Abstract
TGF-β can signal by means of Smad transcription factors, which are quintessential tumor suppressors that inhibit cell proliferation, and by means of Smad-independent mechanisms, which have been implicated in tumor progression. Although Smad mutations disable this tumor-suppressive pathway in certain cancers, breast cancer cells frequently evade the cytostatic action of TGF-β while retaining Smad function. Through immunohistochemical analysis of human breast cancer bone metastases and functional imaging of the Smad pathway in a mouse xenograft model, we provide evidence for active Smad signaling in human and mouse bone-metastatic lesions. Genetic depletion experiments further demonstrate that Smad4 contributes to the formation of osteolytic bone metastases and is essential for the induction of IL-11, a gene implicated in bone metastasis in this mouse model system. Activator protein-1 is a key participant in Smad-dependent transcriptional activation of IL-11 and its overexpression in bone-metastatic cells. Our findings provide functional evidence for a switch of the Smad pathway, from tumor-suppressor to prometastatic, in the development of breast cancer bone metastasis.
Keywords: IL-11, Smad4, TGF-β
TGF-β plays a crucial role as a growth-inhibitory cytokine in many tissues (1, 2). The cytostatic effect of TGF-β is mediated by a serine/threonine kinase receptor complex that phosphorylates Smad2 and Smad3, which then translocate into the nucleus and bind Smad4 to generate transcriptional regulatory complexes (3). SMAD4 (also known as Deleted in Pancreatic Carcinoma locus 4 or DPC4) and, to a lesser extent, SMAD2 suffer mutational inactivation in a proportion of pancreatic and colon cancers (1, 2). However, tumor cells that evade this antiproliferative control by other mechanisms may display an altered sensitivity to TGF-β and undergo tumorigenic progression in response to this cytokine (1, 2). Patients whose pancreatic or colon tumors express TGF-β receptors fare less well than those with low or absent TGF-β receptor expression in the tumor (4). In mouse models of breast cancer, TGF-β signaling promotes lung (5, 6) and bone metastasis (7). In the case of osteolytic bone metastasis by breast cancer cells, it has been proposed that TGF-β released from the decaying bone matrix stimulates neighboring tumor cells, establishing a vicious cycle that exacerbates the growth of the metastatic lesion (8).
The TGF-β signaling mechanisms that foster metastasis in human cancer are an important open question and a subject of debate. Because Smad factors are quintessential tumor suppressors, the basis for the protumorigenic effects of TGF-β has been sought in the Smad-independent signaling pathways that may be triggered by TGF-β. Results obtained by means of overexpression of dominant negative mutant components of the Rho pathway (9, 10) or pharmacologic inhibitors of p38 mitogen-activated protein kinase (11, 12) have implicated these pathways in the proinvasive and metastatic effects of TGF-β in transformed cells. In contrast, results obtained with overexpression of dominant negative mutant forms of Smad2 or Smad3 have argued for an involvement of the Smad pathway in tumor invasion and metastatic spreading (13) and in the formation of metastases by oncogenically transformed cells (12, 14) or tumor-derived cell lines (11) xenografted into immunodeficient mice. However, protein overexpression experiments are open to unwanted interference with other cellular functions. The lack of suitable genetic evidence has precluded a firm ascription of metastatic activity to the Smad pathway.
In the present work, we sought to determine whether the Smad pathway is activated in clinical samples of breast cancer bone metastasis and whether genetic evidence could be obtained for a role of this pathway as a mediator of bone metastasis in a mouse model system. In a search of organ-specific metastasis mechanisms, we recently isolated variants of a human breast cancer cell line that have predilection for metastasis to the bones or to the lungs and adrenal glands in mice (15–17). A bone metastasis gene-expression signature delineated by using this model system includes, among many others genes, IL-11 and connective tissue growth factor (CTGF), which are known targets of TGF-β signaling. Enforced expression of IL-11 and CTGF in these cells increases their osteolytic bone-metastatic activity (17). We have explored the role of the Smad pathway in bone metastasis using this model system.
Materials and Methods
Tumor Sample Analysis. Formalin-fixed paraffin embedded bone metastasis tissues were obtained from therapeutic procedures performed as part of routine clinical management of breast cancer patients at our institution. Hematoxylin and eosin-stained sections were examined for regions that contained tumor cells and stroma, which were further analyzed for phosphorylated Smad2 on serial sections. All studies were conducted under protocols approved by the Memorial Sloan-Kettering Cancer Center Institutional Review Board.
TGF-β1–Smads–Herpes Simplex Virus 1 (HSV1) Thymidine Kinase (tk)/GFP Reporter System. Double-stranded complementary oligonucleotides containing a sequence from the mouse germline Ig-α promoter 5′-AATTCGGCCATGTGGTCAGACACACCTGTCT CCACCACAGCCAGACCACAGGCCAGACATGACGTGGAGGTT-3′ (18) were used to construct the TGF-β1–Smads–HSV1-tk/GFP reporter vector. After annealing of oligonucleotides, the resulting DNA fragment was cloned into the EcoRI and XbaI sites of the dxNFAT-tk/GFP–Neo vector (19) in place of the nuclear factor of activated T cells enhancer element. Thus, the HSV1-tk-EGFP fusion reporter gene was linked to the enhancer elements specific for Smad–Runx transcriptional complexes. The resulting plasmid was transfected into the GPG29 packaging cell line with Lipofectamine 2000 (Invitrogen). The retrovirus-containing medium was collected for 4 consecutive days and stored at –80°C. The retrovirus was then used to transduce MDA-MB-231 cells and their subline SCP3 (15, 17). Selection of stable transfectants was accomplished by adding 1 g/liter G418. Cells containing the TGF-β1–Smads–HSV1-tk/GFP reporter system were further transduced with a second retroviral vector, SFG-tdRFP-cmvFLuc, in which tandem repeat red fluorescent protein (tdRFP) (20) and firefly luciferase (FLuc)-encoding cDNAs were placed under constitutive promoters. RFP-positive cells were sorted by FACS. The retrovirus vector encoding tk–EGFP–luciferase triple fusion proteins has been described in ref. 21.
Bioluminescence Imaging and Analysis. Anesthetized mice were retroorbitally injected with 75 mg/kg d-Luciferin (Xenogen, Alameda, CA) in PBS. Bioluminescence images were acquired by using the IVIS Imaging System (Xenogen) 2–5 min after injection. Acquisition times at the beginning of the time course started at 60 sec and were reduced in accord with signal strength to avoid saturation. Analysis was performed with livingimage software (Xenogen) by measuring photon f lux (measured in photons·sec–1·cm–2·steradian–1) using a region of interest drawn around the bioluminescence signal to be measured. Images were set at the indicated pseudocolor scale to show relative bioluminescent changes over time. Data were normalized to the signal obtained right after xenografting (day 0).
Micropositron-Emission Tomography (MicroPET) Imaging. MicroPET imaging was performed by using 18F-2′-fluoro-2′deoxy-1β-d-arabionofuranosyl-5-ethyl-uracil as the HSV1-tk substrate, as previously described (22). Two hours before whole-body, positron-emission tomography, the mice were administered 18F-2′-fluoro-2′deoxy-1β-d-arabionofuranosyl-5-ethyl-uracil [i.v.; 100 μCi per animal (1 Ci = 37 GBq)]. Imaging was performed on a microPET (Concorde Microsystems, Knoxville, TN), and images were acquired over 15 min under inhalation anesthesia (2% isoflurane).
Supporting Materials and Methods. For details about cell culture and retroviral transduction, plasmids, luciferase reporter assays, immunohistochemistry, transcriptomic profiling and clustering analyses, electrophoretic mobility shift assay, DNA precipitation assay, intracardiac injections, radiographic analysis of bone metastasis, and ELISA assays, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
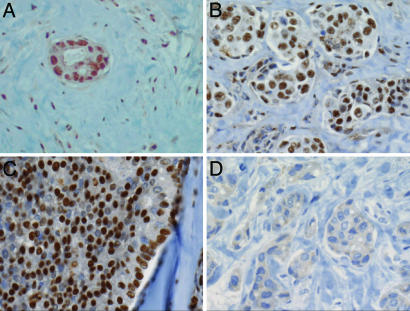
Human Breast Cancer Bone Metastases Contain an Activated Smad Pathway. Receptor-mediated phosphorylation of Smad2 at the C terminus and accumulation of phospho-Smad2 in the nucleus are typical indicators of TGF-β stimulation (3). To determine whether this pathway is active in bone metastasis, metastatic tissues from breast cancer patients were subjected to immunohistochemistry with anti-phosphopeptide antibodies against receptor-phosphorylated Smad2. Bone metastasis tissues from 16 breast cancer patients were obtained from therapeutic procedures performed as part of routine clinical management of these patients at our institution. Twelve of these samples showed prominent anti-phospho-Smad2 staining (Table 1, which is published as supporting information on the PNAS web site), and this staining was concentrated in the nucleus (Fig. 1). Nuclear phospho-Smad2 staining was present both in the tumor cells and cells of the surrounding stroma (e.g., Fig. 1B), suggesting that the entire field was under TGF-β stimulation in these lesions. The other four metastasis samples analyzed showed little or no staining. Thus, a majority of breast cancer bone metastases exhibited evidence of Smad pathway activation.
Fig. 1.
Activated Smad pathway in breast cancer bone metastasis. Examples of intense immunohistochemical staining of receptor-phosphorylated Smad2 in breast cancer bone metastasis samples from different patients. The samples shown were chosen to illustrate the nuclear phospho-Smad2 staining in a metastatic island and the surrounding stroma (A), in a cluster of metastatic islands (B), or in a contiguous metastatic mass (C), as well as a cluster of islands stained with normal rabbit serum as a negative control (D).
Functional Imaging Reveals Smad Signaling in a Bone Metastasis Model. Prompted by these results, we sought evidence for Smad-dependent transcriptional activity in bone metastasis by functional imaging in a mouse xenograft model. This model is based on the MDA-MB-231 cell line, which was derived from the pleural effusions of a breast cancer patient with metastatic disease (23). From parental MDA-MB-231 cells, we isolated various single-cell-derived sublines with distinct organ-specific metastatic behavior (16, 17). The subline SCP2 is highly metastatic to bone via arterial circulation, whereas subline SCP3 is highly metastatic to the adrenal glands. A retroviral reporter vector cis-TGF-β1–Smads–HSV1-tk/GFP was created, in which a fusion protein containing HSV1-tk and GFP was placed under the transcriptional control of a TGF-β-responsive promoter element (Fig. 2A). We chose the TGF-β responsive element from the mouse germline Igα promoter (24, 25). This TGF-β responsive element is recognized by Smad2/3–Smad4 in complex with RUNX family members and responds to TGF-β in many different cell lines (24, 25). RUNX activity in breast cancer cells is implicated in osteolytic bone metastasis (26).
Fig. 2.
Functional imaging of Smad signaling in breast cancer bone metastasis. (A) Schematic representation of the retroviral vectors SFG-tdRFP-cmvFLuc (constitutively expressing tdRFP and FLuc) and cis-TGF-β1–Smads–HSV1-tk/GFP (expressing HSV-tk/GFP fusion protein in response to TGF-β). (B and C) SCP3 transduced with these two vectors were treated with TGF-β or no additions for 24 h and analyzed by fluorescence microscopy (B) or two-color FACS (C). The constitutive tdRFP fluorescence is shown on the ordinate, and the HSV-tk/GFP fusion fluorescence, inducible by TGF-β, is shown on the abscissa. (D and E Upper) In vivo bioluminescence and microPET imaging of metastases in mice. SCP2 (D) and SCP3 (E Upper) cells bearing the SFG-tdRFP-cmvFLuc and cis-TGF-β1–Smads–HSV1-tk/GFP vectors were injected into the left cardiac ventricle and analyzed after 4 weeks (SCP2) or 18 weeks (SCP3). Bioluminescence imaging shows sites of metastases in the skull (D and E) and adrenal gland (E Upper). 18F-2′-fluoro-2′deoxy-1β-D-arabionofuranosyl-5-ethyl-uracil microPET images of tk/GFP reporter activation shows localization of radioactivity to the skull in the coronal and sagittal image planes. No visualization of the adrenal metastasis was seen on microPET imaging. Note the nonspecific accumulation of the tracer in the gastrointestinal tract and bladder attributable to clearance of the tracer. (E Lower) At necroscopy, the head showing the skull and the adrenal metastasis plus kidney were removed and imaged ex vivo for photographic (–) and bioluminescence (+) imaging.
cis-TGF-β1–Smads–HSV1-tk/GFP was transduced into SCP2 and SCP3 cells together with a second retroviral vector SFG-tdRFP-cmvFLuc expressing RFP (20) and FLuc under constitutive promoters (Fig. 2A). The RFP-positive cells expressed green fluorescence in response to TGF-β, demonstrating responsiveness of the HSV1-tk/GFP construct (Fig. 2 B and C). When inoculated into the arterial circulation of immunodeficient mice, SCP2 cells formed aggressive bone metastases, as visualized by luciferase bioluminescence imaging (Fig. 2D). These lesions also expressed tk activity, as determined by microPET (Fig. 2D). SCP3 cells formed small-bone metastases and very large adrenal (Fig. 2E Upper) and lung metastases when injected into the tail vein (17). Interestingly, although the small-bone metastases formed by SCP3 expressed tk activity in the live animals, the large adrenal metastases formed by the same cells did not (Fig. 2E Upper). The location of these lesions was verified by ex vivo bioluminescence of the affected organs after necropsy (Fig. 2E Lower). Of nine mice that were inoculated with the SCP3 cells, five developed adrenal metastasis, none of which showed tk activity by microPET. In contrast, two mice developed bone metastases to the skull and vertebrae, and both of these lesions showed tk activity (Fig. 2E and data not shown). Therefore, TGF-β signaling activity could be readily detected in bone metastases but not in adrenal gland metastases, although all of the metastases were formed by the same cell line. These results suggest that breast cancer cells undergo Smad-dependent transcriptional activation in the bone microenvironment.
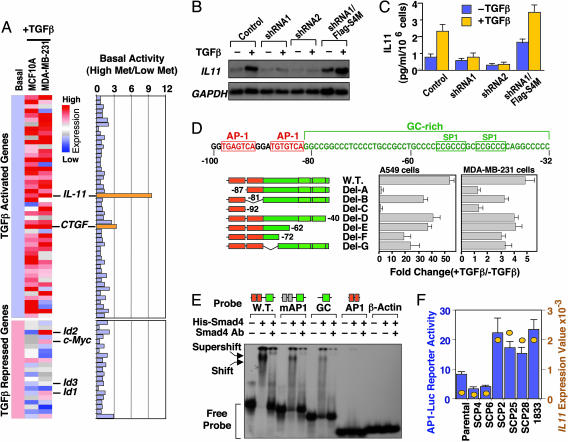
Smad4-Dependent Transcriptional Activation of Candidate Metastasis Genes. We recently identified a set of genes that mediate osteolytic bone metastasis by MDA-MB-231 cells (17). Among these genes, IL-11 was of interest because it has been proposed to play a role in osteoclast differentiation (27) and as a mediator of osteolysis in breast cancer bone metastasis (28, 29). Enforced expression of IL-11 in MDA-MB-231 cells increases their bone-metastatic activity (17). Intriguingly, IL-11 is a TGF-β-inducible gene (17, 30), suggesting a role the prometastatic activity of TGF-β in MDA-MB-231 cells. These cells are defective in TGF-β cytostatic gene responses, including repression of c-myc and Id genes (31), but retain many responses that are common among normal epithelial cells (32), including IL-11 induction (Fig. 3A and Table 2, which is published as supporting information on the PNAS web site). A comparison of the basal expression of TGF-β-responsive genes in various MDA-MB-231 derivatives revealed a sharp (>9-fold) and selective increase in the basal expression of IL-11 in highly bone-metastatic sublines compared with the poorly metastatic sublines and with all of the other TGF-β-responsive genes (Table 3, which is published as supporting information on the PNAS web site; summarized in Fig. 3A). A smaller increase was observed in the basal expression of CTGF, which is another TGF-β-responsive gene implicated in bone metastasis by MDA-MB-231 cells (17) (Table 3). In the present study, we chose IL-11 to probe the role of Smad signaling in the induction of a candidate metastasis gene in this model system.
Fig. 3.
Smad4 and AP1-dependent transcriptional activation of IL-11 by TGF-β. (A) Basal expression levels of 50 TGF-β-activated genes and 21 TGF-β-repressed genes in MCF-10A and MDA-MB-231 cells were normalized to the same level. (Left) Responses of these genes to TGF-β in each cell line are represented by different shades of red (degrees of activation) or blue (degrees of repression) in the dendrogram. (Right) The ratio of basal expression levels of these 71 genes in highly metastatic versus weakly metastatic MDA-MB-231 cells. Genes of interest are highlighted. (B) SCP25 and its derivatives (see Fig. 5B) were incubated in the absence or presence of TGF-β for 2 h. Total RNA was subjected to Northern blot analysis with the indicated probes. (C) SCP25 and its derivatives were treated with or without TGF-β for 24 h. IL-11 production in the media was determined with an ELISA assay. Data are the average of triplicate determinations ± SD. (D)(Upper) Nucleotide sequence of the minimal TGF-β-responsive region of the IL-11 promoter. Nucleotide sequence positions are indicated relative to the transcription start site. Two AP1 sites (red boxes) and a GC-rich sequence (green) containing two SP1 sites (green boxes) are indicated. (Lower)(Center and Right) A549 (Center) and MDA-MB-231 (Right) cells were transfected with the indicated IL-11 reporter constructs, treated with or without TGF-β for 16–20 h before lysis, and analyzed for luciferase activlty. Data are the average of triplicate determinations ± SD. (Left) The schematic representation of each promoter construct. (E) [γ32-P]ATP end-labeled probes matching to the wild-type IL-11 proximal promoter region, this region with mutant AP1 sites (mAP1), or the indicated fragments of this region were subjected to electrophoretic mobility shift analysis with recombinant full-length His-Smad4 protein. Antibody against Smad4 was added as indicated to create supershifts. The β-actin promoter was used as a negative control. Schematic representations of the probes are shown above the gel. (F) Various MDA-MB-231 sublines were transfected with 1 μg of 4xAP1-Luc reporter plasmid and analyzed for luciferase activity 2 days after transfection. Data are the average of triplicate determinations ± SD. The absolute values of IL 11 mRNA level as detected by an Affymetrix U133A GeneChip were plotted in the same graph (yellow circles). The scales for the luciferase activity and for IL-11GeneChip expression values are shown to the left and right of the graph, respectively.
Several observations suggested that IL-11 induction by TGF-β is an immediate gene response. IL-11 induction by TGF-β was rapid, peaking at 2 h and gradually declining thereafter (Fig. 5A, which is published as supprotive information on the PNAS web site), and the protein synthesis inhibitor cycloheximide did not block this response (data not shown). TGF-β stimulation induced the binding of Smad2/3 and Smad4 to the IL-11 promoter in chromatin immunoprecipitation experiments (17). To determine whether the Smad pathway is required for IL-11 induction and bone metastasis, we analyzed MDA-MB-231 SCP sublines that were depleted of Smad4 by means of RNA interference. Compared with parental cells or in vivo-selected bone-metastatic populations, which are heterogeneous, SCPs are derived from single cells and, therefore, are more homogenous in genetic makeup (15, 17). Three bone-metastatic sublines (SCP2, SCP25, and SCP28) were engineered to stably express the short-hairpin RNA (shRNA) probes Smad4-shRNA1 or Smad4-shRNA2, which target different regions of the Smad4 mRNA. Expression of Smad4-shRNA1 reduced Smad4 protein levels by 70–90% in all three SCPs, whereas Smad4-shRNA2 almost completely eliminated Smad4 production (Fig. 5B). As a control, we engineered a Smad4 vector (pBabe-hygro-FLAG-Smad4M) containing two silent mutations in the sequence targeted by Smad4-shRNA1 and an N-terminal FLAG epitope distinguishing the exogenous product from endogenous Smad4. Transduction of this retrovirus ensured expression of Smad4 in cells containing Smad4-shRNA1 (Fig. 5B).
As determined by Northern blot analysis, the IL-11 response to TGF-β was very weak in cells expressing Smad4-shRNA1 and undetectable in cells expressing Smad4-shRNA2 (Fig. 3B for SCP25; data not shown for SCP2 and SCP28). Expression of Smad4M restored the TGF-β response in Smad4-shRNA1-expressing cells. A similar response pattern was observed at the level of IL-11 protein secretion, as determined by ELISA (Fig. 3C and data not shown for SCP2 and SCP28). Thus, Smad4 is essential for TGF-β activation of IL-11 expression. RNA interference-mediated depletion of Smad4 also inhibited the CTGF response to TGF-β in these cells (data not shown).
Activator Protein-1 (AP1) and Smad Inputs into the TGF-β Responsive IL-11 Promoter Region. To further investigate the role of Smad factors in the IL-11 response to TGF-β, we focused on a 158-bp region immediately upstream of the TATA box in the IL-11 promoter. This region (–100 to +58) was previously mapped by deletion analysis of the IL-11 –728/+58 promoter as the region that mediates the TGF-β response in human epithelial and carcinoma cells (30, 33). We confirmed these result by using the same constructs in MDA-MB-231 cells, A549 human bronchial carcinoma cells, and HaCaT human keratinocytes (data not shown). A reporter construct under the control of the minimal IL-11 promoter [pIL-11(–100)-Luc] (30) was unresponsive to TGF-β in the Smad4-deficient breast cancer cell line MDA-MB-468 (34), which was used here to assess the role of Smad4 in IL-11 promoter activation by TGF-β (Fig. 5C). Expression of exogenous Smad4 enabled TGF-β induction of this promoter, and this effect was further enhanced by cotransfection of Smad2 or Smad3 (Fig. 5C), arguing that Smads mediate transcriptional activation from this promoter region.
This 100-bp region includes two AP1 binding sites, which are critical for IL-11 transcription (30, 33), and an adjacent GC-rich (92% GC) sequence with two putative SP1 sites (Fig. 3D). No canonical Smad-binding element (AGAC sequence) is present in this region. However, Smads can bind to GC-rich sequences in certain promoters (3). Deletion analysis of the IL-11 promoter region by means of a reporter construct indicated that the response to TGF-β minimally requires the 5′ AP1 site and an adjacent GC-rich sequence (Fig. 3D). The 3′AP1 site, which conforms less well to the consensus AP1 binding sequence, contributed little to the promoter response to TGF-β (Fig. 3D). In electrophoretic mobility shift assays, recombinant Smad4 bound to the wild-type minimal IL-11 promoter probe, resulting in the formation of a complex that could be shifted by addition of an anti-Smad4 monoclonal antibody (Fig. 3E). Mutation or deletion of the AP1 sites decreased but did not abolish Smad4 binding to the probe, whereas the AP1 sites alone did not bind Smad4 (Fig. 3E). The binding of endogenous Smad and AP1 factors to this region was assessed by means of oligonucleotide precipitation assays. MDA-MB-231 cells were incubated with or without TGF-β for 2 h, lysed, and precipitated with biotinylated dsDNA probes. Immunoblotting of DNA-bound factors demonstrated TGF-β-dependent binding of endogenous Smad3 and Smad4 to the wild-type IL-11 minimal promoter region and TGF-β-independent binding of the endogenous AP1 component JunB to this region (Fig. 5D). Deletion or mutation of the AP1 sites eliminated binding of JunB and weakened Smad binding.
Consistent with a role of AP1 in the IL-11 response to TGF-β in the breast cancer cells, the AP1 activator 12-O-tetradecanoylphorbol-13-acetate (35) increased the basal level of IL-11 expression as well as the level upon TGF-β stimulation, whereas the AP1 inhibitor curcumin (35) abolished the activation of IL-11 by TGF-β (Fig. 5E). As determined with an AP1 reporter construct (4xAP1-luciferase), the basal level of AP1 activity was significantly higher in the highly metastatic sublines SCP2, SCP25, SCP28, and 1833 than in poorly metastatic sublines SCP4 and SCP6 or parental MDA-MB-231 cells (Fig. 3F). The level of AP1 activity in these cell populations was closely correlated with the basal level of IL-11 expression (Fig. 3F; refer to Table 3). No change in 4xAP1-luciferase activity was observed after 4 h of TGF-β treatment (data not shown). Collectively, these results suggest that TGF-β-activated Smad proteins bind to the GC-rich region in the proximal IL-11 promoter. This binding is strengthened by the presence of a proximal AP1 site, and transcriptional activation results from cooperation between Smad3 and AP1. These observations also indicate a role of AP1 in the hyperactivity of IL-11 in bone-metastatic MDA-MB-231 cells.
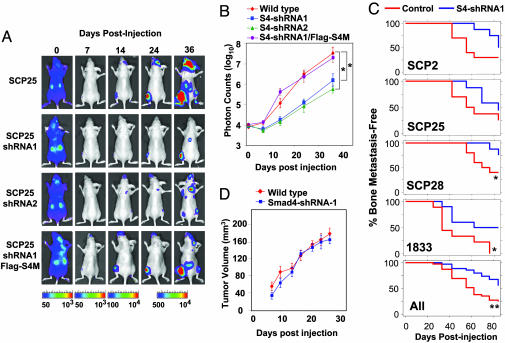
Smad4 Contribution to Breast Cancer Bone Metastasis Formation. Having shown that the TGF-β response of a bone metastasis gene in these cells required Smad function, we tested the contribution of Smad signaling to the metastatic process itself. Wild-type, Smad4-knockdown, and FLAG-Smad4M versions of the various SCP cell lines were infected with a retroviral vector expressing HVS1-tk/GFP/luciferase triple fusion protein (21). The cells were inoculated into the left cardiac ventricle of immunodeficient mice to allow the formation of bone metastasis. As determined by bioluminescence imaging of luciferase activity, the inoculated cells became immediately distributed throughout the entire animal followed by extensive clearing within 1 week (Fig. 4A). Accumulation of luciferase signal was clear 14 days after injection and became more intense over the following weeks. To quantify the rate of metastatic growth in bone, a region of interest was drawn around the bone metastases signals near the joint of the affected hind limbs, and the normalized photon counts of each metastasis were plotted (Fig. 4B). A linear correlation between the intensity of the bioluminescence and tumor burden is obtained by using this method (36). Suppression of Smad4 activity by two different shRNA constructs caused a significant reduction in the growth rate of bone-metastatic lesions (Fig. 4 A and B). Restoration of Smad4 function by the shRNA-insensitive Smad4M construct restored the wild-type rate of metastatic growth (Fig. 4 A and B). These results were consistently observed in similarly modified SCP2 and SCP28 cells (data not shown).
Fig. 4.
Smad4 mediation of breast cancer bone metastasis. Wild-type and genetically modified SCP25 was labeled with the TGL reporter and 1 × 105 cells were injected into the left cardiac ventricle of five mice for each cell line. At the indicated days after xenografting, bioluminescence images were acquired and quantified. (A) Representative mice from each group are shown in the supine position. The intensity of the signal from days 24 and 36 are on equivalent scales, whereas days 0, 7, and 14 are each on separate scales because of increasing signal strength and to avoid signal saturation. (B) The normalized photon counts from the bone metastases in the hindlimbs were measured over the indicated time course. (C) Kaplan–Meier curves showing the incidence of bone metastasis by indicated wild-type and Smad4-knockdown MDA-MB-231 sublines. For each cell sample, 105 tumor cells were inoculated into the left cardiac ventricle of 10 nude mice. Metastasis was scored as the time to first appearance of a visible bone lesion by x-ray imaging of the whole mouse. The percentages of animals in each group and in all groups combined that were free of detectable bone metastases are plotted. *, < 0.05; **, P < 0.01; calculated by log rank test. (D) Tumor cells (106) were injected s.c. into nude mice. s.c. tumor growth was monitored and quantified by caliper measurements. No significant difference was found between wild-type and Smad4-knowdown cells.
Formation of overt osteolytic bone metastases was monitored by weekly full-body x-ray imaging of the mice. Smad4 depletion consistently reduced the rate of bone metastasis formation in all three MDA-MB-231 SCPs and in the in vivo-selected bone-metastatic population, 1833 (17) (Fig. 5C). A significant level of metastatic activity still remained after Smad4 depletion, which is consistent with the TGF-β-independent involvement of other genes in these lesions (17) and may additionally be due to the incomplete elimination of Smad4 by RNA interference. Smad4 knockdown did not decrease the growth rate of the SCPs or 1833 cells in culture (data not shown) or their ability to form s.c. tumors in mice (Fig. 5D), arguing that the Smad4-dependent growth of these tumors is specifically stimulated by the bone microenvironment.
Discussion
We provide clinical, genetic, and functional evidence suggesting that the Smad tumor suppressor pathway may become prometastatic in breast cancer. Our results show the presence of receptor-phosphorylated Smad2 in a majority of bone metastasis samples from a cohort of breast cancer patients treated at our institution. By means of noninvasive functional imaging, we show that MDA-MB-231 human breast cancer cells growing as bone metastases in mice are engaged in Smad-dependent transcription. Furthermore, RNA interference-mediated depletion of Smad4 inhibited bone metastasis in this mouse xenograft model. Previous evidence implicating the Smad2/3 pathway in metastasis was based on overexpression of mutant Smad constructs (12–14). However, because protein overexpression approaches are prone to unwanted interference with other cellular functions, the involvement of Smad signaling in metastasis remained the subject of debate against observations implicating Smad-independent pathways (9–12). The present results clearly argue that signaling through the Smad pathway can facilitate breast cancer bone metastasis.
IL-11 and CTGF were among 43 genes whose expression was elevated in MDA-MB-231 subpopulations selected in vivo for high bone-metastatic activity (17). The expression of endogenous IL-11 and CTGF in these highly metastatic isolates is further increased by TGF-β addition. Exogenous overexpression of IL-11 and CTGF mediates osteolytic metastatic activity in MDA-MB-231 xenografts (17). Therefore, we chose IL-11 as a model metastasis gene in the present studies. We provide evidence that IL-11 induction by TGF-β involves Smad2/3 and Smad4, which cooperate with a previously described AP1 input (30, 33). Our results also show that Smad4 is essential for IL-11 induction by TGF-β. Interestingly, an increase in the level of AP1 activity appears to be responsible for the elevated expression of IL-11 in highly bone-metastatic MDA-MB-231 subpopulations. In separate studies, we have determined that the induction of CTGF by TGF-β is also a Smad-dependent process. But it does not involve a cooperation of Smads with AP1 (our unpublished observations).
The present results provide insights into the mechanism of activation of IL-11 in this model system. It is important to note that IL-11 (and CTGF) can only be considered candidate bone metastasis genes at present. IL-11 is known to stimulate osteoclastic bone resorption in vitro (27–29), but it has complex effects on bone formation, including positive effects (37). Its role as a mediator of bone metastasis in humans remains to be established. Other TGF-β-responsive secretory factors may also be involved in this process. Chief among these is parathyroid hormone-related peptide (PTHrP), whose role as a mediator of breast cancer metastasis has been shown in several studies using MDA-MB-231 cells (7, 38). However, unlike the induction of IL-11 and CTGF by TGF-β, the increase in PTHrP secretion in these cells occurs without an increase in PTHrP mRNA levels (our unpublished work).
The intrinsic genomic instability of tumor cell populations allows for the selection of functions that favor growth in a given environment. Thus, a bone-metastatic lesion will harbor functions that the bone environment selects for. We speculate that prometastatic Smad-mediated gene responses can emerge once this pathway becomes uncoupled from tumor-suppressor effects. If at that point a Smad pathway can provide metastatic functions to cancer cells, it likely will be selected as a prometastatic force. Certain Smad-responsive genes could provide an advantage to cancer cells in a TGF-β-rich bone microenvironment. Therefore, an increase in the basal expression of these genes coupled with their further induction by bone-derived TGF-β would favor tumor growth in the bone. Our results are consistent with this possibility. By implicating the Smad pathway in the osteolytic vicious cycle of breast cancer metastasis (8), our results additionally call further attention to the possibility of therapeutically targeting this pathway (10, 39) in TGF-β-rich metastatic sites.
Supplementary Material
Acknowledgments
We would like to thank Y.-C. Yang (Case Western Reserve University) for reagents and A. Minn and L. Jayaraman for advice. We acknowledge the use of the Genomics Core Facility and the Flow Cytometry Core Facility at Memorial Sloan-Kettering Cancer Center. This research is supported by the W. M. Keck Foundation and National Institutes of Health Grant P01-CA94060 (to J.M.) and by National Institutes of Health Grant P50-CA86438 (to R.B.) and U.S. Army Medical Research Grant DAMD17-02-0484 (to W.L.G.). Y.K. was the recipient of a postdoctoral fellowship from the Irvington Institute for Immunological Research. G.P.G. was supported by National Institutes of Health Medical Scientist Training Program Grant GM07739, a fellowship from the Katherine Beineke Foundation, and Department of Defense Breast Cancer Research Program Predoctoral Traineeship Award W81XWH-04-1-0334. J.M. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: J.M. designed research; Y.K., W.H., S.T., G.P.G., I.S., C.-R.C., and K.M.-T. performed research; R.B. and W.L.G. contributed new reagents/analytic tools; and Y.K. and J.M. wrote the paper.
Abbreviations: CTGF, connective tissue growth factor; HSV1, herpes simplex virus 1; tk, thymidine kinase; RFP, red fluorescent protein; tdRFP, tandem repeat RFP; FLuc, firefly luciferase; AP1, activator protein-1; PET, positron-emission tomography; shRNA, short-hairpin RNA; PTHrP, parathyroid hormone-related peptide.
References
- 1.Derynck, R., Akhurst, R. J. & Balmain, A. (2001) Nat. Genet. 29, 117–129. [DOI] [PubMed] [Google Scholar]
- 2.Siegel, P. M. & Massague, J. (2003) Nat. Rev. Cancer 3, 807–821. [DOI] [PubMed] [Google Scholar]
- 3.Shi, Y. & Massague, J. (2003) Cell 113, 685–700. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe, T., Wu, T. T., Catalano, P. J., Ueki, T., Satriano, R., Haller, D. G., Benson, A. B., III, & Hamilton, S. R. (2001) N. Engl. J. Med. 344, 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel, P. M., Shu, W., Cardiff, R. D., Muller, W. J. & Massague, J. (2003) Proc. Natl. Acad. Sci. USA 100, 8430–8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muraoka-Cook, R. S., Kurokawa, H., Koh, Y., Forbes, J. T., Roebuck, L. R., Barcellos-Hoff, M. H., Moody, S. E., Chodosh, L. A. & Arteaga, C. L. (2004) Cancer Res. 64, 9002–9011. [DOI] [PubMed] [Google Scholar]
- 7.Yin, J. J., Selander, K., Chirgwin, J. M., Dallas, M., Grubbs, B. G., Wieser, R., Massague, J., Mundy, G. R. & Guise, T. A. (1999) J. Clin. Invest. 103, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mundy, G. R. (2002) Nat. Rev. Cancer 2, 584–593. [DOI] [PubMed] [Google Scholar]
- 9.Bhowmick, N. A., Ghiassi, M., Bakin, A., Aakre, M., Lundquist, C. A., Engel, M. E., Arteaga, C. L. & Moses, H. L. (2001) Mol. Cell. Biol. 12, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumont, N. & Arteaga, C. L. (2003) Cancer Cell 3, 531–536. [DOI] [PubMed] [Google Scholar]
- 11.Kakonen, S. M., Selander, K. S., Chirgwin, J. M., Yin, J. J., Burns, S., Rankin, W. A., Grubbs, B. G., Dallas, M., Cui, Y. & Guise, T. A. (2002) J. Biol. Chem. 277, 24571–24578. [DOI] [PubMed] [Google Scholar]
- 12.Tian, F., Byfield, S. D., Parks, W. T., Stuelten, C. H., Nemani, D., Zhang, Y. E. & Roberts, A. B. (2004) Cancer Res. 64, 4523–4530. [DOI] [PubMed] [Google Scholar]
- 13.Oft, M., Akhurst, R. J. & Balmain, A. (2002) Nat. Cell. Biol. 4, 487–494. [DOI] [PubMed] [Google Scholar]
- 14.Tian, F., DaCosta Byfield, S., Parks, W. T., Yoo, S., Felici, A., Tang, B., Piek, E., Wakefield, L. M. & Roberts, A. B. (2003) Cancer Res. 63, 8284–8292. [PubMed] [Google Scholar]
- 15.Minn, A. J., Kang, Y., Serganova, I., Gupta, G. P., Giri, D. D., Doubrovin, M., Ponomarev, V., Gerald, W. L., Blasberg, R. & Massague, J. (2005) J. Clin. Invest. 115, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minn, A. J., Gupta, G. P., Siegel, P. M., Bos, P. D., Shu, W., Giri, D. D., Viale, A., Olshen, A. B., Gerald, W. L. & Massagué, J. (2005) Nature 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, Y., Siegel, P. M., Shu, W., Drobnjak, M., Kakonen, S. M., Cordon-Cardo, C., Guise, T. A. & Massague, J. (2003) Cancer Cell 3, 537–549. [DOI] [PubMed] [Google Scholar]
- 18.Jakubowiak, A., Pouponnot, C., Berguido, F., Frank, R., Mao, S., Massague, J. & Nimer, S. D. (2000) J. Biol. Chem. 275, 40282–40287. [DOI] [PubMed] [Google Scholar]
- 19.Ponomarev, V., Doubrovin, M., Lyddane, C., Beresten, T., Balatoni, J., Bornman, W., Finn, R., Akhurst, T., Larson, S., Blasberg, R., et al. (2001) Neoplasia 3, 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponomarev, V., Doubrovin, M., Serganova, I., Vider, J., Shavrin, A., Beresten, T., Ivanova, A., Ageyeva, L., Tourkova, V., Balatoni, J., et al. (2004) Eur. J. Nucl. Med. Mol. Imaging 31, 740–751. [DOI] [PubMed] [Google Scholar]
- 22.Serganova, I., Doubrovin, M., Vider, J., Ponomarev, V., Soghomonyan, S., Beresten, T., Ageyeva, L., Serganov, A., Cai, S., Balatoni, J., et al. (2004) Cancer Res. 64, 6101–6108. [DOI] [PubMed] [Google Scholar]
- 23.Cailleau, R., Young, R., Olive, M. & Reeves, W. J., Jr. (1974) J. Natl. Cancer Inst. 53, 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanai, J., Chen, L. F., Kanno, T., Ohtani-Fujita, N., Kim, W. Y., Guo, W. H., Imamura, T., Ishidou, Y., Fukuchi, M., Shi, M. J., et al. (1999) J. Biol. Chem. 274, 31577–31582. [DOI] [PubMed] [Google Scholar]
- 25.Pardali, E., Xie, X. Q., Tsapogas, P., Itoh, S., Arvanitidis, K., Heldin, C. H., ten Dijke, P., Grundstrom, T. & Sideras, P. (2000) J. Biol. Chem. 275, 3552–3560. [DOI] [PubMed] [Google Scholar]
- 26.Javed, A., Barnes, G., Pratap, J., Antkowiak, T., Gerstenfeld, L. C., van Wijnen, A. J., Stein, J. L., Lian, J. B. & Stein, G. S. (2005) Proc. Natl. Acad. Sci. USA 102, 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girasole, G., Passeri, G., Jilka, R. L. & Manolagas, S. C. (1994) J. Clin. Invest. 93, 1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotiriou, C., Lacroix, M., Lespagnard, L., Larsimont, D., Paesmans, M. & Body, J. J. (2001) Cancer Lett. 169, 87–95. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, H., Tumber, A. & Hill, P. A. (2004) Int. J. Cancer 109, 653–660. [DOI] [PubMed] [Google Scholar]
- 30.Tang, W., Yang, L., Yang, Y. C., Leng, S. X. & Elias, J. A. (1998) J. Biol. Chem. 273, 5506–5513. [DOI] [PubMed] [Google Scholar]
- 31.Chen, C. R., Kang, Y. & Massague, J. (2001) Proc. Natl. Acad. Sci. USA 98, 992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, Y., Chen, C. R. & Massague, J. (2003) Mol. Cell 11, 915–926. [DOI] [PubMed] [Google Scholar]
- 33.Bamba, S., Andoh, A., Yasui, H., Makino, J., Kim, S. & Fujiyama, Y. (2003) Am. J. Physiol. 285, G529–G538. [DOI] [PubMed] [Google Scholar]
- 34.Schutte, M., Hruban, R. H., Hedrik, L., Cho, K. R., Nadasdy, G. M., Weinstein, C. L., Bova, G. S., Isaacs, W. B., Cairns, P., Nawroz, H., et al. (1996) Cancer Res. 56, 2527–2530. [PubMed] [Google Scholar]
- 35.Eferl, R. & Wagner, E. F. (2003) Nat. Rev. Cancer 3, 859–868. [DOI] [PubMed] [Google Scholar]
- 36.Contag, C. H., Spilman, S. D., Contag, P. R., Oshiro, M., Eames, B., Dennery, P., Stevenson, D. K. & Benaron, D. A. (1997) Photochem. Photobiol. 66, 523–531. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi, Y., Watanabe, S., Ishii, G., Takeda, S., Nakayama, K., Fukumoto, S., Kaneta, Y., Inoue, D., Matsumoto, T., Harigaya, K. & Fujita, T. (2002) J. Biol. Chem. 277, 49011–49018. [DOI] [PubMed] [Google Scholar]
- 38.Guise, T. A., Yin, J., Taylor, S. D., Kumagai, Y., Dallas, M., Boyce, B. F., Yoneda, T. & Mundy, G. R. (1996) J. Clin. Invest. 1544–1549. [DOI] [PMC free article] [PubMed]
- 39.Yingling, J. M., Blanchard, K. L. & Sawyer, J. S. (2004) Nat. Rev. Drug Discovery 3, 1011–1022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.