Abstract
We have analyzed by ex vivo ELISPOT the anti-vaccinia cytotoxic T lymphocyte responses of peripheral blood mononuclear cells from humans vaccinated with Dryvax vaccine. More than 6,000 peptides from 258 putative vaccinia ORFs predicted to bind the common molecules of the HLA A1, A2, A3, A24, B7, and B44 supertypes were screened with peripheral blood mononuclear cells of 31 vaccinees. A total of 48 epitopes derived from 35 different vaccinia antigens were identified, some of which (B8R, D1R, D5R, C10L, C19L, C7L, F12, and O1L) were recognized by multiple donors and contain multiple epitopes recognized in the context of different HLA types. The antigens recognized tend to be >100 residues in length and are expressed predominantly in the early phases of infection, although some late antigens were also recognized. Viral genome regulation and virulence factor were recognized most frequently, whereas few structural proteins were immunogenic. Finally, most epitopes were highly conserved among vaccinia virus Western Reserve, variola major and modified vaccinia Ankara, supporting their potential use in vaccine and diagnostic applications.
Keywords: immunodominance, smallpox vaccination, virulence factors
Recent concern that variola virus could be used as a biological weapon has led to a renewed interest in smallpox vaccination programs (1-5). Because of the safety concerns with the currently used Dryvax vaccine, efforts are underway to develop safer vaccine candidates, including highly attenuated strains of vaccinia virus (VACV), such as NYVAC and modified vaccinia Ankara (MVA). VACV and variola virus are 90% homologous, and it is assumed that cross-reactive epitopes for T and B cells play a role in the protection supplied by vaccinia immunization (6). Cellular immunity is believed to be most crucial for clearance of poxvirus infections, whereas antibody responses appear to be most crucial for protection from subsequent reinfection (7).
Despite the success of VACV in smallpox eradication, the targets of cellular immunity at the cytotoxic T lymphocyte (CTL) and HTL level are largely unknown, and the correlates of protection after vaccinia immunization are uncertain. Experiments with recombinant VACVs suggested that late genes are poorly immunogenic for CTLs, and, in a recent study in mice, four of five newly identified epitopes derived from early viral genes (8, 9). In contrast, antibody responses are directed against late-expression structural proteins. Some of these antigens (B5R and L1R) have been considered for the development of subunit recombinant vaccines. Recent studies have defined five T cell epitopes restricted by murine MHC molecules (9), and three epitopes restricted by the human class I MHC molecule, HLA A*0201 (10-13). A more extensive definition of the antigens and the epitopes contained within VACV is necessary to (i) enable adequate immunogenicity testing of new vaccine candidates, (ii) help elucidate the correlates of protection offered by the current vaccine, and (iii) understand human cellular immune responses to complex viruses. To identify viral epitopes recognized by human CTL we employ a multistep process that begins with computational prediction of peptides that have the capacity of binding various HLA molecules (14, 15).
An important issue in human responses to VACV and other complex viruses is the extent to which immunodominance limits the response. In many mouse-viral models, CTL responses focus on a limited number of epitopes (sometimes one) (16, 17). In the case of VACV in H-2b mice, it appears that a large fraction of the response is specific for a single epitope (9). This issue is relevant to immune escape and bioterrorist threats. If the response elicited by vaccination were to target a single or few immunodominant epitopes or antigens, mutant viruses lacking such targets could arise, either by natural evolution or deliberate manipulation, against which the current vaccine would be ineffective.
Materials and Methods
Peptide Synthesis. Peptides were synthesized and radiolabeled by the chloramine T method as described in refs. 18 and 19.
MHC Peptide Binding Assays. Quantitative assays to measure the binding affinity of peptides to various HLA molecules are based on the inhibition of binding of a radiolabeled standard peptide and were performed as described in refs. 14, 18, and 19.
Bioinformatic Analyses. Each predicted ORF of the VACV Western Reserve strain (VACV-WR) was analyzed by using previously described algorithms (14). Peptides predicted to bind with an IC50 < 100 nM were selected for study. To reduce the number of peptides identified in large ORFs, the top 10 candidates per supertype per antigen and the 40 best scoring candidates, regardless of supertype, were selected. For the B44 supertype, peptides were selected on the basis of the presence of the specified primary anchor motif.
Characteristics of Study Population. Healthy males and females between 18 and 59 years of age were used in this study. Exclusion criteria were body weight of <45.4 kg and established pregnancy. All recruited donors had received a vaccinia virus (Dryvax) vaccination as a prophylactic measure either because of their potential exposure to vaccinia in a laboratory or hospital setting or because of their enrollment into military and health worker vaccination programs. Institutional Review Board approval and appropriate consent were obtained for this study.
Peripheral Blood Mononuclear Cell (PBMC) Isolation, HLA Typing, and Viruses. PBMCs were isolated from heparinized blood by gradient centrifugation with a Histopaque-1077 (catalogue no. H8889, Sigma) (20), and the cells were cryopreserved in liquid nitrogen in 10% DMSO/FBS. Each donor's PBMCs were typed (Forensic Analytical Molecular Genetics, San Francisco) for HLA-A, -B, -C, and -DR by high-resolution PCR. VACV-WR (9, 21) was obtained from Bernard Moss (National Institute of Allergy and Infectious Diseases, Bethesda).
Ex Vivo Primary ELISPOT Assay. In initial experiments, 2 × 105 PBMCs per well were incubated in the presence of 2, 1, and 0.5 multiplicities of infection of VACV-WR, a control pool of peptides (5 μg/ml per peptide) from commonly encountered pathogens (Epstein-Barr virus, CMV, and influenza A virus) (22, 23), or relevant HLA supertype VACV-derived peptide pools. Peptide pools that gave positive responses [>20 spot-forming cells (SFCs) for every 106 cells, stimulation index ≥ 2 and P ≤ 0.05], were deconvoluted by subsequent testing of the PBMCs against individual peptides at a final concentration of 5 μg/ml.
The ELISPOT assays were performed exactly as described in ref. 24. Responses against an irrelevant peptide (HIVgag77-85, SLYN-TVATL) were subtracted from the experimental values. To assess statistical significance, a one-tailed Student t test was performed in which the triplicate values of each condition were compared with those of the negative controls.
Results
Study Donor Population. A total of 58 donors vaccinated by arm scarification with Dryvax within 1 year of the blood draw were recruited. Of the 58 donors, 21 were first-time vaccinees, and the remainders received booster vaccinations. All individuals reported to PMA controls, and no significant differences were noted between first and booster vaccination donors. The population comprised 31% males and 69% females, with an average age of 41 years. The ethnic distribution of the study population was 86% Caucasian, 5.3% Hispanic, 3.5% African American, 3.5% Asian, and 1.7% American Indian (Table 10, which is published as supporting information on the PNAS web site). We focused on the most common HLA class I supertypes (A1, A2, A3, A24, B7, and B44). One or more of these supertypes has been shown to be present in virtually 100% of the worldwide population, irrespective of ethnicity (15). Based on high-resolution HLA typing, each donor could be assigned to one or more HLA supertype group (15). All donors expressed at least one of the HLA molecules of the six supertypes analyzed (Table 11, which is published as supporting information on the PNAS web site).
The frequencies observed for each supertype were compared with expected frequencies (Table 11), and good agreement was seen in most instances. The A24 supertype was somewhat underrepresented, whereas the B7, A1, and A3 supertypes were somewhat overrepresented in our study population, likely reflecting the relative underrepresentation of individuals with Asian ethnicity and overrepresentation of Caucasians in the volunteer pool.
Bioinformatic Analysis. In total, 6,055 peptides were selected with an average of 1,009 peptides per supertype and synthesized. The supertype for which the fewest peptides were predicted was B7, reflecting the fact that this supertype is associated with a stringent requirement for the relatively infrequent proline residue in position 2 of its peptide ligands. At the other end of the spectrum, ≈1,600 peptides were predicted in the case of the A3 supertype, likely reflecting the relative permissiveness (Table 11).
Identification of Vaccinia-Derived CTL Epitopes. As a preliminary step to the identification of the VACV-WR-derived epitopes, we identified donors whose PBMCs had a vaccinia response of >100 SFC per 106 cells. This criterion was necessary to ensure sufficient numbers of CTL that would be available for the study. Seven to 11 different donors were analyzed, with peptide pools for each supertype. Individual pools were scored positive if the response matched the following conditions: stimulation index ≥ 1.4, SFC for every 106 cells > 20, and P < 0.05 when pool (triplicates) and negative control (sextuplicates) wells were compared. A total of 16 donors (four to eight donors for each supertype) yielded positive pools (Tables 1, 2, 3, 4, 5, 6).
Table 1. Summary of identified A1 supertype epitopes.
| Donors
|
||||||
|---|---|---|---|---|---|---|
| Peptide | Sequence | V173 | V147 | V153 | V160 | V158 |
| C19L(29–38) | VSVNNVCHMY | 55 | ||||
| C10L(297–305) | SQSDTVFDY | 60 | ||||
| C10L(298–306) | QSDTVFDYY | 340 | ||||
| C12L(97–106) | VTDTNKFDNY | 52 | ||||
| VWR050(259–267) | CMLTEFLHY | 63 | ||||
| D1R(156–164) | FTIDFKLKY | 165 | ||||
| D12L(11–19) | GTHVLLPFY | 42 | 362 | |||
| B8R(139–147) | DMCDIYLLY | 208 | 41 | 590 | ||
| B8R(153–162) | FGDSKEPVPY | 38 | 38 | |||
| B8R(262–271) | FLSMLNLTKY | 56 | ||||
| C19L(104–112) | QSITRSLIY | 40 | ||||
| VACV-WR-infected PBMCs | N/A | 801 | 2,570 | 2,767 | 2,217 | 883 |
The HLA types for each donor used included the following. V173: A*2601, A*2902, B*2705, and novelB44; V147: A*0101, A*2902, B*0702, and B*5501; V153: A*1101, A*2601, B*0702, and B*0702; V160: A*0101, A*3002, B*0801, and B*4402; V158: A*0301, A*3002, B*0702, and B*4402. The number of SFCs per 106 cells is indicated.
Table 2. Summary of identified A2 supertype epitopes.
| Donors
|
|||||||
|---|---|---|---|---|---|---|---|
| Peptide | Sequence | V163 | V148 | V157 | V172 | V103 | V111 |
| C7L(74–82) | KVDDTFYYV | 102 | 92 | ||||
| N2L(93–101) | YVNAILYQI | 328 | |||||
| F12L(286–295) | NLFDIPLLTV | 198 | |||||
| F12L(404–412) | FLTSVINRV | 63 | |||||
| VWR082(18–26) | ILDDNLYKV | 415 | 337 | 120 | 205 | ||
| E2L(249–257) | KIDYYIPYV | 290 | 95 | ||||
| E9L(107–115) | FLNISWFYI | 102 | |||||
| O1L(247–255) | GLNDYLHSV | 25 | 93 | 62 | |||
| I4L(720–128) | SMHFYGWSL | 102 | |||||
| A6L(172–180) | ILSDENYLL | 148 | |||||
| D3R(342–350) | FLVIAINAM | 195 | |||||
| A26L(177–186) | YLYTEYFLFI | 63 | |||||
| A36R(1–9) | MMLVPLITV | 30 | |||||
| A55R(78–86) | YIYGIPLSL | 30 | 100 | ||||
| VACV-WR-infected PBMCs | 103 | 1,127 | 2,383 | 343 | 930 | 530 | |
The HLA types for each donor used included the following. V163: A*0201, A*0301, B*0702, and B*1501; V148: A*0201, A*0201, B*1302, and B*1501; V157: A*0201, A*0301, B*5701, and B*1501; V172: A*0201, A*0301, B*0702, and B*4402; V103: A*0201, A*0301, B*1501, and B*1501; V111: A*0101, A*0201, B*0702, and B*0702. The number of SFCs per 106 cells is indicated.
Table 3. Summary of identified A3 supertype epitopes.
| Donors
|
||||
|---|---|---|---|---|
| Peptide | Sequence | V153 | V158 | V177 |
| C12L(93–102) | KVLHVTDTNK | 60 | ||
| C9L(193–201) | ATSLDVINY | 21 | ||
| C7L(31–40) | KLKIISNDYK | 138 | ||
| C5L(158–166) | KVMFVIRFK | 26 | 63 | |
| I3L(116–124) | AVYGNIKHK | 192 | ||
| G8R(65–73) | IVFNLPVSK | 127 | ||
| J6R(332–340) | NQVKFYFNK | 108 | ||
| D1R(152–161) | KTKNFTIDFK | 35 | ||
| D5R(670–678) | YLLVKWYRK | 68 | ||
| A8R(79–88) | AVKDVTITKK | 160 | ||
| A31R(86–94) | VTSSGAIYK | 86 | ||
| B5R(154–163) | GTIAGGVCYY | 73 | ||
| B14R(74–83) | AVFKDSFLRK | 28 | ||
| VACV-WR-infected PBMCs | 2,767 | 883 | 626 | |
The HLA types for each donor used included the following. V153: A*1101, A*2601, B*0702, and B*0702; V158: A*0301, A*3002, B*0702, and B*4402; V177: A*2402, A*3303, B*0801, and B*1301. The number of SFCs per 106 cells is indicated.
Table 4. Summary of identified A24 supertype epitopes.
| Donors
|
|||||
|---|---|---|---|---|---|
| Peptide | Sequence | V165 | V177 | V136 | V109 |
| D5R(349–357) | VWINNSWKF | 37 | 161 | 45 | |
| D5R(663–672) | RYRFAFLYLI | 51 | |||
| C6L(54–63) | RYYDGNIYE | 33 | |||
| VACV-WR-infected PBMCs | 248 | 626 | 423 | 283 | |
The HLA types for each donor used included the following. V165: A*2402, A*2402, B*0702, and B*1501; V177: A*2402, A*3303, B*0801, and B*1301; V136: A*2301, A*2902, B*5101, and B*5201; V109: A*0301, A*2407, B*0101, and B*1501. The number of SFCs per 106 cells is indicated.
Table 5. Summary of identified B7 supertype epitopes.
| Donor
|
||
|---|---|---|
| Peptide | Sequence | V153 |
| C1L(102–111) | KPKPAVRFAI | 22 |
| F4L(6–14) | APNPNRFVI | 37 |
| O1L(335–344) | RPMSLRSTII | 77 |
| J6R(303–311) | MPAYIRNTL | 230 |
| D1R(686–694) | HPRHYATVM | 28 |
| VACV-WR-infected PBMCs | 2,767 |
The HLA types for the V153 donor were A*1101, A*2601, B*0702, and B*0702. The number of SFCs per 106 cells is indicated.
Table 6. Summary of identified B44 supertype epitopes.
| Donors
|
|||
|---|---|---|---|
| Peptide | Sequence | V113 | V152 |
| C3L(120–128) | GESKSYCEL | 50 | |
| G2R(181–189) | DELVDPINY | 42 | |
| B8R(110–118) | TEYDDHINL | 87 | |
| VACV-WR-infected PBMCs | 2,757 | 858 | |
The HLA types for each donor used included the following. V113: A*0101, A*6801, B*0801, and B*4001; V147: A*0301, A*0301, B*0702, and B*3701. The number of SFCs per 106 cells is indicated.
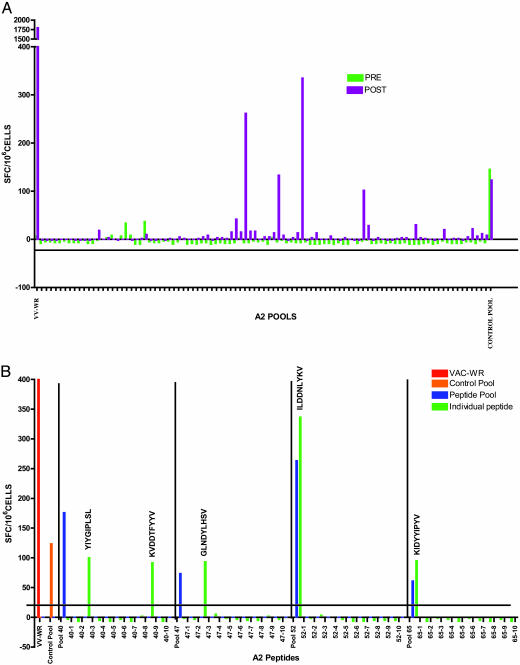
Examples of results from the screening of peptide pools against PBMCs from a vaccinated HLA-A*0201 donor is shown in Fig. 1A. The net SFC per 106 cells obtained in response to stimulation with each of the 91 pools of predicted A2 supertype binding peptides is shown. Controls included the total response to VACV-WR-infected cells (far left column), and responses to the control pool of peptides from commonly encountered pathogens (far right column). Prevaccination blood samples were also tested. Four positive peptide pools were identified by using postimmunization PBMCs. As expected, the prevaccination samples were negative except for the pool of epitopes from commonly encountered pathogens.
Fig. 1.
Response of human volunteers before and after vaccination. (A) Example of the testing of A2 supertype peptide pools in a HLA-A*0201 donor. The responses are expressed in net SFC for every 106 cells. The criteria of positivity were stimulation index ≥ 1.4, SFC for every 106 cells > 20, and P < 0.05 between pool and negative control. (B) Example of identification of specific vaccinia epitopes by deconvolution of positive pools in the same donor showed in A
Next, positive pools were deconvoluted to identify the specific peptides responsible for the activity detected within the pool. An example of the deconvolution of positive pools leading to the identification of specific vaccinia epitopes is shown in Fig. 1B.Inthe case of the four positive pools from the donor used in Fig. 1A, five epitopes were identified after deconvolution (Fig. 1B). In all cases, the response to the pool was similar to the response observed with the individual epitopes. By using this methodology for multiple donors and all supertypes, we were able to identify a total of 49 different epitopes. In the case of the A1, A2, and A3 supertypes, 11 or more different epitopes were identified for each supertype, whereas three to five epitopes were identified in the case of the A24, B7, and B44 supertypes.
Tables 1, 2, 3, 4, 5, 6 detail the results from the deconvolution experiments, listing for each supertype the epitopes identified, their sequence and antigen of origin, the donor in which the responses were observed, and the magnitude of the response(s) (SFC per 106 PBMCs). As a control, the response observed to whole-virus-infected PBMCs is also shown. Of 21 donor/supertype combinations, a slight majority (12 of 21) recognized more than one epitope, with seven donors recognizing four or more epitopes. Nine of the epitopes were recognized by more than one individual. One epitope, C7L(74-82), recognized by two different A2 donors, was previously identified by Terajima et al. (11).
Magnitude of Responses to the Identified Epitopes Compared with Total Vaccinia Responses. In a few instances, sufficient cells were available to allow testing of the PBMCs from a given donor with pools from more than one supertype. For example, in the case of donor 153 (A1, A3, and B7 supertype), peptide pools specific for all three supertypes could be tested, thus providing some insight into the complexity of the immune response to vaccination in a given individual (Table 7). We observed responses to 11 different epitopes ranging in magnitude from 21 to 230 SFCs, with a total of 752 SFC per 106 PBMCs. One epitope was identified from the A1 supertype pools, five from the B7 supertype and five from the A3 supertype. When the total response observed against all of the identified epitopes was compared with the response observed against virus-infected PBMCs, it was found that the sum of the antipeptide responses were 60.9% of the antivirus response.
Table 7. Detail of responses observed in PBMCs from donor V153.
| HLA Type | Supertype | Epitopes | Response SFC per 106 Cells |
|---|---|---|---|
| A*2601 | A1 | B8R(139–147) | 41 |
| A*1101 | A3 | A31R(85–94) | 83 |
| A31R(86–94) | 86 | ||
| B5R(154–163) | 73 | ||
| B14R(74–83) | 28 | ||
| C9L(193–201) | 21 | ||
| C5L(158–166) | 26 | ||
| B*0702 | B7 | C1L(102–111) | 22 |
| J6R(303–311) | 230 | ||
| D1R(686–694) | 28 | ||
| F4L(6–14) | 37 | ||
| O1L(335–344) | 77 |
For donor V153, the total epitope count was 752, the total VACV-WR-infected PBMC count was 1,234, and the ratio of epitopes per VACV-WR-infected PMBCs was 60.9%.
HLA Binding Capacity of the Identified Epitopes. We inferred HLA restriction of responding PBMCs based on the nature of the peptides tested. Although the scarcity of cells precluded us from performing formal HLA restriction analysis at the cellular level, we addressed the issue of HLA restriction further by testing the capacity of the identified epitopes to bind to the relevant HLA supertype molecules in vitro. Tables 12-17, which are published as supporting information on the PNAS web site, list the binding affinity of the 49 identified epitopes to common molecules of the corresponding HLA supertypes. Of a total of 53 HLA-epitope combinations, 52 (96.6%) were associated with good or intermediate binding affinity (≤500 nM), and, in one additional combination, weak binding was detected (500-5,000 nM). It was also noted that 75% of the epitopes identified bound 50% or more of the HLA molecules tested in their respective supertype.
Structural Analysis of the Antigens Recognized After Vaccination. Overall, the 49 epitopes identified were encoded by a total of 35 different protein antigens. Three antigens (D1R, D5R, and B8R) encoded three or more epitopes each, restricted by at least two different HLA molecules. Four additional antigens encoded two epitopes each. These data suggest that a subset of ORFs is relatively frequently recognized. The data also show that the VACV-WR-specific responses are directed against a rather large number of different ORFs. In fact, considering that 35 different ORFs were recognized and that a total of 258 potential ORFs were studied, it can be concluded that class I restricted responses can recognize at least 13.6% of the proteins encoded by the vaccinia virus genome.
The 35 ORFs recognized were analyzed with respect to stage of expression (early/intermediate vs. late) (21, 25-29) and function (structural protein, virulence factor, and regulation). We also analyzed the percent homology of the vaccinia epitopes with the comparable peptides in the genomes of variola major and MVA (Tables 8 and 9; see also Table 18, which is published as supporting information on the PNAS web site).
Table 8. Structural characteristics for VACV-WR antigens recognized by human cellular responses.
| Average size
|
Time of expression
|
Functional categories
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORFs | >100 aa | ≤100 aa | Total | Early/intermediate | Late | Total | Virulence factors | Structural | Viral regulation | Total |
| Recognized | 35 | 0 | 35 | 16 | 4 | 20 | 5 | 1 | 12 | 18 |
| Others | 148 | 75 | 223 | 40 | 54 | 94 | 16 | 29 | 38 | 83 |
| Total | 183 | 75 | 258 | 56 | 58 | 114 | 21 | 30 | 50 | 101 |
For size data, P = 2.08127 × 10–6; for time of expression data, P = 0.00184; and, for functional categories data, P = 0.0020.
Table 9. Homology of VACV-WR epitopes with variola major India and MVA sequences.
| Percent homology | Variola major India | MVA |
|---|---|---|
| 100 | 29 | 29 |
| 80–99 | 13 | 4 |
| 51–79 | 5 | 10 |
| ≤50 | 2 | 6 |
| Total | 49 | 49 |
Based on size, we expected larger antigens to be intrinsically richer in epitopes. Indeed, we found that all epitopes were contained in relatively large proteins (>100 residues), and no epitopes were found in the smaller antigens (<100 residues) (Table 8).
Experiments using temporal regulation of influenza hemagglutinin expression in VACV recombinants suggested that VACV late proteins are poorly immunogenic for CD8+ T cells (30). We found that, although the prediction was correct (P = 0.00184), four different late proteins were also recognized after vaccination, indicating that this correlation is not absolute (Table 8).
Although the immunogenicity of virion structural proteins might be expected to be enhanced by their relatively high levels of expression and delivery to professional antigen presentation cells during viral penetration (25, 31), we found that only one of 30 structural proteins was immunogenic (0.8%, P = 0.0020) (Table 8). By contrast, 12 of 50 proteins involved in genome replication (24%) and five of 21 (23.8%) virulence factors were immunogenic. In addition, we identified epitopes encoded by 10 genes of unknown function, providing the initial evidence for the expression of these putative ORFs. Finally, none of the ≈50 genes labeled as A or B in the VACV Copenhagen genome and which overlap other ORFs were recognized, suggesting that most of these genes are not likely to be expressed.
In Table 9, we list the percent identity of the epitopes with their closest homolog in variola major and MVA proteomes. An identical sequence is present in variola virus for 29 of the 49 epitopes (59%), and a highly homologous (80% or more) match was found in another 27% of the cases. However, for 14% of the epitopes, a much less homologous match at the level of predicted protein sequence was identified (80% or less). Similar results were noted in the case of MVA (67% of the epitopes being 80% or more homologous).
Discussion
We identified VACV-derived epitopes recognized in humans after vaccination with the Dryvax vaccine.
We report 48 previously uncharacterized epitopes restricted by various HLA molecules, thus greatly increasing the number of epitopes available for further study (10, 11). Importantly, because several epitopes restricted by molecules other than A2 were identified, our results effectively enable the study of human poxvirus cellular immunity in the general population in a non-ethnically biased fashion.
When we added the responses directed against individual epitopes in a given donor, we found that they on average accounted for approximately one-third of the response observed against vaccinia-infected PBMCs. This estimate should, however, be taken with caution. A number of factors could contribute to overestimation of CTL responses to individual peptides. To activate T cells, we used antigen-presenting cells bearing far greater amounts of peptide class I complexes than are typically generated by virus-infected cells. This concentration might reveal low avidity T cells or T cells specific for mimotope-like determinants not detected with virus-infected stimulator cells (26). The total anti-VACV response is likely to be an underestimate due to nonoptimal presentation by infected antigen-presenting cells.
Several factors might contribute to epitopes being missed by our approach. First, some epitopes not identified herein may be low-affinity MHC binders that were excluded from our study. Second, we only studied the top 10 predicted binders per antigen and thus, especially for large antigens, might have overlooked some of the epitopes. Third, some epitopes might not carry any discernable motif and/or might be missed by our predictive algorithm altogether. Fourth, epitopes may be restricted by HLA alleles falling either outside the supertypes tested or within a supertype but having distinct peptide-binding properties. Finally, some epitopes may be the result of unexpected translation initations, posttranslational modifications, or other events, such as protein splicing, which cannot be predicted by approaches based on primary sequence data (27, 28). These possibilities suggest that combining a variety of epitope mapping methods may yield the most comprehensive epitopic map.
We identified a total of 35 different vaccinia protein antigens recognized by human class I HLA-restricted responses or at least 13.6% of the putative VACV ORFs. This observation underscores the breadth of the immune response to VACV in humans. Furthermore, in most cases, no single epitope dominated the response. If the response elicited by vaccination were to target a single or a few immunodominant epitope(s) or antigen(s), mutant viruses lacking such targets could arise by natural evolution or deliberate manipulation, against which the current vaccine would be ineffective. Our results suggest that this scenario is not likely and reaffirm confidence in the current vaccine.
Our results should not be interpreted as demonstrating that no immunodominance at all exists in responses to VACV in humans. At the protein level, certain antigens, such as D1R, D5R, B8R, C10L, C19L, C7L, F12L, and O1L, were recognized by multiple donors and contained multiple epitopes restricted by multiple HLA molecules. Strikingly, the virulence factor B8R was independently and previously identified as the source of the immunodominant epitope recognized in mice of the H-2b haplotype (9).
In the case of HLA A*0201, a dominant epitope [VWR082(18-26)] was observed. Four of six donors recognized this epitope, and, in all four cases, the response to this epitope was greater than the response to all other A2-restricted epitopes recognized by the same donor. Furthermore, this epitope was also identified by vaccinia-immunized HLA-A2 transgenic mice (32).
Although eight of the 49 epitopes were recognized in two or more donors, in general, different individuals expressing the same HLA molecule appeared to recognize different epitopes, and the concordance of epitope recognition amongst study subjects shows that a HLA type was low. This effect may be related to the differences in the other HLA molecules that are coexpressed in the individual that share a particular restriction element. Recent experiments in murine systems indeed suggest that the type of MHC molecule coexpressed in a given individual can have a positive or negative influence on the breadth of the T cell repertoire restricted by a given MHC type (32). It is possible that this effect is related to the positive or negative selection modulated by the coexpressed MHC types.
Immunogenic gene products were >100 residues in size and were heavily biased toward early expression. In addition, we found an intriguing bias toward virulence factors, which might be related to their high level of synthesis early in the course of infection. If antigenic peptides are derived largely from defective ribosomal products (29), then translation rate should have a great influence on immunogenicity when T cells are activated by direct priming, e.g., by VACV-infected dendritic cells. There is no evidence that VACV encodes genes that target antigen presentation by class I MHC (33). However, VACV infection causes an efficient shutdown of most host protein synthesis, which is likely to inhibit de novo antigen presentation by the time that VACV late genes are expressed. This finding has relevance in the context of selection of potential candidate antigens for new poxvirus subunit vaccines.
We have recently performed analyses similar to those described herein in HLA A*0201, A*1101, and B7*0702 transgenic mice, leading to the identification of 21 VACV-derived epitopes. Of the epitopes identified in the transgenic mice, only three were among the 30 different epitopes identified in the present study restricted by the same MHC types. A number of factors could contribute to this discrepancy: (i) the influence on T cell receptor repertoire of the different nonrestricting HLA alleles and murine MHC alleles coexpressed in the two experimental systems, (ii) the different strains of VACV (VACV-WR versus Dryvax) and different quantities and routes of administration (i.p. injection versus dermal scarification) with which HLA transgenic mice were potentially immunized (9), (iii) potential responses detected in mouse splenocytes versus human PBMC, and (iv) changes in the number of peptide-class I complexes generated by antigen-presenting cells due to the different processing machinery of murine and human cells.
Finally, we wish to comment on the potential practical implications of our efforts. Of relevance for the utilization of vaccinia and related viruses as smallpox vaccines, we found that >70% of the epitopes identified in the current study are highly conserved in variola major and MVA. Accordingly, the epitopes identified in the present study can be used to monitor class I-restricted cellular immunity induced by different candidates of new and potentially safer smallpox vaccines. Second, this work demonstrates the feasibility of using the combined bioinformatics synthetic peptide approach to identifying epitopes in large viruses that are recognized by T cells restricted by any human class I allele.
Supplementary Material
Acknowledgments
This study was supported by National Institute of Health Contract HHSN266200400024C and National Institute of Health Grant RO1-AI-56268. D.C.T. was supported by Howard Florey Centenary Fellowship 224273 from the Australian National Health and Medical Research Council.
Author contributions: C.O., F.K., J.S., S.S., H.M.G., and A.S. designed research; C.O., F.K., J.G., and J.S. performed research; T.P., D.C.T., J.R.B., S.S., and J.W.Y. contributed new reagents/analytic tools; C.O., H.-H.B., B.P., V.P., J.S., and H.M.G. analyzed data; and C.O., V.P., D.C.T., H.M.G., and J.W.Y. wrote the paper.
Abbreviations: VACV, vaccinia virus; VACV-WR, VACV Western Reserve; MVA, modified vaccinia Ankara; CTL, cytotoxic T lymphocyte; PBMC, peripheral blood mononuclear cell; SFC, spot-forming cell.
References
- 1.Henderson, D. A. (1999) Science 283, 1279-1282. [DOI] [PubMed] [Google Scholar]
- 2.Gani, R. & Leach, S. (2001) Nature 414, 748-751. [DOI] [PubMed] [Google Scholar]
- 3.Meltzer, M. I., Damon, I., LeDuc, J. W. & Millar, J. D. (2001) Emerg. Infect. Dis. 7, 959-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Toole, T., Mair, M. & Inglesby, T. V. (2002) Clin. Infect. Dis. 34, 972-983. [DOI] [PubMed] [Google Scholar]
- 5.Smith, S. A. & Kotwal, G. J. (2002) Crit. Rev. Microbiol. 28, 149-185. [DOI] [PubMed] [Google Scholar]
- 6.Esposito, J. J. & Fenner, F. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 2885-2991.
- 7.Belyakov, I. M., Earl, P., Dzutsev, A., Kuznetsov, V. A., Lemon, M., Wyatt, L. S., Snyder, J. T., Ahlers, J. D., Franchini, G., Moss, B. & Berzofsky, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9458-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coupar, B. E., Andrew, M. E., Both, G. W. & Boyle, D. B. (1986) Eur. J. Immunol. 16, 1479-1487. [DOI] [PubMed] [Google Scholar]
- 9.Tscharke, D. C., Karupiah, G., Zhou, J., Palmore, T., Irvine, K. R., Haeryfar, S. M., Williams, S., Sidney, J., Sette, A., Bennink, J. R. & Yewdell, J. W. (2005) J. Exp. Med. 201, 95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drexler, I., Staib, C., Kastenmuller, W., Stevanovic, S., Schmidt, B., Lemonnier, F. A., Rammensee, H. G., Busch, D. H., Bernhard, H., Erfle, V. & Sutter, G. (2003) Proc. Natl. Acad. Sci. USA 100, 217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terajima, M., Cruz, J., Raines, G., Kilpatrick, E. D., Kennedy, J. S., Rothman, A. L. & Ennis, F. A. (2003) J. Exp. Med. 197, 927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder, J. T., Belyakov, I. M., Dzutsev, A., Lemonnier, F. & Berzofsky, J. A. (2004) J. Virol. 78, 7052-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Nicola, M., Carlo-Stella, C., Mortarini, R., Baldassari, P., Guidetti, A., Gallino, G. F., Del Vecchio, M., Ravagnani, F., Magni, M., Chaplin, P., et al. (2004) Clin. Cancer Res. 10, 5381-5390. [DOI] [PubMed] [Google Scholar]
- 14.Doolan, D. L., Southwood, S., Freilich, D. A., Sidney, J., Graber, N. L., Shatney, L., Bebris, L., Florens, L., Dobano, C., Witney, A. A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 9952-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sette, A. & Sidney, J. (1999) Immunogenetics 50, 201-212. [DOI] [PubMed] [Google Scholar]
- 16.Vitiello, A., Yuan, L., Chesnut, R. W., Sidney, J., Southwood, S., Farness, P., Jackson, M. R., Peterson, P. A. & Sette, A. (1996) J. Immunol. 157, 5555-5562. [PubMed] [Google Scholar]
- 17.van der Most, R. G., Sette, A., Oseroff, C., Alexander, J., Murali-Krishna, K., Lau, L. L., Southwood, S., Sidney, J., Chesnut, R. W., Matloubian, M. & Ahmed, R. (1996) J. Immunol. 157, 5543-5554. [PubMed] [Google Scholar]
- 18.Sidney, J., Southwood, S., Oseroff, C., Del Guercio, M. F., Sette, A. & Grey, H. (1998) in Current Protocols in Immunology (Wiley, San Diego), pp. 18.3.1-18.3.19. [DOI] [PubMed]
- 19.Sidney, J., Southwood, S., Mann, D. L., Fernandez-Vina, M. A., Newman, M. J. & Sette, A. (2001) Hum. Immunol. 62, 1200-1216. [DOI] [PubMed] [Google Scholar]
- 20.Kanof, M. E., Smith, P. D. & Zola, H. (1997) in Current Protocols in Immunology, eds. Coligan, J. E., Kruisbeek, A. M., Margulies, D. H., Shevach, E. M. & Strober, W. (Wiley, San Diego), Vol. 2, pp. 7.1.1-7.1.5. [Google Scholar]
- 21.McCraith, S., Holtzman, T., Moss, B. & Fields, S. (2000) Proc. Natl. Acad. Sci. USA 97, 4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothbard, J. B., Lechler, R. I., Howland, K., Bal, V., Eckels, D. D., Sekaly, R., Long, E. O., Taylor, W. R. & Lamb, J. R. (1988) Cell 52, 515-523. [DOI] [PubMed] [Google Scholar]
- 23.Currier, J. R., Kuta, E. G., Turk, E., Earhart, L. B., Loomis-Price, L., Janetzki, S., Ferrari, G., Birx, D. L. & Cox, J. H. (2002) J. Immunol. Methods 260, 157-172. [DOI] [PubMed] [Google Scholar]
- 24.Tangri, S., Ishioka, G. Y., Huang, X., Sidney, J., Southwood, S., Fikes, J. & Sette, A. (2001) J. Exp. Med. 194, 833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonini, C., Lee, S. P., Riddell, S. R. & Greenberg, P. D. (2001) J. Immunol. 166, 5250-5257. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Y., Rubtsov, A., Heiser, R., White, J., Crawford, F., Marrack, P. & Kappler, J. W. (2005) Proc. Natl. Acad. Sci. USA 102, 2476-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwab, S. R., Li, K. C., Kang, C. & Shastri, N. (2003) Science 301, 1367-1371. [DOI] [PubMed] [Google Scholar]
- 28.Hanada, K., Yewdell, J. W. & Yang, J. C. (2004) Nature 427, 252-256. [DOI] [PubMed] [Google Scholar]
- 29.Princiotta, M. F., Finzi, D., Qian, S. B., Gibbs, J., Schuchmann, S., Buttgereit, F., Bennink, J. R. & Yewdell, J. W. (2003) Immunity 18, 343-354. [DOI] [PubMed] [Google Scholar]
- 30.Bronte, V., Carroll, M. W., Goletz, T. J., Wang, M., Overwijk, W. W., Marincola, F., Rosenberg, S. A., Moss, B. & Restifo, N. P. (1997) Proc. Natl. Acad. Sci. USA 94, 3183-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yewdell, J. W., Bennink, J. R. & Hosaka, Y. (1988) Science 239, 637-640. [DOI] [PubMed] [Google Scholar]
- 32.Pasquetto, V., Bui, H.-H., Giannino, R., Mizra, F., Sidney, J., Oseroff, C., Tscharka, D. C., Irvine, K., Bennink, J. R., Peters, B., et al. (2005) J. Immunol., in press. [DOI] [PubMed]
- 33.Blake, N. W., Kettle, S., Law, K. M., Gould, K., Bastin, J., Townsend, A. R. & Smith, G. L. (1995) J. Gen. Virol. 76, 2393-2398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.