Abstract
We previously demonstrated an increased degradation of mRNAs in mutants of Saccharomyces cerevisiae having blocks in nuclear export. The degradation activity, designated DRN (degradation of mRNA in the nucleus), requires Cbc1p, a nuclear cap-binding protein, and Rrp6p, a nuclear exosome component. Microarray procedures were used to determine the half-lives of mRNAs from normal and mutant strains, leading to the tentative identification of hundreds of normal mRNAs that were notably stabilized when either CBC1 or RRP6 were deleted. Northern blot analysis of representative mRNAs confirmed the diminished degradation. One representative of this group, SKS1 mRNA, was also shown by a cytological procedure to be preferentially retained in the nucleus compared with typical mRNAs. We suggest that all normal mRNAs are subjected to degradation by DRN, but the degree of degradation is determined by the degree of nuclear retention. Furthermore, these mRNAs particularly susceptible to DRN were also diminished by overproduction of Cbc1p, demonstrating a regulatory role for CBC1. This conclusion was corroborated by finding an inverse relationship of the CBC1 and SKS1 mRNA levels in normal strains grown under different conditions.
Keywords: cap-binding protein, CBC1, mRNA degradation, nuclear exosome, RRP6
Messenger RNA stability is an important factor in the control of gene expression (1, 2). The major mRNA turnover pathway in yeast takes place in the cytoplasm and is initiated by deadenylation which is followed by the decapping reaction and 5′→3′ exonucleolytic degradation (3, 4). The deadenylation and decapping, carried out by the Ccr4p/Pop2p (5) and Dcp1p/Dcp2p complexes (6, 7), respectively, are considered to be the rate-limiting steps of mRNA degradation. After the deadenylation and decapping, mRNAs undergo rapid 5′→3′ degradation by Xrn1p, a 5′→3′ exonuclease that takes place in the cytoplasm (8). Although 5′→3′ degradation by Xrn1p appears the predominant mode of decay in yeast (9), mRNAs can also be degraded from the 3′ →5′ direction by the cytoplasmic exosome, in association with the Ski complex, a pathway that plays a minor role (4). In contrast to yeast, the exosome-mediated 3′ degradation seems to be the predominant pathway in mammalian cells (4). In addition to the default mRNA turnover pathway, specialized surveillance pathways also exist in the cytoplasm to ensure the fidelity of the transcripts that are undergoing translation (10, 11), including the following: degradation of mRNAs harboring premature termination codon (PTCs), known as nonsense-mediated decay (NMD) (12); and degradation of transcripts lacking termination codon, known as nonstop decay (NSD) (10, 11).
In addition to these cytoplasmic pathways, recent evidence suggests that the nuclear exosome (13) participates in a nuclear mRNA quality control mechanism that degrades unspliced (14), unadenylated (15), or 3′-extended mRNAs (16). In addition to aberrant mRNAs, normal mRNAs are also reported to undergo decay in the nucleus by a degradation system designated DRN (degradation of mRNA in the nucleus) (17). Das et al. (17) determined that this DRN system requires Cbc1p, a nuclear cap-binding protein, and Rrp6p, a nuclear exosome component. Furthermore, they suggested that DRN acts on all normal mRNAs, with the degree of degradation dependent on the degree of nuclear retention. However, natural substrates particularly susceptible to DRN were not identified, and it was reported that deletion of Cbc1p does not affect the stability of normal mRNAs, such as represented by ACT1, CYC1, and CYH2 (17).
In this study, we used genome-wide microarray analysis to identify normal mRNAs whose stability was particularly enhanced by deletion of Cbc1p and Rrp6p. We have shown that a group of mRNAs became significantly stabilized in cbc1-Δ and rrp6-Δ strains compared with normal strains. The results were further confirmed with Northern blot analysis. In addition, we used a cytological procedure to demonstrate the preferential retention of SKS1 mRNA in the nucleus, a representative from this class whose half-life and steady-state level were both increased in cbc1-Δ strain. Furthermore, we have also demonstrated that overexpressing Cbc1p led to diminution of these mRNAs particularly susceptible to DRN, thus establishing a regulatory role for CBC1. Also, microarray analysis revealed an inverse relationship of the CBC1 and SKS1 mRNA levels in normal strains grown under different conditions. We suggest that DRN, the Cbc1p-dependent mRNA degradation system, functions as a minor pathway to control eukaryotic gene expression.
Materials and Methods
Strains and Plasmids. The strains of Saccharomyces cerevisiae used in this study are listed in Table 1. The U1A-GFP expression plasmids and RNA expression plasmids designated pAB2830 (CEN LEU), pAB2832 (CEN URA), and pAB2834 (CEN URA), respectively, were provided by P. Silver (Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Cambridge, MA) (18). The plasmids expressing SKS1–U1A reporter mRNA, pAB3124, was constructed with pAB2834 by replacing the PGK1 promoter-PGK1 ORF region (2.3 kb) with SKS1 promoter-SKS1 ORF (2.5 kb), by using XbaI and BamHI restriction sites and replacing PGK1 3′ UTR region with SKS1 downstream 1-kb region by using SpeI and NotI restriction sites.
Table 1. List of yeast strains used in this study.
| Strain designation | Genotype | Abbreviated genotype | Reference |
|---|---|---|---|
| B-9037 | MATa cyc1-512 trp2-1 ura3-52 | Normal | 19 |
| B-9038 | MATa cyc1-512 cbc1::URA3 trp2-1 ura3-52 | cbc1-Δ | 19 |
| B-11872 | MATa cyc1-512 rrp6::URA3 trp2-1 | rrp6-Δ | 19 |
| B-11599 | MATα leu2-3,113 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 | nupl16-Δ | 17 |
| B-13398 | MATα leu2-3,113 his3-11,15 ade2-1 ura3-1 trp1-1 can1-10 0 nup116::HIS3 cbc1::URA3 | nup116-Δ cbc1-Δ | 17 |
| B-15405 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 | Normal | Open Biosystems B-4742 |
Analysis of mRNA Steady-State Levels and Stability. The stability of the various mRNAs and pre-mRNAs were determined by the inhibition of transcription with thiolutin (4 μg/ml) at 30°C unless mentioned otherwise, as described (19). Briefly, the drug was added to exponentially growing culture of A600 = 1.0, followed by collecting 40-ml portions of the cultures at different time points from 0 min to 50 min or as indicated. Steady-state samples were collected without adding thiolutin from the same phase of the growth cycle. Cells were briefly centrifuged at 4°C and quickly frozen in dry ice followed by isolation of total RNA as described (19). Northern blot analyses of different mRNAs were conducted as outlined (20). Briefly, 20 μg of total RNA for each sample was separated by electrophoresis, transferred onto nylon membrane (DuPont/NEN) and hybridized with 32P-labeled double-stranded probe, which was generated by exo– Klenow in the presence of random octadeoxynucleotides. The levels of specific mRNAs were quantified by storage PhosphorImager analysis (model 425E; Molecular Dynamics) and normalized against the signal of SCR1 RNA, which is an RNA polymerase III transcript (21), and which was presumably stable under the condition of our experiments. The decay rates and half-lives were estimated with the sigmaplot 6.0 regression analysis program, by using a single exponential decay formula, y = 100 e–bx.
Microarray Sample Handling and Data Processing. Cells were grown in yeast extract/peptone/dextrose (YPD) medium to A600 = 1.0 followed by treatment of 4 μg/ml thiolutin. Cells were collected at the following three time points: before treatment, 5 min after treatment, and 20 min after treatment. Total RNA was isolated by using the method described above and labeled by two rounds of in vitro transcription with T7-Oligo(dT) Promoter Primer Kit (Affymetrix), following the procedure recommended by the manufacturer. The fluorescent-labeled cRNA was hybridized to the Affymetrix Yeast S98 gene chip and scanned by a Hewlett Packard GeneArray Scanner. The fluorescent signals corresponding to hybridization intensities were analyzed with affymetrix microarray suite 5.0 software (www.affymetrix.com). The chips, hybridized to the time 0 sample of each strain, were normalized with global scaling normalization to conduct the steady-state level of each transcript. The software subtracted the signal for each mismatched oligonucleotide (MM) from the signal for each perfectly matched oligonucleotide (PM). The adjusted signals for each probe pair (PM-MM) are averaged across each probe pair set, excluding the probe pairs that give signals 3 standard deviations or more from the mean. A present call “P,” marginal call “M,” or absent call “A” was assigned to each probe set representing the ratio between signal reported and the detection threshold. For half-life determination, the data from three time points were normalized so that the half-life of ACT1 mRNA is 50 min and the normalization factors were applied to the whole data sets.
Fluorescent Localization of Individual mRNA. GFP RNA imaging was carried out essentially as described by Brodsky and Silver (22). Strains harboring one of the RNA expression plasmids and the U1A-GFP expression plasmid were grown in selective 2% glucose medium overnight and reinoculated at 1:100 in selective medium containing 2% raffinose and grown to A600 = 0.5. A total of 0.1% galactose was added to induce the expression of U1A-GFP. Cells were allowed to grow for 50 min and then were concentrated by centrifugation and resuspended in selective medium containing 2% glucose. The cells were grown for an additional 1 h to maximize the GFP signal. Long time recovery was necessary to reduce the background when a higher amount of galactose is used for induction. The cells were stained with 5 μg/ml Hoechst 33342 for 1 min and quickly washed with glucose medium. Other desirable stress conditions were then applied before imaging. Images of cells were obtained by using a Leica TCS NT confocal microscope equipped with UV, Ar, Kr-Ar, and He-Ne lasers.
Results
Genome-Wide mRNA Decay Rates. DRN was previously demonstrated to act on several kinds of aberrant mRNAs (ref. 19 and B.D. and F.S., unpublished results), as well as on normal mRNAs that are retained in the nucleus in export-defective mutants nup116-Δ (17). However, the natural substrate for this pathway was not previously identified. In an attempt to uncover natural cellular mRNAs undergoing degradation by DRN under normal physiological condition, we have performed genome-wide screens for mRNAs whose steady-state levels and half-lives were enhanced in cbc1-Δ and rrp6-Δ mutant strains. Affymetrix Yeast S98 GeneChips were used to determine decay rates of mRNAs from normal and cbc1-Δ strains. Both strains contained cyc1-512, a mutation that is suppressed by cbc1-Δ and rrp6-Δ (19). The decay rate of each individual mRNA in these strains was determined from hybridization intensities at three time points after treatment with thiolutin, assuming that the degradation is a single-order reaction. The steady-state levels and half-lives of the entire data set are in Table 4, which is published as supporting information on the PNAS web site. Candidates susceptible to DRN were identified in the screen by comparing half-lives and steady-state levels of all of the mRNAs from the normal and cbc1-Δ strains. The microarray assays revealed ≈500 mRNAs with increased half-lives in the cbc1-Δ strain. The following four stringent criteria were applied to reveal the most reliable candidates and to exclude unreliable data and possible false positives. (i) Each time point value used for calculating a half-life must have a valid “present” call assigned by Affymetrix mas 5.0 software application. (ii) The half-life value resulting from linear regression must have a positive value. (iii) The correlation coefficient of the linear regression must be >0.5. (iv) Both the steady-state level and the half-life in cbc1-Δ strain must increase by at least 2-fold with respect to normal strain. The application of these four criteria ultimately eliminated most of these mRNAs and revealed only six mRNAs having enhanced stability and abundance in the cbc1-Δ and rrp6-Δ mutant strains (Table 2), including cyc1-512 mRNAs, which were previously shown to be stabilized by cbc1-Δ (19). Although our stringent criteria undoubtedly eliminated some mRNAs that were truly susceptible to DRN, our initial goal was not to identify all such mRNAs, but to identify representatives with a high degree of certainty.
Table 2. mRNAs that significantly exhibited increased abundance and half-life in the cbc1-Δ strain.
| Fold change
|
||||
|---|---|---|---|---|
| ORF | Gene | Half-life | Steady-state level | Description |
| YPL026C | SKS1 | 2.38 | 2.37 | Serine/threonine protein kinase homologous to Ran1p |
| YOL005C | RPB11 | 2.52 | 2.11 | RNA polymerase II subunit |
| YLR194C | — | 2.07 | 2.20 | Hypothetical protein |
| YLR046C | — | 2.01 | 2.50 | Strong similarity to Rta1p and Rtm1p |
| YJR048W | cyc1-512* | 2.22 | 3.15 | Mutant iso-1-cytochrome c (cyc1-512)* |
| YHR148W | IMP3 | 2.36 | 2.02 | U3 snoRNA-associated protein |
These mRNAs were uncovered after applying the filters as described in the text.
cyc1-512 strains were used for the Microarray assays.
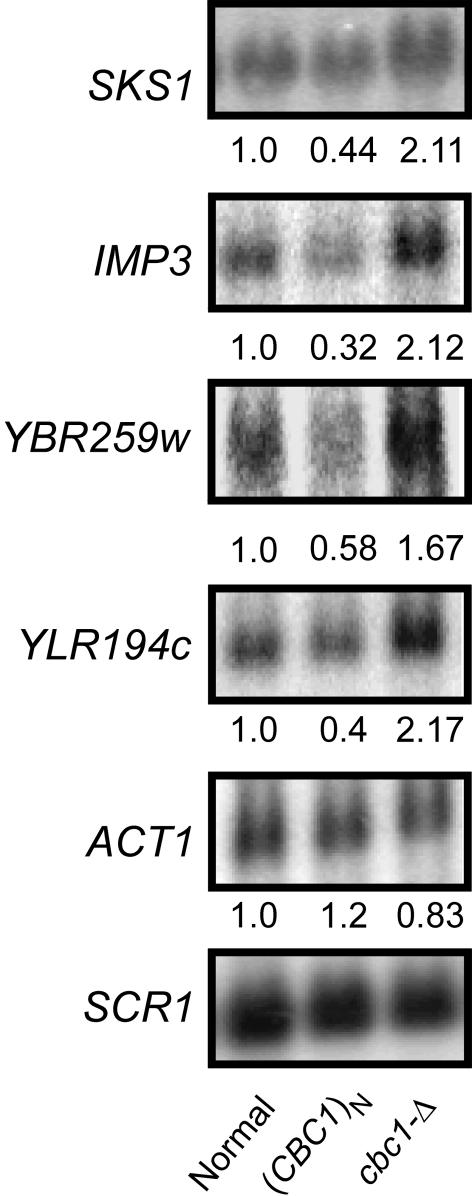
cbc1-Δ and rrp6-Δ Mutations Stabilize SKS1 and IMP3 mRNA. The results obtained from the microarray assays were confirmed by further analyzing the steady-state levels of the four mRNAs, SKS1, IMP3, YBR259w, and YLR194c, from a normal strain, a cbc1-Δ strain, and a strain overproducing CBC1. Northern blot analysis revealed that the steady-state levels of all four of these mRNAs were approximately 2-fold higher in the cbc1-Δ strain compared with the normal strain (Fig. 1). In contrast, the steady-state level of ACT1 mRNA, representing a typical mRNA, remained the same in the cbc1-Δ and normal strains, as reported (19) (Fig. 1).
Fig. 1.
CBC1 regulates the abundance of a subclass of mRNAs. The normal strain (Normal, B-15524; B-9037 with the empty vector pAB524), the Cbc1p overproducing strain [(CBC1)N, B-15525; B-9037 with the CBC1 pAB3198 vector], and the cbc1-Δ strain (cbc1-Δ; B-11558 with the empty vector pAB524) were grown at 30°C to an A600 = 1.0 and cells were harvested. Total RNAs were extracted, electrophoresed, and blotted, and the steady-state levels of the specific mRNAs were determined by Northern blot analysis by using random priming probe specific to the SKS1, IMP3, YBR259w, YLR194c (Table 1), ACT1, and SCR1 RNAs. The blot was hybridized sequentially at 68°C for 2 h with a given probe, washed, and exposed to PhorphorImager screen for 14–20 h, and then the hybridized probe was stripped off before the next one was hybridized and washed in the same way. The values below each signal correspond to the PhosphorImager quantitation of the respective mRNAs. The signal values of each mRNA were normalized to SCR1 RNA and then expressed as the percentage of the normal strain, which was arbitrarily assigned a value of 1.0.
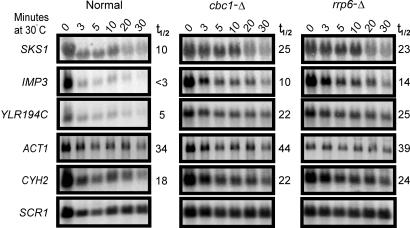
To determine whether diminished rates of degradation were responsible for the enhanced levels of mRNAs, the decay rates of these mRNAs were analyzed in the normal and cbc1-Δ strains by blocking transcription with thiolutin as described (19). As expected, the increased steady-state levels of these representative mRNAs in cbc1-Δ strain were due to diminished degradation, and not, for example, to changes in the rates of transcription. The half-life of SKS1 mRNA was 10 min in the normal strain and was increased to ≈25 min in the cbc1-Δ strain (Fig. 2 and Table 3, 30°C). The IMP3 and YLR194c mRNAs also were similarly stabilized by cbc1-Δ, with a 3- to 4-fold increase in half-life (Fig. 2 and Table 3, 30°C). Most importantly, rrp6-Δ also stabilized all of these mRNAs to about the same extent as cbc1-Δ (Fig. 2 and Table 3, 30°C). Also, as reported (19), neither cbc1-Δ nor rrp6-Δ stabilized CYH2, ACT1, and CYC1 mRNAs (19), three representatives of typical mRNAs (Fig. 2). Therefore, these results confirmed the microarray data and substantiated the view that certain mRNAs are natural substrates of DRN, requiring the action of Cbc1p and Rrp6p.
Fig. 2.
SKS1, IMP3, and YLR194C mRNAs are stabilized by deletion of Cbc1p and Rrp6p. Normal, cbc1-Δ, and rrp6-Δ cells were grown at 30°C to an A600 = 1.0 and then treated with 4 μg/ml thiolutin, and samples were removed from the culture at various times after the treatment as indicated at the top of each panel. Total RNAs were extracted from each sample, electrophoresed, and blotted, and the decay of the specific mRNA was determined by Northern blot analysis. The blot was sequentially hybridized with random priming probe specific to the SKS1, IMP3, YLR194C, ACT1, and CYH2 as noted. The half-life in min of each mRNA is shown to the right of the panel. The signal values were normalized to SCR1 RNA.
Table 3. Half-life (min) of different mRNAs in different normal and nup116-Δ strains.
| 30°C
|
25°C
|
37°C
|
|||||
|---|---|---|---|---|---|---|---|
| cbc1-Δ | cbc1-Δ | ||||||
| mRNA | Normal | cbc1-Δ | rrp6-Δ | nup116-Δ | nup116-Δ | nup116-Δ | nup116-Δ |
| SKS1 | 10 | 25 | 23 | 20 | 40 | 8 | 20 |
| IMP3 | <3 | 10 | 14 | 12 | 28 | <5 | 12 |
| YLR194C | 5 | 22 | 25 | 15 | 32 | 9 | 26 |
| ACT1 | 34 | 44 | 39 | 42 | 45 | 14 | 52 |
| CYH2 | 18 | 22 | 24 | 35 | 38 | 10 | 28 |
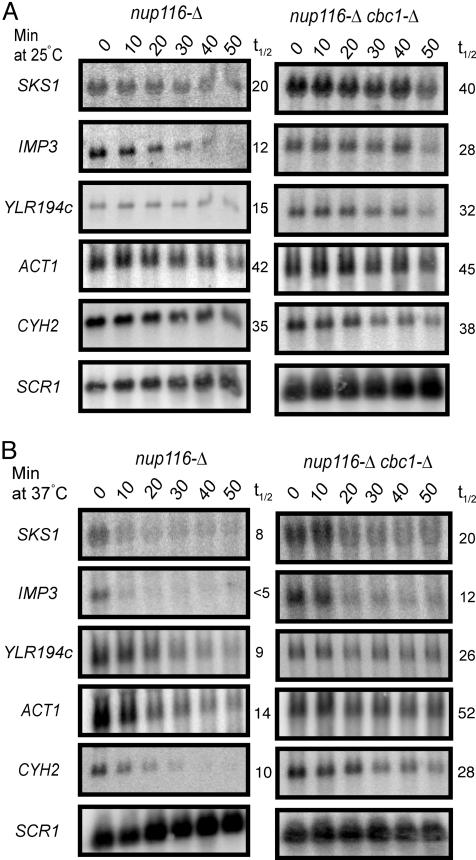
The Rapid Cbc1p-Dependent Degradation of SKS1 mRNA Is Due to Preferential Nuclear Retention. The observation that cbc1-Δ and rrp6-Δ stabilized these special classes of mRNAs suggested that their degradation takes place in the nucleus because Cbc1p and Rrp6p were known to localize primarily in the nucleus (15, 19, 23) and both Cbc1p and Rrp6p were implicated to participate in DRN, a nuclear pathway of mRNA decay (17). The hypothesis that this Cbc1p-dependent degradation of certain normal mRNAs takes place in the nucleus was further tested by determining the stability of representative normal mRNAs from a nup116-Δ temperature-sensitive mRNA export-defective strain, which prevents the export of all mRNAs in the nucleus upon shifting the strain to 37°C (17). In this experiment, the nup116-Δ and nup116-Δ cbc1-Δ cells were grown at 25°C till midlog phase. Half of the culture was maintained at 25°C and the other half was shifted to 37°C; both cultures were grown for 1 additional hour, which completely shuts off nuclear export and depletes the remaining mRNA in the cytosol (19). Thiolutin was then added to both the permissive control cultures and restrictive experimental cultures to inhibit transcription. The decay rate of specific mRNAs was then followed by Northern blot analysis as described by Das et al. (19).
As reported (17), typical mRNAs, such as CYC1 (data not shown), CYH2, and ACT1 mRNAs, did not show any significant difference in stability in nup116-Δ and nup116-Δ cbc1-Δ strains at permissive condition of 25°C (Table 3, 25°C) because their export continues at this condition and they are rapidly exported and thus escaped from the action of DRN. Thus, these results with nup116-Δ strains at 25°C resemble the results with NUP116+ strains (Table 3, 30°C). In contrast, typical mRNAs in the nup116-Δ strains at restrictive condition of 37°C exhibited rapid degradation because of their nuclear accumulation as the export is blocked, and therefore they undergo rapid decay by DRN, which is diminished by nup116-Δ cbc1-Δ (Table 3, 25°C and 37°C). Most importantly, SKS1 and the other special mRNAs exhibit rapid decay both at permissive and restrictive conditions, and this rapid decay was stabilized by nup116-Δ cbc1-Δ (Table 3, 25°C and 37°C). This result therefore suggests that, unlike the typical mRNAs, as exemplified by CYC1, CYH2, or ACT1, the decay rates of the special mRNAs are independent of any export defect at all conditions, indicating that these susceptible mRNAs, such as SKS1 mRNA, are rapidly degraded because of their preferential nuclear retention, a property that distinguishes this type of mRNAs from typical mRNAs. The half-lives of the susceptible mRNAs, SKS1, IMP3, and YLR194c, and the typical mRNAs, ACT1 and CYH2, from various strains are summarized in Fig. 3 and Table 3.
Fig. 3.
cbc1-Δ stabilizes mRNAs retained in the nucleus by the nup116-Δ mutation at both permissive (A) and restrictive (B) temperatures. The cells were grown at 25°C to an A600 of 1.0. One-half of the culture was grown at 25°C while the other half was shifted to 37°C. Both of them were incubated at the respective temperatures for 1 h before their transcription was shut off by treating the cells with 4 μg/ml thiolutin. Total RNA was extracted from the cells harvested at various times after transcription inhibition as indicated, electrophoresed, and blotted, and the decay of the specific mRNA was determined by Northern blot analysis as described (17). The blot was hybridized with random priming probe specific to the SKS1, IMP3, YLR194c, ACT1, and CYH2 as noted. The half-life of each mRNA is shown to the right of the panel. The signal values were normalized to the SCR1 RNA signal.
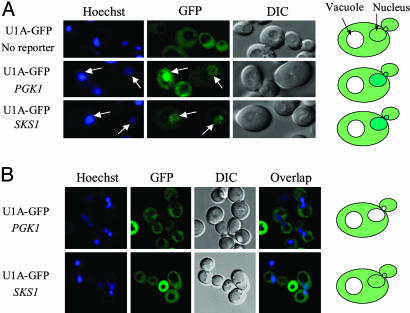
SKS1 mRNA Is Preferentially Retained in the Nucleus. The hypothesis that SKS1 mRNA is in fact preferentially retained in the nucleus was demonstrated by cytological examination of subcellular distributions of this mRNA. A GFP-fused U1A RNA-binding protein, designated U1A-GFP, was used to determine the distribution of one reporter SKS1 mRNA according to the procedure described (18, 22). The U1A protein binds to the U1A hairpin structure, which does not exist in yeast. Sixteen repeats of U1A hairpin were inserted into the plasmid carrying SKS1 and PGK1 gene between the coding region and the 3′ UTR. Both the PGK1 and the SKS1 reporter plasmids were cotransformed separately with the plasmid expressing U1A-GFP under the GAL1 promoter. The binding of the U1A-GFP protein to reporter mRNAs was examined under 10% ethanol stress condition, a condition which is known to cause accumulation of all mRNAs, except heat shock mRNAs, in the nucleus (18). As expected, after galactose induction, both PGK1 and SKS1 reporter mRNAs accumulated the GFP signal in the nucleus under 10% ethanol stress, whereas the GFP signal is uniformly distributed within the cells that lack a reporter (Fig. 4A). Under the normal growth condition, the GFP signal in a strain carrying the PGK1 mRNA reporter seemed to be excluded from the nucleus, which was localized by Hoechst staining (Fig. 4B). This distribution pattern is consistent with the poly(A) RNA distribution reported by Hector et al. (24), indicating that the PGK1 mRNA, representing normal mRNAs, was quickly exported to the cytoplasm and therefore seems to be excluded from the nucleus. In contrast, no apparent nuclear exclusion was observed in the strain carrying the SKS1 mRNA reporter at different levels of the GFP signal. Instead, the GFP distribution seemed to be localized in both nucleus and cytoplasm (Fig. 4B), indicating that a larger fraction of SKS1 mRNA is located in the nucleus due to inefficient export. The difference between the location of PGK1 mRNA and SKS1 mRNA under normal conditions is best explained by a slower export rate of the SKS1 mRNA.
Fig. 4.
A greater portion of SKS1 mRNA is located in the nucleus. Cells containing U1A-GFP expression plasmids and RNA reporter plasmids as indicated were grown in 2% raffinose medium to an A600 = 0.5. U1A-GFP expression was induced with 0.1% galactose medium for 50 min. (A) Cells were allowed to grow 60 min in 2% glucose medium. DNA was stained by a brief exposure to 5 μg/ml Hoechst, washed with the glucose medium, and then stressed with the glucose medium containing 10% ethanol. Images were taken 30 min later. No nuclear accumulation was found even after 60 min in strains lacking a reporter. (B) The cell was washed and grown in 2% glucose medium for 1.5 h. Live cell images were taken after briefly staining the cell with 5 μg/ml Hoechst. The nuclear exclusion was found for PKG1 reporter but not for SKS1 reporter.
Overproduction of Cbc1p Diminishes the mRNAs Susceptibility to DRN. The observation of the inverse relationship between the level of Cbc1p and the level of mRNAs susceptible to DRN in the normal CBC1 strain and the cbc1-Δ mutant was extended by determining the levels of SKS1, IMP3, YBR259w, and YLR194c mRNAs in a strain that overproduced Cbc1p from a multi-copy plasmid. The results demonstrated that the overproduction of Cbc1p diminished the levels of these mRNAs below the normal levels. The diminution varied from 25% to 54% with different mRNAs (Fig. 1). In contrast, the level of ACT1 mRNA, representing a typical mRNA, was not dependent on the level of Cbc1p. The findings with the CBC1, cbc1-Δ, and (CBC1)N strains clearly demonstrate an inverse relationship of the level of Cbc1p with the level of a special class of mRNAs, which are particularly susceptible to DRN. These results establish that Cbc1p has the potential to play a regulatory role by degrading those normal mRNAs that are preferentially retained in the nucleus.
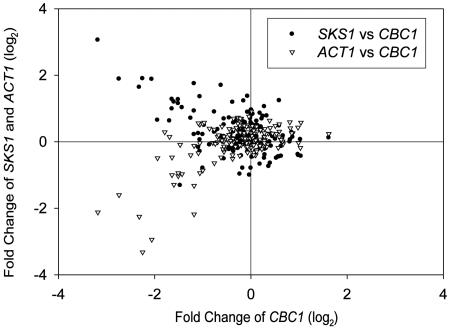
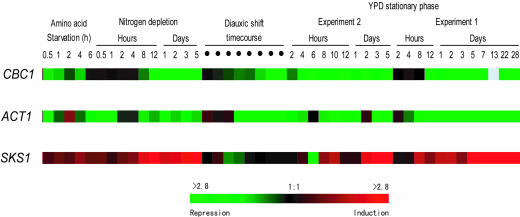
The view that Cbc1p serves a regulatory function in wild-type strains was also inferred from the analysis of the genome-wide expression profiles of CBC1 (www.yeastgenome.org) (25). Microarray analysis indicated that the level of the CBC1 transcript can vary >4-fold with different growth, starvation, and stress conditions (Figs. 5 and 6). Most importantly, a parallel analysis revealed that the SKS1 mRNA level exhibited an inverse relationship with the CBC1 mRNA level in response to various conditions (Fig. 5), thus implying that the causal relationship determined with CBC1, cbc1-Δ, and (CBC1)N strains (Fig. 1) is manifested in wild-type strains. On the other hand, the ACT1 mRNA level was not affected by CBC1 mutations (Fig. 1), although the levels of both mRNAs seemed to some extent to be directly related in normal strains under various environmental conditions (Fig. 5). All of these results, taken together, strongly suggest a regulatory role of DRN in controlling the abundance of a special class of mRNAs, thus representing another means of regulating eukaryotic gene expression.
Fig. 5.
The log2 values of fold changes of SKS1 and ACT1 transcript (y axis) level in response to environmental change were plotted against that of CBC1 transcript (x axis). The values were obtained from comparisons of the expression profile of CBC1, SKS1, and ACT1 mRNA in response to environmental changes (see Fig. 6). Microarray-based expression data are from the Saccharomyces Genome Database (www.yeastgenome.org) (25).
Fig. 6.
The expression of SKS1, ACT1, and CBC1 under stress conditions as indicated. The induction or repression levels were represented with the color scale. (The values were obtained as described in Fig. 5.)
Discussion
Gene expression in eukaryotic cells is regulated at many levels. Although the regulation achieved at the level of transcription is the most common and efficient way to control the abundance of mRNAs, this control also can be achieved posttranscriptionally. The study presented here clearly establishes that the stability of a group of mRNAs in S. cerevisiae is regulated in part by a Cbc1p-dependent nuclear degradation system (DRN) at the posttranscriptional level.
DRN was initially identified as an mRNA decay system that degrades aberrant mRNAs, such as cyc1-512 mRNA (19), and normal mRNAs artificially retained in the nucleus. DRN requires the nuclear exosome component Rrp6p, a nuclear 3′→5′ exoribonuclease, and the nuclear cap-binding complex protein, Cbc1p, which participates in a variety of processes involving mRNA biogenesis (17, 19). DRN also utilizes the nuclear 5′→3′ exoribonuclease RAT1/RAI1 complex as a minor pathway. We have suggested that all mRNAs are degraded by this system, but the extent of susceptibility varies with the degree of nuclear accumulation. In this study, we have identified normal mRNAs particularly susceptible to DRN and shown that these mRNAs are regulated by the level of Cbc1p. Thus, DRN has a new biological function other than mRNA surveillance.
Furthermore, using SKS1 as representative of mRNAs highly susceptible to DRN in normal strains, we have demonstrated that the enhanced degradation is due to preferential nuclear retention. On the other hand, cbc1-Δ and rrp6-Δ strongly affect the decay rates of typical mRNAs only in strains defective in mRNA export.
Although all of the factors that govern nuclear retention have not been identified, the DRN may play a regulatory role on certain mRNA life spans. It is worthwhile to point out that the identified mRNAs whose stability is susceptible to DRN exhibited a 2–3 times longer half-life in cbc1-Δ strain compared with normal strain, suggesting that disabling DRN does not totally diminish the degradation of these mRNAs. Instead, DRN seems to be an additional means of regulation in addition to normal mRNA turnover occurring in the cytoplasm. Although the mRNAs are very rapidly degraded when they are retained in the nucleus, their half-lives are much longer in normal strain, indicating that the degradation rate of DRN is much higher than normal mRNA turnover. The normal mRNAs probably are very rapidly transported out of nucleus so that they escape from the degradation activity whereas some mRNAs, such as SKS1 mRNA, stay longer in the nucleus and are more susceptible to DRN.
Although the abundance of an mRNA is primarily determined by the rates of transcription and cytoplasmic degradation, apparently, DRN also plays a role in controlling gene expression. The expression profiles in response to environmental changes allow us to speculate on the physiological significance of regulation by CBC1. Various degrees of glucose starvation (25) are one of the major stress conditions resulting in an inverse relationship in the levels of CBC1 and SKS1 mRNAs (Fig. 5). In this regard, it should be mentioned that Sks1p is a putative serine/threonine protein kinase required for long-term adaptation of low glucose for snf3-null strains that are deficient in high-affinity glucose transport (26). This finding suggests a role for CBC1 in regulating genes involved in growth in media low in glucose. On the other hand, the physiological relationships of other proteins encoded by mRNAs particularly susceptible to DRN (Table 2) are not that obvious. For example, IMP3 is an essential gene encoding a protein, which is component-specific to the U3 snoRNP, and which is involved in 18S rRNA biogenesis (27), whereas RPB11 encodes a subunit of RNA polymerase II (28). Perhaps CBC1 regulates certain genes involved in general growth. However, no simple inverse relationship of IMP3 and RPB11 mRNAs with CBC1 mRNA was revealed, as was found with SKS1 and CBC1 mRNA (Fig. 5). Apparently the abundance of IMP3 and RPB11 mRNAs is controlled by other parameters besides CBC1.
Although the molecular mechanism by which DRN acts on normal mRNAs is unknown, we favor the view that all mRNAs associated with the nuclear cap-binding complex, which includes Cbc1p (CBP80) and Cbc2p (CBC20), have two fates. First, they are subjected to export and second, if retained, they are directed to site of degradation within the nucleus, where they are degraded by Rrp6p and to some extent by Rat1p (17). Export of typical mRNAs is favored over degradation and therefore, is only marginally affected by DRN. We also suggest that these normal mRNAs are differentially retained in the nucleus because of some intrinsic properties of their structures. In extreme cases of retention, such as with SKS1 mRNA, Cbc1p regulates these mRNAs through the action of DRN, thus providing another level of functional control in eukaryotic gene regulation.
Supplementary Material
Acknowledgments
We thank Dr. Nataliya Shulga (Department of Biology, University of Rochester) for assistance in the use of the confocal microscope, Ms. Linda Salamone (Functional Genomics Center, University of Rochester) for processing Microarray analysis, and Dr. Edward Pagani (Pfizer Inc.) for providing thiolutin. This work was supported by National Institutes of Health Grant R01 GM12702 (to F.S.). Affymetrix chips and processing were provided in part by The Nathan Shock Center for Excellence in Aging Research Microarray Core, National Institutes of Health Grant P30 AG18254.
Author contributions: L.K., B.D., and F.S. designed research; L.K. and B.D. performed research; L.K., B.D., and F.S. analyzed data; and L.K., B.D., and F.S. wrote the paper.
Abbreviation: DRN, degradation of mRNA in the nucleus.
References
- 1.Caponigro, G. & Parker, R. (1996) Microbiol. Rev. 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy, J. E. (1998) Microbiol. Mol. Biol. Rev. 62, 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decker, C. J. & Parker, R. (1993) Genes Dev. 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell, P. & Tollervey, D. (2000) Curr. Opin. Genet. Dev. 10, 193–198. [DOI] [PubMed] [Google Scholar]
- 5.Daugeron, M. C., Mauxion, F. & Seraphin, B. (2001) Nucleic Acids Res. 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunckley, T. & Parker, R. (1999) EMBO J. 18, 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaGrandeur, T. E. & Parker, R. (1998) EMBO J. 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, A. W. (1997) Mol. Cell. Biol. 17, 6122–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner, E. & Lykke-Andersen, J. (2002) J. Cell Sci. 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- 10.van Hoof, A., Staples, R. R., Baker, R. E. & Parker, R. (2000) Mol. Cell. Biol. 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frischmeyer, P. A., van Hoof, A., O'Donnell, K., Guerrerio, A. L., Parker, R. & Dietz, H. C. (2002) Science 295, 2258–2261. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, C. I., Bhattacharya, A., Wang, W. & Peltz, S. W. (2001) Gene 274, 15–25. [DOI] [PubMed] [Google Scholar]
- 13.Briggs, M. W., Burkard, K. T. & Butler, J. S. (1998) J. Biol. Chem. 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- 14.Bousquet-Antonelli, C., Presutti, C. & Tollervey D. (2000) Cell 102, 765–775. [DOI] [PubMed] [Google Scholar]
- 15.Burkard, K. T. & Butler, J. S. (2000) Mol. Cell. Biol. 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torchet, C., Bousquet-Antonelli, C., Milligan, L., Thompson, E., Kufel, J. & Tollervey, D. (2002) Mol. Cell 9, 1285–1296. [DOI] [PubMed] [Google Scholar]
- 17.Das, B., Butler, J. S. & Sherman, F. (2003) Mol. Cell. Biol. 23, 5502–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodsky, A. S. & Silver, P. A. (2000) RNA 6, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das, B., Guo, Z., Russo, P., Chartrand, P. & Sherman, F. (2000) Mol. Cell. Biol. 20, 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo, P., Li, W. Z., Hampsey, D. M., Zaret, K. S. & Sherman, F. (1991) EMBO J. 10, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felici, F., Cesareni, G. & Hughes, J. M. (1989) Mol. Cell. Biol. 9, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodsky, A. S. & Silver, P. A. (2002) Methods 26, 151–155. [DOI] [PubMed] [Google Scholar]
- 23.Gorlich, D. & Mattaj, I. W. (1996) Science 271, 1513–1518. [DOI] [PubMed] [Google Scholar]
- 24.Hector, R. E., Nykamp, K. R., Dheur, S., Anderson, J. T., Non, P. J., Urbinati, C. R., Wilson, S. M., Minvielle-Sebastia, L. & Swanson, M. S. (2002) EMBO J. 21, 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagnoli, P. & Bisson, L. F. (1998) Yeast 14, 359–369. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. J. & Baserga, S. J. (1999) Mol. Cell. Biol. 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myer, V. E. & Young, R. A. (1998) J. Biol. Chem. 273, 27757–27760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.