Abstract
Chronic lymphocytic leukemia (CLL) is the most common human leukemia and is characterized by predominantly nondividing malignant B cells overexpressing the antiapoptotic B cell lymphoma 2 (Bcl2) protein. miR-15a and miR-16-1 are deleted or down-regulated in the majority of CLLs. Here, we demonstrate that miR-15a and miR-16-1 expression is inversely correlated to Bcl2 expression in CLL and that both microRNAs negatively regulate Bcl2 at a posttranscriptional level. BCL2 repression by these microRNAs induces apoptopsis in a leukemic cell line model. Therefore, miR-15 and miR-16 are natural antisense Bcl2 interactors that could be used for therapy of Bcl2-overexpressing tumors.
Keywords: microRNAs, translation, leukemia
B cell lymphoma 2 (BCL2) is a central player in the genetic program of eukaryotic cells favoring survival by inhibiting cell death (1). Overexpression of Bcl2 protein has been reported in many types of human cancers, including leukemias, lymphomas, and carcinomas (2). In follicular lymphomas and in a fraction of diffuse BCLs, the mechanism of BCL2 activation was found to be the translocation t(14,18)(q32;q21), which places the BCL2 gene under the control of Ig heavy chain enhancers, resulting in deregulated expression of the gene (3, 4). B cell chronic lymphocytic leukemia (CLL) is the most frequent adult leukemia in the Western world (5), and the malignant, mostly nondividing, B cells of CLL overexpress Bcl2 (6). However, with the exception of <5% of cases in which the BCL2 gene is juxtaposed to Ig loci (7), no mechanism has been discovered to explain BCL2 deregulation in CLL.
We and others have previously reported that microRNAs (miRNAs) are a class of genes involved in human tumorigenesis (refs. 8–15 and reviewed in refs. 16 and 17). In animals, single-stranded miRNA binds specific mRNA through sequences that are imperfectly complementary to the target mRNA, mainly to the 3′ UTR. The bound mRNA remains untranslated, resulting in reduced levels of the corresponding protein or can be degraded, resulting in reduced levels of the corresponding mRNA (18, 19). Deletions and translocations involving two miRNAs, miR-15a and miR-16-1, located in a cluster at 13q14.3, and their down-regulation was found in ≈65% of B cell CLL patients (8). A germ-line mutation in miR-16-1/miR-15a primary precursor located 7 bp after the 3′ end of miR-16-1 caused low levels of miRNA expression in vitro and in vivo and was associated with deletion of the normal allele (20). The presence of pathogenic mutations in miR-15a/miR-16-1 indicate that these genes are involved in CLL, and that at least some miRNAs can function as tumor suppressor genes. To decipher the miR-15a and miR-16-1 involvement in CLL we pursued the identification of main targets with importance for human tumorigenesis.
Here, we report the identification of a mechanism of regulation of BCL2 expression in hematopoietic cancer cells consisting of posttranscriptional down-regulation by miR-15 and miR-16. This interaction has an important functional consequence: the activation of the intrinsic apoptosis pathway.
Methods
Patient Samples. For the expression study we used 26 CLL samples obtained after informed consent from patients diagnosed with CLL at the CLL Research Consortium institutions. Briefly, blood was obtained from CLL patients, and mononuclear cells were isolated through Ficoll/Hypaque gradient centrifugation (Amersham Pharmacia Biotech) and processed for RNA extraction according to described protocols. Two normal pools each containing CD5+ cells from two different normal individuals were used as normal controls. CD5+ B cells were prepared from tonsillar lymphocytes. Briefly, tonsils were obtained from patients in the pediatric age group undergoing routine tonsillectomies, after informed consent. Purified B cells were prepared by rosetting T cells from mononuclear cells with neuraminidase-treated sheep erythrocyte. To obtain CD5+ B cells, purified B cells were incubated with anti-CD5 mAb followed by goat anti-mouse Ig conjugated with magnetic microbeads as described. CD5+ B cells were positively selected by collecting the cells retained on the magnetic column MS by the Mini MACS system (Miltenyi Biotec, Auburn, CA). The degree of purification of the cell preparations was >95%, as assessed by flow cytometry.
Westen Blottings for BCL2. The levels of Bcl2 protein were quantified by using the mouse monoclonal anti-BCL2 antibody (DakoCytomation, Carpinteria, CA) and confirmed by using mouse monoclonal anti-BCL2 antibody purchased from Santa Cruz Biotechnology using standard procedures for Western blotting. The normalization was done with mouse monoclonal anti-actin antibody (Sigma). The band intensities were quantified with imagequanttl (Nonlinear Dynamics, Durham, NC).
RNA Extraction, Northern Blots, and miRNACHIP Experiments. Procedures were performed as described (8, 11, 21). Briefly, labeled targets from 5 μg of total RNA were used for hybridization on each miRNACHIP microarray chip containing 368 probes in triplicate, corresponding to 245 human and mouse miRNA genes. Raw data were normalized and analyzed in genespring software, version 7.2 (Silicon Genetics, Redwood City, CA). Expression data were median-centered by using both the genespring normalization option or the global median normalization of the bioconductor package (www.bioconductor.org) without any substantial difference. Statistical comparisons were done with both the genespring ANOVA tool and sam software (Significance Analysis of Microarray, wwwstat.stanford.edu/∼tibs/SAM/index.html). The microarray data were confirmed by Northern blottings for mir-16-1 and mir-15a as reported (11).
DNA Constructs. mir-16-1/mir-15a expression plasmids were constructed with an 832-bp genomic sequence including both mir-16-1 and mir-15a, one WT (mir-16-1-WT) and the other containing the +7(CtoT) substitution (mir-16-1-MUT), by ligating the relevant sequence in a sense orientation into a mammalian expression vector, pSR-GFP-Neo (OligoEngine, Seattle). All sequenced constructs were transfected in 293T cells by using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). The expression of both constructs was assessed by Northern blots as described. Western blotting for the GFP levels was used to show the equal efficiency of transfection with the pRS-neo-GFP constructs.
For luciferase reporter experiments a 3′ UTR segment of 546 bp of the 3′UTR of the BCL2 gene was amplified by PCR from human genomic DNA and inserted into the pGL3 control vector (Promega), using the XbaI site immediately downstream from the stop codon of luciferase. The following primer set was used to generate specific fragments: BCL2-UTRF2, 5′-CTAGTCTAGAGCCTCAGGGAACAGAATGATCAG-3′ and BCL2-UTRR2, 5′-CTAGTCTAGAAAGCGTCCACGTTCTTCATTG3′. We also generated two inserts with deletions of 9 bp (3′M1) and 5 bp (3′M2), respectively from the site of perfect complementarity by using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene). WT and mutant inserts were confirmed by sequencing.
Transfection Assays. The human megakaryocytic cell line MEG-01 was grown in 10% FBS in RPMI medium 1640, supplemented with 1× nonessential amino acid and 1 mmol sodium pyruvate at 37°C in a humified atmosphere of 5% CO2. The cells were cotransfected in 12-well plates by using siPORT neoFX (Ambion, Austin, TX) according to the manufacturer's protocol with 0.4 μg of the firefly luciferase report vector and 0.08 μg of the control vector containing Renilla luciferase, pRL-TK (Promega). For each well 10 nM mir-16-1-sense and mir-15a-sense (Dharmacon Research, Lafayette, CO) and anti-miR-16-1 and anti-mir-15a precursor miRNA inhibitor (Ambion) were used. Firefly and Renilla luciferase activities were measured consecutively by using dual-luciferase assays (Promega) 24 h after transfection.
Apoptosis Assays. Apoptosis assays were performed in duplicate experiments on transfected MEG-01 cells either with miR-15a/miR-16-1 vectors or the specific oligonucleotide RNAs. We used the Apoptotic DNA Ladder Kit (Roche Diagnostics). Immunoblot analysis was performed by standard protocols (22) with rabbit polyclonal anti-caspase-8 (Chemicon International, Temecula, CA), rabbit polyclonal anti-caspase-9 (Santa Cruz Biotecnology), rabbit polyclonal anti-apoptotic protease activating factor 1 (APAF-1) (Pharmingen), and mouse polyclonal antipoly(ADP-ribose) polymerase (PARP). For the TUNEL assay, the in situ cell death detection kit, TMR red, was used as described by the manufacturer (Roche Diagnostics).
Results
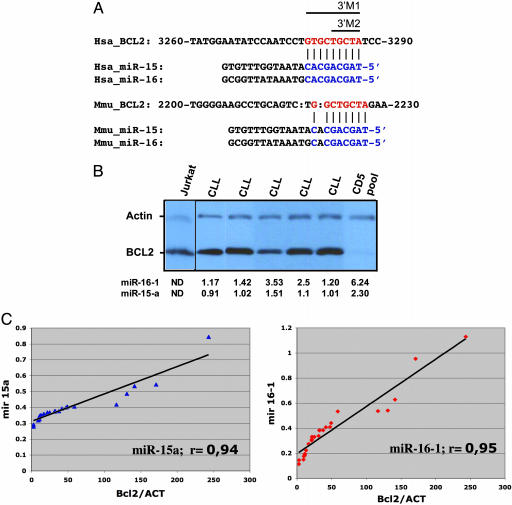
The Levels of miR-15a and miR-16-1 Are Inversely Correlated with BCL2 Protein Expression in CLL Cells. By analyzing homology between these two miRNAs and the BCL2 mRNA sequence we found that the first nine nucleotides from the 5′ ends of both miRNAs are complementary to bases 3287–3279 of the Bcl2 cDNA (clone NM_000633, Fig. 1A). This complementarity was also identified for the members of a second cluster, miR-15b/miR-16-2, from chromosome 3q26 and was conserved for the Bcl2::miR-15 and Bcl2::miR-16 Mus musculus putative interactors. To evaluate this putative interaction, we first checked for the existence of a correlation between the expression levels of miR-15a and miR-16-1 and the Bcl2 protein levels in CLL cells and normal CD5+ lymphocytes. In normal CD5+ B lymphocytes the cluster from chromosome 13q14 is highly expressed, whereas the miR-15b and miR-16-2 precursors are barely detected by Northern blots (8). By miRNA-CHIP and Western blottings we analyzed a set of 30 samples, composed of 26 CLL samples and normal CD5+ lymphocytes from tonsils of four different normal individuals. Whereas in normal CD5+ lymphoid cells the levels of both miRNAs were high and the Bcl2 protein was expressed at low levels, in the majority of leukemic cells both miR-15a and miR-16-1 were expressed at low levels and Bcl2 was overexpressed (Fig. 1B). Moreover, in all leukemia samples we found a highly concordant inverse correlation between the miRNAs and Bcl2 levels (Fig. 1C). Thus, in CLL cases we observed a concordant down-regulation of miR-15a and miR-16-1 and overexpression of Bcl2 protein.
Fig. 1.
Bcl2 protein expression is inversely correlated with miR-15a and miR-16-1 miRNAs expression in CLL patients. (A) The unique site of complementarity miR::mRNA is conserved in human and mouse and is the same for all four human members of the family. The sites of target mutagenesis are indicated (3′M1 and 3′M2). (B) In CLL patients the levels of Bcl2 protein are inversely correlated with miR-15a and miR-16-1 expression. Five different CLL cases are presented, and the normal cells were pools of CD5+ B lymphocytes. The T cell leukemia Jurkat was used as control for Bcl2 protein expression. For normalization we used β-actin. The numbers represent normalized expression on miRNACHIP. ND, not determined. (C) The coefficients of correlation in the full set of 26 samples is ≈95% for both miR-15a and miR-16-1. The normalized Bcl2 expression is on abscissa vs. miR-15a (Left) and miR-16-1 (Right) levels by miRNA chip on ordinates. ACT, β-actin.
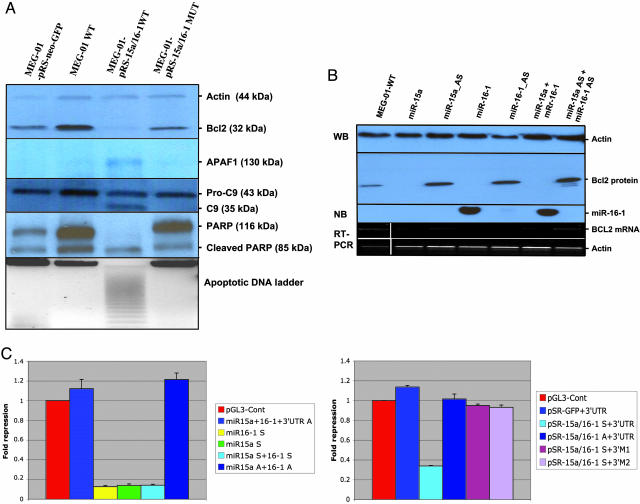
BCL2 Is a Target of Posttranscriptional Repression by miR-15 and miR-16. Then, we investigated the effects of transfection of the full cluster miR-15a/miR-16-1 on Bcl2 expression. We used a cell line with high levels of Bcl2 and no expression of these miRNAs. Because of the lack of cell lines derived from CLL patients, we selected MEG-01, a leukemia-derived cell line with the deletion of one allele and alteration of the other miR-15a/16-1 locus and no expression of miR-15a and miR-16-1 genes. We constructed a pSR-GFP-Neo expression vector by inserting an 832-bp genomic sequence that contains both miRNAs (pSR-miR-15/16-WT). The vector was transiently transfected into MEG-01 cells. The levels of Bcl2 were highly reduced (to ≈7% normalized expression) in comparison with WT MEG-01 cells or cells transfected with the empty vector (which shows similar levels of Bcl2) (Fig. 2A). To further confirm this effect, we transfected the same expression vector containing a +7(CtoT) germ-line substitution (pSR-miR-15/16-MUT) detected in cases of familial CLL (20) that strongly reduces the expression of both miRNAs. As expected, the Bcl2 levels were comparable (75% normalized expression) with that in WT cells or control-empty vector-transfected cells (Fig. 2A).
Fig. 2.
BCL2 is a target of miR-15 and miR-16.(A) Transfection of miR-15a/miR-16-1 cluster in MEG-01 BCL2+ leukemia cells is followed by a significant reduction in protein levels. Data were confirmed in duplicate experiments. The pSR-mir15/16-WT-transfected cells shows cleavage of APAF-1 (a cytochrome c interactor), pro-caspase 9 (intrinsic pathway), and PARP (a final effector of various apoptotic pathways). (B) Transfection with RNA oligos miR-15 and miR-16 separately or combined significantly reduce Bcl2 protein levels. Normalization was performed with β-actin. The Northern blot (NB) showed the miR-16-sense transfection efficiency. The same results were obtained for the other three oligos used (data not showed). The mRNA levels of the BCL2 gene in the same cells are shown and normalized against β-actin mRNA expression. WB, Western blot. (C) The 3′ UTR of BCL2 enables miR-15/16 regulation. Relative repression of firefly luciferase expression was standardized to a transfection control, Renilla luciferase. pGL-3 (Promega) was used as the empty vector. (Left) miR-15-a and miR-16-1 oligoRNAs (sense and antisense) were used for transfections. (Right) pSR-mir15/16-WT was used. Two different types of 3′ UTR mutants were constructed, one without all 9 bp of miRNA::mRNA interaction (3′M1) and the other with a deletion of the first 5 bp in the same complementarity region (3′M2). All of the experiments were performed twice in triplicate (n = 6).
Then we investigated whether both miRNAs affect Bcl2 protein expression or this effect is restricted to only one of them. We have transiently transfected miR-15a and miR-16-1 sense and antisense oligo RNAs into the same MEG-01 cell line. As shown in Fig. 2B, separate transfections of miR-15a-sense and miR-16-1-sense, respectively, completely abolished BCL2 expression, whereas transfection of antisense RNAs did not. Similar data were obtained by cotransfection with both sense RNAs or both antisense RNAs, confirming that both miR-15a and miR-16-1 influence Bcl2 protein expression.
Translocations affecting the BCL2 gene increase both messenger and protein levels (4). Because it was recently shown that miRNAs can down-regulate a specific target by affecting mRNA translation or mRNA stability (19), we tested at which level BCL2 was down-regulated by miR15a/16-1 interaction. We have evaluated mRNA levels by amplifying BCL2 cDNA from cells transfected with the pSR-miR-15/16-WT or with specific miR-15a and miR-16-1 RNA oligos by RT-PCR. No differences in the levels of BCL2 expression were observed between any of these samples and control cells (untransfected or transfected with the empty vector) (Fig. 2B). This result demonstrates that miR-15a and miR-16-1 do not affect mRNA stability and regulate Bcl2 expression at the posttranscriptional level.
Down-regulation of Bcl2 protein levels could be explained by a direct effect (miRNA::mRNA complementarity) or by an indirect interaction. It can be argued that miR-15a and miR-16-1 interact with other unknown targets that, in turn, down-regulates Bcl2 levels. To solve this issue, we fused a 536-bp 3′UTR sequence of human BCL2 interacting with the two miRNAs to a luciferase reporter gene. The presence of an interaction miRNA::mRNA would reduce the firefly luciferase activity (normalized to Renilla luciferase activity of the transfection control plasmid). The data presented in Fig. 2C argue for a direct effect, with significant repression of luciferase activity, of both miRNAs compared with control vectors. We also performed a control experiment with two types of mutated target mRNA sequences lacking all nine (3′M1) or five (3′M2) complementary bases of BCL2 cDNA. As expected, both mutants completely abolish the interaction between miR-15a and miR-16-1 and the 3′UTR of BCL2 (Fig. 2C). These data indicate that both miRNAs directly interact with the 3′UTR of BCL2.
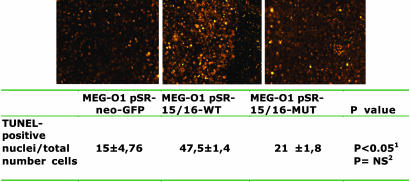
BCL2 Repression by miR-15a and miR-16-1 Induces Apoptosis in the MEG-01 Cell Line. Bcl2 is an antiapoptotic gene that is involved in an evolutionary conserved pathway crucial to apoptosis and programmed cell death: Bcl2 blocks the mitochondrial release of cytochrome c and inhibits the activation of caspase 9 by the cytoplasmic scaffolding protein Apaf-1 (23). We used three different assays to identify the biological effects of Bcl2 repression by miR-15 and miR-16. First, we showed apoptotic DNA fragments on transfected MEG-01 cells. Only the cells transfected with pSR-miR-15/16-WT and none of the controls (nontransfected WT cells, pSR-Empty, or pSR-mir-16-1-MUT transfected cells) showed the typical DNA ladder image (Fig. 2A). Then, to understand the specific apoptotic pathway that is activated, we used immunoblot analysis. The pSR-miR-15a/16-1-WT-transfected MEG-01 cells showed the activation of intrinsic apoptosis pathway as identified by activation of the APAF-1–caspase9–PARP pathway. None of the controls, including the MUT-transfected cells, showed changes in the levels of these proteins (Fig. 2A). The extrinsic pathway (caspase 8) was not activated (data not shown). The same results were also reproduced in MEG-01 cells transfected with oligoRNAs miR-15 and miR-16 (data not shown). Finally, to confirm the data at a single-cell level, we used the TUNEL assay (Fig. 3). Statistically significant more apoptotic cells were found in the MEG-01 pSR-15/16-WT-transfected cells vs. pSR-neo-GFP cells, but no difference was found between the levels of apoptosis in the latter cells and that transfected with the mutated version inducing low levels of expression of the miR-15a/miR-16-1 transcript. These data confirm that BCL2 down-regulation by miR-15 and miR-16 triggers apoptosis and that the levels of expression of these two miRNAs are important for this mechanism.
Fig. 3.
Apoptotic evaluation determined through comparison of TUNEL-positive apoptotic nuclei and total number of cells. Corresponding images to the numeric counts are presented. All values are mean ± SD for three different sets of 100 cells for each type of sample. NS, not significant. Superscript 1 indicates P values between empty vector and miR-15/16 WT. Superscript 2 indicates P values between empty vector and miR-15/16 mutated.
Discussion
In the present study we identify a mechanism of BCL2 gene expression regulation that can fully explain the inverse correlation between the expression of Bcl2 protein and miRNAs miR-15a and miR-16-1 in B cell CLL cells. Bcl2 overexpression by miRNAs miR-15a and miR-16-1 down-regulation seems to be the main regulatory mechanism involved in the pathogenesis of the major fraction of human B cell CLL. Because down-regulation of miR-15a and miR-16-1 was also reported in cases of diffuse large BCLs (15), we can postulate that the significance of this mechanism may extend to other human malignancies. In the human genome four members of the mir15/16 family are present and all have the same 9-bp Bcl2-complementarity sequence. This functional redundancy indicates the existence of a very fine mechanism to regulate Bcl2 expression. Recently, several target prediction software programs, such as targetscan (24), pictar (25), and miranda (26) identified 22 different miRNAs having as putative target BCL2, with mir15/mir16 among the highest ranks (Table 1, which is published as supporting information on the PNAS web site). These data indicate that other miRNAs could participate in the regulation of Bcl2 protein expression in various cell types and argue in favor of the proposed combinatorial miRNA target regulation, in which different combinations of miRNAs are expressed in different cell types and may coordinately regulate cell-specific target genes (25). Much more, this interaction was conserved in several species, including mouse, rat, dog, and chicken, confirming the importance of this mechanism of Bcl2 regulation during philogenetic evolution.
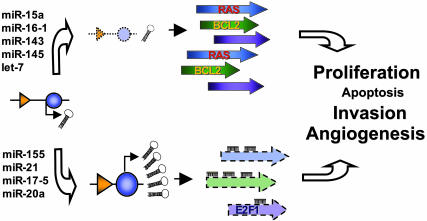
To date only a few miRNA::mRNA interactions with importance for cancer pathogenesis have been proved (27). It was elegantly demonstrated that the let-7 miRNA family regulates RAS oncogenes and that let-7 expression is lower in lung tumors than in normal lung tissue, whereas RAS protein has an inverse variation (14). Furthermore, enforced expression of the miR-17-92 cluster from chromosome 13q32-33 in conjunction with c-myc accelerates tumor development in a mouse BCL model (28). Two miRNAs from the same cluster, miR-17-5p and miR-20a, negatively regulate the E2F1 transcription factor (29), a gene proved to function as a tumor suppressor in some experimental systems (30). Putting these data together with our findings, we can expand an early proposed model of miRNA as a cancer player (10). A miRNA located in a deleted region or down-regulated in a particular human cancer (a “tumor suppressor” miRNA) can have, in fact, oncogene-like effects if the main targets for that specific cell types are oncogenes. A miRNA located in an amplified region or overexpressed in a particular human cancer (an “oncogene” miRNA) can have, in fact, tumor suppressor-like effects if the main targets for those specific cell types are tumor suppressors (Fig. 4).
Fig. 4.
miRNAs as oncogenes and/or tumor suppressors: two different looks of the same genes. A miRNA located in a deleted region or down-regulated in a particular human cancer (a tumor suppressor miRNA) can have, in fact, oncogene-like effects if the main targets for that specific cell types are oncogenes. The absence of the miRNA will induce overexpression of the oncogenic targets, the same effect as the target amplification or activation (as was proved for let-7 and RAS in lung cancers). The same miRNA, but in a different cell type, can have as main target a tumor suppressor and therefore its deletion will increase the levels of the suppressor protein, practically protecting from malignant transformation. A miRNA located in an amplified region or overexpressed in a particular human cancer (an oncogene miRNA) can have, in fact, tumor suppressor-like effects if the main targets for that specific cell types are tumor suppressors. The abundance of the miRNA will produce down-regulation of the suppressor targets, the same effect as the loss of heterozygosity directly affecting the target gene. The same miRNA, but in a different cell type, can have as main target an oncogene, and therefore its overexpression will decrease the levels of the oncogenic protein, practically protecting the cell from malignant transformation. To completely define this puzzle, it has to be considered that, depending on cell type, the same miRNAs (as proved for miR-17-5 and miR-20a) can behave as oncogene (as shown in ref. 28) or tumor suppressor (as shown in ref. 29). Furthermore, each miRNA has more than one target, and each target has more than one interacting miRNA in a specific cell type, giving rise to a very complex and intricate regulatory network. The miRNA genes are shown in blue, and the corresponding promoters are shown in orange. For simplicity, only one allele was shown and only a few mRNA–miRNA interactions are represented.
The data presented in this study are of considerable therapeutic significance because miR-15 and miR-16 are natural antisense Bcl2 interactors that could be used for therapy in tumors overexpressing Bcl2. In the MEG-01 cells transfected with pSR-miR-15/16-WT we have observed apoptosis and the cleavage of pro-caspase 9 and PARP, meaning that the reduction of Bcl2 protein levels by miRNAs is sufficient to initiate the apoptotic process. These results are encouraging in the light of promising results regarding the therapeutic potential of antisense Bcl2 as a chemosensitizer for cancer therapy (31).
In conclusion, our results reveal that the mir15/16 family negatively regulates BCL2 expression and promotes apoptosis. miR-15a and miR-16-1 contribute to malignant transformation by up-regulating Bcl2 similarly to what happens in follicular lymphomas, but by a different mechanism.
Supplementary Material
Acknowledgments
This work was supported by Program Project Grants P01CA76259, P01CA81534, and P30CA56036 from the National Cancer Institute (to C.M.C. and T.J.K.), a Kimmel Scholar award (to G.A.C.), a grant from the Italian Ministry of Public Health (to M.N. and S.V.), a grant from the Italian Ministry of University Research (to M.N. and S.V.), a grant from the Italian Association for Cancer Research (to M.N. and S.V.), and Fondo per gli Investimenti della Ricerca di Base Grant RBNE01N4Z9_004 (to S.Z.).
Author contributions: C.M.C. designed research; A.C., G.A.C., M. Fabbri, and M.V.I. performed research; M.S., S.E.W., R.A., S.Z., M.D., L.R., H.A., C.-g.L., T.J.K., and M.N. contributed new reagents/analytic tools; and M. Ferracin and S.V. analyzed data.
Abbreviations: CLL, chronic lymphocytic leukemia; miRNA, microRNA; BCL2, B cell lymphoma 2; APAF-1, apoptotic protease activating factor 1; PARP, poly(ADP-ribose) polymerase.
References
- 1.Cory, S. & Adams, J. M. (2002) Nat. Rev. 2, 647–656. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Beato, M., Sanchez-Aguilera, A. & Piris, M. A. (2003) Blood 101, 1220–1235. [DOI] [PubMed] [Google Scholar]
- 3.Tsujimoto, Y., Finger, L. R., Yunis, J., Nowell, P. C. & Croce, C. M. (1984) Science 226, 1097–1099. [DOI] [PubMed] [Google Scholar]
- 4.Tsujimoto, Y., Cossman, J., Jaffe, E. & Croce, C. M. (1985) Science 228, 1440–1443. [DOI] [PubMed] [Google Scholar]
- 5.Chiorazzi, N., Rai, K. R. & Ferrarini, M. (2005) N. Engl. J. Med. 352, 804–815. [DOI] [PubMed] [Google Scholar]
- 6.Kitada, S., Andersen, J., Akar, S., Zapata, J. M., Takayama, S., Krajewski, S., Wang, H. G., Zhang, X., Bullrich, F., Croce, C. M., et al. (1998) Blood 91, 3379–3389. [PubMed] [Google Scholar]
- 7.Adachi, M., Tefferi, A., Greipp, P. R., Kipps, T. J. & Tsujimoto, Y. (1990) J. Exp. Med. 171, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. (2002) Proc. Natl. Acad. Sc. USA 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michael, M. Z., O'Connor, S. M., van Holst Pellekaan, N. G., Young, G. P. & James, R. J. (2003) Mol. Cancer Res. 1, 882–891. [PubMed] [Google Scholar]
- 10.Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M. & Croce, C. M. (2004) Proc. Natl. Acad. Sci. USA 101, 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin, G. A., Liu, C. G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C. D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzler, M., Wilda, M., Busch, K., Viehmann, S. & Borkhardt, A. (2004) Genes Chromosomes Cancer 39, 167–169. [DOI] [PubMed] [Google Scholar]
- 13.Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., et al. (2004) Cancer Res. 64, 3753–3756. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K. L., Brown, D. & Slack, F. J. (2005) Cell 120, 635–647. [DOI] [PubMed] [Google Scholar]
- 15.Eis, P. S., Tam, W., Sun, L., Chadburn, A., Li, Z., Gomez, M. F., Lund, E. & Dahlberg, J. E. (2005) Proc. Natl. Acad. Sci. USA 102, 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McManus, M. T. (2003) Semin. Cancer Biol. 13, 253–258. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, R. I. & Shiekhattar, R. (2005) Cancer Res. 65, 3509–3512. [DOI] [PubMed] [Google Scholar]
- 18.Bartel, D. P. (2004) Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 19.Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S. & Johnson, J. M. (2005) Nature 433, 769–773. [DOI] [PubMed] [Google Scholar]
- 20.Calin, G. A., Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S., Iorio, M. V., Visone, R., Sever, N. I., Fabbri, M., et al. (2005) N. Engl. J. Med., in press. [DOI] [PubMed]
- 21.Liu, C.-G., Calin, G. A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, D. C., Shimizu, M., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel, F. M., Brent, R., Kingston, R. E., Moor, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1989) Current Protocols in Molecular Biology (Wiley, New York).
- 23.Cory, S., Huang, D. C. S. & Adams, J. M. (2003) Oncogene 22, 8590–8607. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, B. P., Burge, C. B. & Bartel, D. P. (2005) Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- 25.Krek, A., Grun, D., Poy, M. N., Wolf, R., Rosenberg, L., Epstein, E. J., MacMenamin, P., da Piedade, I., Gunsalus, K. C., Stoffel, M. & Rajewsky, N. (2005) Nat. Genet. 37, 495–500. [DOI] [PubMed] [Google Scholar]
- 26.John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C. & Marks, D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croce, C. M. & Calin, G. A. (2005) Cell 122, 6–7. [DOI] [PubMed] [Google Scholar]
- 28.He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S. W., Hannon, G. J. & Hammond, S. M. (2005) Nature 435, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. (2005) Nature 435, 839–843. [DOI] [PubMed] [Google Scholar]
- 30.Weinberg, R. A. (1996) Cell 85, 457–459. [DOI] [PubMed] [Google Scholar]
- 31.Kim, H., Emi, M., Tanabe, K. & Toge, T. (2004) Cancer 101, 2491–2502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.