordinate (28). The reaction remains limited by the final ADP release step, which is sped up very little because of a small change in the relative free energies between AM*D and the ADP release transition state. If the motor is working against a backward force (red curve, mimicking a leading head), the free energy increases as a function of reaction coordinate. The forward transition to AM*D becomes significantly slower and the reverse transition is favored compared with the release of ADP from the AM*D state. If the initial state is populated, the weakly bound myosin head might release from the actin because of an off-pathway dissociation of the AMD to A + MD (blue arrow). In a two-headed walking motor, the lead head experiences an increasing backward force as it tries to swing its lever arm because of the attachment to the stationary trailing head (black dashed line), locking the lead head in the initial AMD state.
Further evidence for a weakly bound lead head comes from single-molecule studies on processive two-headed myosin V (20). At a superstall backward force of 5 pN, they observed backward steps at a rate of 7 s–1. They propose that these steps involve unbinding of the lead head, which is subsequently pulled back to rebind at a new trail position. At higher forces of 10 pN, the backward stepping rate increases to 30 s–1 (M. Rief, personal communication). Extrapolating back to the forces applied directly onto a lead head in this study, according to the relation ki(F) = k0i exp(F di/kBT) (21) predicts an unbinding rate of ≈2 s–1 (k0i = 1.2 s–1, di = 1.3 nm) at 2 pN.
The effect on the head is far more extensive if a six- or four-IQ lever arm is present. The greatly shortened one-IQ lever arm shows far less inhibition with backward load, which could simply be because of the shorter arm exerting less net force on the head. Previous studies have shown that the lever arm serves to convert a small conformational change in the head into a large movement of the attached cargo (5–7), but the lever arm may also act as a force amplifier, transmitting force between the two heads. A 2-pN force applied at the end of the shorter one-IQ lever arm would result in a reduced influence on the catalytic head, allowing the equilibrium depicted in Fig. 1B to populate the poststroke/tightly bound state more than in the six-IQ head. The result is a net reduction in the rate of release of ADP caused by a lower proportion of A·M*·ADP, but with this state populated sufficiently to allow some release of ADP, which locks the molecule in a strongly bound state that can be unbound from actin only by binding a new ATP.
Strain-dependent ADP release kinetics have been observed in smooth muscle myosin II (22). A substep associated with ADP release is sped up in response to a forward force and slowed down in response to a backward force on the myosin. This substep is thought to play a role in maintaining force economically in smooth muscle by trapping the molecule in a strongly bound state in response to steady load.
An ADP release-associated substep has also been observed in myosin V (23). In contrast to our conclusions, previously published models for processive stepping of myosin V proposed that strain coordinates the two heads by acceleration of the ADP release rate from the rear head (23, 24). In processive runs, Veigel et al. (23) observed a stepping rate of ≈14 s–1 at 100 μM ATP for a myosin V HMM construct, consistent with other observations. In single-headed interactions or single-step interactions with a two-headed construct, they observed an apparent deceleration of the ADP release rate to ≈5 s–1. Given the average step size of 20–25 nm and the approximate stiffness of the dumbbell of 0.015–0.02 pN·nm–1, the construct would experience an average 0.3- to 0.5-pN backward force. This force is less than the 0.75- to 3-pN force that greatly inhibits ADP release in the four- and six-IQ S1 constructs used in this study, although it may be sufficient to slow down the effective ADP release rate, as observed with the one-IQ S1 construct. Our results with myosin V do not show acceleration in ADP release rate with forward pulling, nor a specific reduction in ADP release rate in response to backward pulling, which is consistent with the critical gating step occurring in the lead head before ADP release.
The one situation that does appear to cause an increase in ADP release rate is off-axis strain, as seen in our experiments with processively walking four-IQ HMM molecules, where we observe an increased rate of ADP release from ≈15 to ≈30 s–1. These results support the conclusions from a recent study on two-headed myosin V by Rosenfeld and Sweeney (25), which reveals that the kinetics of the first actin encounter of two-headed myosin V are different from for the steady-state situation, with the first encounter showing an increased ADP release rate from ≈15 to ≈30 s–1. Such a difference is reasonable, because both heads are in an ADP.Pi state before the initial encounter, whereas only the lead head is in that state during the subsequent steady-state steps. The Rosenfeld and Sweeney data can be interpreted as follows. During the initiation of processive movement, the two heads simultaneously bind to actin in an ADP.Pi state, simultaneously release Pi (200 s–1), and simultaneously undergo an accelerated weak-to-strong transition. At that point, ADP release from the rear head is accelerated (30 s–1), whereas ADP release from the lead head is prevented (gated) until the rear head detaches. In the absence of excess ATP, the rear head is prevented from detaching, and dissociation of ADP from the lead head occurs at a rate of 0.3 s–1. However, for the steps after the first step, no acceleration of ADP release is observed from the rear head, although ADP release is prevented in the lead head until the rear head detaches. This difference in steady-state and initial ADP release rates from the rear head implies that the geometry of the initial encounter is different from subsequent encounters.
It is reasonable to assume that the geometry of the initial encounter of two-headed myosin V with actin is different from subsequent encounters during steady-state walking, because in the former case both heads begin in prepowerstroke states. The initial binding event for two-headed myosin V could, for example, generate off-axis strain caused by the two heads binding to actin with the two heads spaced closer than the 36-nm actin pseudorepeat, similar to what we have observed with processively walking four-IQ HMM.
One possible explanation for these data is that the ADP-releasing isomerization involves a lever arm movement with a significant orthogonal component (relative to the actin filament) that allows off-axis strain to accelerate the isomerization, whereas on-axis strain cannot. Another possibility is that the lever arm compliance is asymmetric and is greater if pulled forward on-axis than if pulled off-axis. However the data presented here demonstrate that even a forward-directed strain of 2 pN on a single, actin-attached, ADP-containing myosin V head does not accelerate ADP release from that head. Thus the lead head cannot accelerate ADP release from a rear head if the force is applied parallel to the actin filament axis, and gating of the myosin V heads during processive movement does not involve acceleration of ADP release from a rear head. This model favors the need for an orthogonal component to the strain to accelerate ADP release.
In summary, our data support a model in which strain coordinates two heads by preventing the lead head from releasing ADP. We observe that backward force induces a state in a one-headed myosin V that is not affected by nucleotide concentration and is tightly associated with actin but bound less strongly than a rigor head. The lever arm of the lead head in a two-headed processively walking molecule would be strained backward by its attachment to the rear head and it is our assumption that the external force applied on a single-headed molecule induces the same biochemical state. In this state, the lead head is unable to release ADP because it is not free to form the full strongly bound attachment state to actin. This finding is consistent with the model proposed for myosin VI (26) and myosin V (27) processivity in which the leading head is weakly bound to actin in an ADP state.
Author contributions: T.J.P. and J.A.S. designed research; T.J.P. performed research; T.J.P. and H.L.S. contributed new reagents/analytic tools; T.J.P., H.L.S., and J.A.S. analyzed data; and T.J.P., H.L.S., and J.A.S. wrote the paper.
See Commentary on page 13719.
References
- 1.Cheney, R. E., O'Shea, M. K., Heuser, J. E., Coelho, M. V., Wolenski, J. S., Espreafico, E. M., Forscher, P., Larson, R. E. & Mooseker, M. S. (1993) Cell 75, 13–23. [DOI] [PubMed] [Google Scholar]
- 2.Vale, R. D. (2003) Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- 3.Mehta, A. D., Rock, R. S., Rief, M., Spudich, J. A., Mooseker, M. S. & Cheney, R. E. (1999) Nature 400, 590–593. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto, T., Amitani, I., Yokota, E. & Ando, T. (2000) Biochem. Biophys. Res. Commun. 272, 586–590. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto, T., Wang, F., Schmitz, S., Xu, Y., Xu, Q., Molloy, J. E., Veigel, C. & Sellers, J. R. (2003) J. Biol. Chem. 278, 29201–29207. [DOI] [PubMed] [Google Scholar]
- 6.Purcell, T. J., Morris, C., Spudich, J. A. & Sweeney, H. L. (2002) Proc. Natl. Acad. Sci. USA 99, 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schott, D. H., Collins, R. N. & Bretscher, A. (2002) J. Cell Biol. 156, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildiz, A., Forkey, J. N., McKinney, S. A., Ha, T., Goldman, Y. E. & Selvin, P. R. (2003) Science 300, 2061–2065. [DOI] [PubMed] [Google Scholar]
- 9.Walker, M. L., Burgess, S. A., Sellers, J. R., Wang, F., Hammer, J. A., 3rd, Trinick, J. & Knight, P. J. (2000) Nature 405, 804–807. [DOI] [PubMed] [Google Scholar]
- 10.Burgess, S., Walker, M., Wang, F., Sellers, J. R., White, H. D., Knight, P. J. & Trinick, J. (2002) J. Cell Biol. 159, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De La Cruz, E. M., Wells, A. L., Rosenfeld, S. S., Ostap, E. M. & Sweeney, H. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rief, M., Rock, R. S., Mehta, A. D., Mooseker, M. S., Cheney, R. E. & Spudich, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock, R. S., Rice, S. E., Wells, A. L., Purcell, T. J., Spudich, J. A. & Sweeney, H. L. (2001) Proc. Natl. Acad. Sci. USA 98, 13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney, H. L., Rosenfeld, S. S., Brown, F., Faust, L., Smith, J., Xing, J., Stein, L. A. & Sellers, J. R. (1998) J. Biol. Chem. 273, 6262–6270. [DOI] [PubMed] [Google Scholar]
- 15.Altman, D., Sweeney, H. L. & Spudich, J. A. (2004) Cell 116, 737–749. [DOI] [PubMed] [Google Scholar]
- 16.Yengo, C. M., De la Cruz, E. M., Safer, D., Ostap, E. M. & Sweeney, H. L. (2002) Biochemistry 41, 8508–8517. [DOI] [PubMed] [Google Scholar]
- 17.Yengo, C. M. & Sweeney, H. L. (2004) Biochemistry 43, 2605–2612. [DOI] [PubMed] [Google Scholar]
- 18.Rosenfeld, S. S., Houdusse, A. & Lee Sweeney, H. (2005) J. Biol. Chem. 280, 6072–6079. [DOI] [PubMed] [Google Scholar]
- 19.Hannemann, D. E., Cao, W., Olivares, A. O., Robblee, J. P. & De La Cruz, E. M. (2005) Biochemistry 44, 8826–8840. [DOI] [PubMed] [Google Scholar]
- 20.Clemen, A. E., Vilfan, M., Jaud, J., Zhang, J., Barmann, M. & Rief, M. (2005) Biophys. J. 88, 4402–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell, G. I. (1978) Science 200, 618–627. [DOI] [PubMed] [Google Scholar]
- 22.Veigel, C., Molloy, J. E., Schmitz, S. & Kendrick-Jones, J. (2003) Nat. Cell. Biol. 5, 980–986. [DOI] [PubMed] [Google Scholar]
- 23.Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. (2002) Nat. Cell Biol. 4, 59–65. [DOI] [PubMed] [Google Scholar]
- 24.Baker, J. E., Krementsova, E. B., Kennedy, G. G., Armstrong, A., Trybus, K. M. & Warshaw, D. M. (2004) Proc. Natl. Acad. Sci. USA 101, 5542–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenfeld, S. S. & Sweeney, H. L. (2004) J. Biol. Chem. 279, 40100–40111. [DOI] [PubMed] [Google Scholar]
- 26.De La Cruz, E. M., Ostap, E. M. & Sweeney, H. L. (2001) J. Biol. Chem. 276, 32373–32381. [DOI] [PubMed] [Google Scholar]
- 27.Mehta, A. (2001) J. Cell Sci. 114, 1981–1998. [DOI] [PubMed] [Google Scholar]
- 28.Howard, J. (2001) Mechanics of Motor Proteins and the Cytoskeleton (Sunauer, Sunderland, MA).