Abstract
One approach to resolving the complexities of chondrogenesis is to examine simplified systems in vitro. We analyzed cartilage differentiation by human adult stem cells from bone marrow stroma. Marrow stromal cells were cultured as micromass pellets for 21 days in serum-free medium containing transforming growth factor (TGF)-β3, dexamethasone, and bone morphogenetic protein (BMP)-6. Assays for pulse-labeled [3H]DNA and for total DNA indicated that there was little proliferation and a progressive loss of cells in the pellets. There were continuous increases in mRNAs for cartilage matrix (proteoglycans and COL2, -9, -10, and -11), receptors [fibroblast growth factor 2 (FGFR2) and parathyroid hormone-related peptide receptor (PTHrP-R)], and transcription factors (SOX5, -6, and -9) as demonstrated by histochemical and microarray assays. Reverse transcription–PCR assays for 11 mRNAs confirmed the microarray data. SOX4, vascular endothelial growth factor (VEGF), and matrix metalloproteinase 14 (MMP14) increased at day 1 and decreased thereafter, suggesting roles early in chondrogenesis. Also, forkhead, CD10, and MMP13 increased up to day 7 and decreased thereafter, suggesting roles in an intermediate stage of chondrogenesis. In addition, two collagens (COL3A1 and COL16A1), a signaling molecule (WNT11), a homeobox homolog (BAPX1), a receptor (IL-1R1), an IGFs modulator (IGFBP5), and a mettaloproteinase (MMP16) increased progressively up to about day 14, suggesting roles later in chondrogenesis. Our results indicate that the simplicity of the system makes it possible to define in detail the cellular and molecular events during chondrogenesis.
Formation of the vertebrate skeleton through endochondral bone formation is one of the most complex processes in biology. It begins with the migration of undifferentiated mesenchymal cells from the lateral plate mesoderm to sites destined to become bone. The cells undergo a condensation step and then form a cartilaginous scaffold or “mold” that defines the morphology of the bone. Cells at the center synthesize an extracellular matrix that is rich in type II collagen, proteoglycans, and related macromolecules. They then hypertrophy and undergo apoptosis as the matrices are gradually replaced by an invasion of blood vessels, followed by the appearance of osteoblasts that synthesize bone matrix (1).
One approach to resolving the complexities of chondrogenesis is to examine simplified systems in vitro. This approach has been in part accomplished through use of organ cultures of embryonic limbs and long bones (1–3). It has also been in part accomplished by disassociation of embryonic mesenchymal cells that are pelleted to form micromass cultures that then differentiate into cartilaginous modules (4, 5). However, the cells in embryonic limbs and embryonic mesenchyme may have been preprogrammed for chondrogenesis in the lateral plate mesoderm and imprinted by a number of in vivo signals such as circulating levels of retinoic acid, which can in itself direct the patterning of cartilage. In the present report, we have examined molecular events in a simplified system in which adult stem cells from bone marrow stroma (6, 7) are differentiated into cartilage in vitro (8, 9). The system has the advantage that the initial cells are early progenitors that can differentiate into multiple lineages including osteoblasts, adipocytes, myotubes, and neural cells. It also has the advantage that the culture conditions can be precisely defined and adequate numbers of cells generated to examine the time sequence of cellular and molecular events in detail.
Materials and Methods
Isolation and Cultures of Human Marrow Stromal Cells (MSCs).
To isolate human MSCs, 2- to 10-ml bone marrow aspirates were taken from the iliac crest of normal adult donors after informed consent and under a protocol approved by an institutional review board. Nucleated cells were isolated with a density gradient (Ficoll-Paque, Pharmacia) and resuspended in complete culture medium (αMEM, GIBCO/BRL; 20% FBS, lot-selected for rapid growth of MSCs, Atlanta Biologicals, Norcross, GA; 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine, GIBCO/BRL). All of the nucleated cells were plated in 25 ml medium in a culture dish and incubated at 37°C with 5% CO2. After 24 h, nonadherent cells were discarded, and adherent cells were thoroughly washed twice with PBS. The cells were incubated for 5–7 days, harvested with 0.25% trypsin and 1 mM EDTA for 5 min at 37°, and replated at six cells/cm2 in an intercommunicating system of culture flasks (6,320 cm2; Cell Factory, Nunc). After 12 days, the cells (passage 1) were harvested with trypsin/EDTA, suspended at 1 × 106 cells/ml in 5% DMSO and 30% FBS, and frozen in 1-ml aliquots in liquid nitrogen. To expand a culture, a frozen vial of MSCs was thawed, plated in a 58-cm2 culture dish, and incubated for 4 days (passage 2). The cells were harvested and diluted for further expansion by plating at initial densities of 6 or 50 cells/cm2 in a 175-cm2 culture dish. The cells were harvested after 7 days (passage 3). In this study, MSCs derived from two females were used.
Pellet Culture.
For chondrocyte differentiation, a pellet culture system was used (8). Approximately 200,000 MSCs (passage 3) were placed in a 15-ml polypropylene tube (Falcon), and centrifuged to pellet. The pellet was cultured at 37°C with 5% CO2 in 500 μl of chondrogenic media that contained 500 ng/ml bone morphogenetic protein-6 (BMP-6; R & D Systems; ref. 9) in addition to high-glucose DMEM supplemented with 10 ng/ml transforming growth factor (TGF)-β3, 10−7 M dexamethasone, 50 μg/ml ascorbate-2-phosphate, 40 μg/ml proline, 100 μg/ml pyruvate, and 50 mg/ml ITS+ Premix (Becton Dickinson; 6.25 μg/ml insulin, 6.25 μg/ml transferrin, 6.25 ng/ml selenious acid, 1.25 mg/ml BSA, and 5.35 mg/ml linoleic acid). The medium was replaced every 3 to 4 days for 21 days. For microscopy, the pellets were embedded in paraffin, cut into 5-μm sections, and stained with toluidine blue sodium borate.
RNA Collection.
Total RNA was prepared from 2 million undifferentiated MSCs at day 0, from 10 pellets at 1 day, and from 30 pellets each at 7, 14, and 21 days. Pellets incubated 7 days or longer were digested with 3 mg/ml collagenase, 1 mg/ml hyaluronidase, and 0.25% trypsin for about 3 h at 37°C. Total RNA was extracted by using RNAqueous Kit (Ambion, Austin, TX).
Microarray.
Experimental procedures for GeneChip microarray were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA; ref. 10). In brief, 5 μg of total RNA was used to synthesize double-stranded DNA (Superscript Choice System/GIBCO/BRL Life Technologies, Rockville, MD). The DNA was purified by using phenol/chloroform extraction with Phase Lock Gel (Eppendorf) and concentrated by ethanol precipitation. In vitro transcription was performed to produce biotin-labeled cRNA by using a BioArray HighYield RNA Transcription Labeling Kit (Enzo Diagnostics) according to the manufacturer's instructions. Biotinylated cRNA was cleaned with an RNeasy Mini Kit (Qiagen, Chatsworth, CA), fragmented to 50 to 200 nucleotides, and hybridized 16 h at 45°C to Affymetrix HG-U95A array, which contains approximately 12,000 human genes. After being washed, the array was stained with streptavidin-phycoerythrin (Molecular Probes). Staining signal was amplified by biotinylated anti-streptavidin (Vector Laboratories) and by second staining with streptavidin-phycoerythrin, and then scanned on a Hewlett–Packard GeneArray Scanner. The expression data were analyzed by using Affymetrix microarray suite v4.0 and Affymetrix data mining tool v2.0. Signal intensities of all probe sets were scaled to the target value of 2,500. All experiments were done in duplicate, and average signal intensities of the experiment pairs were used in fold change calculations (11).
Fold changes (FC) were calculated by using a formula that is used in Affymetrix microarray suite software. In brief, signal intensity (SI) of the comparison gene was subtracted from the SI of the baseline gene (day 0). The resulting difference in signal intensity (DSI) was then divided by the smaller of either the SI of the comparison or the baseline genes. To avoid false FC values between −1 and +1, the following corrections were made: If the DSI was positive, +1 was added to the initial value of the FC. If the DSI was negative, 1 was subtracted from the initial value. To eliminate extremely high FC values in cases in which one of the SI values was smaller than the highest noise level of the two subject arrays multiplied by correction factor 2.8, that particular SI was replaced by the corrected noise level in FC calculation. Fold changes greater than five were shown.
Reverse Transcription (RT)-PCR.
RNA was converted to cDNA and amplified by the Titan One Tube RT-PCR System (Roche Molecular Biochemicals), according to the manufacturer's recommendations. RT was performed by a 30-min incubation at 50°C, followed by a 2-min incubation at 94°C to inactivate the RT. PCR amplification conditions for the resulting cDNAs was performed by 35 cycles of 94°C for 30 s, 58°C for 45 s, and 68°C for 45 s, in which the 68°C step was increased by 5 s every cycle after 10 cycles. The reaction products were resolved by electrophoresis on a 2% agarose gel and visualized with ethidium bromide. PCR primers were as follows: COL2A1 (forward) 5′-TTCAGCTATGGAGATGACAATC-3′, COL2A1 (reverse) 5′-AGAGTCCTAGAGTGACTGAG-3′ (472 bp); COL10A1 (forward) 5′-CACCAGGCATTCCAGGATTCC-3′, COL10A1 (reverse) 5′-AGGTTTGTTGGTCTGATAGCTC-3′ (825 bp); cartilage oligomeric protein (COMP) (forward) 5′-TGGGCCCGCAGATGCTTC-3′, COMP (reverse) 5′-AGGTTTGTTGGTCTGATAGCTC-3′ (474 bp); link protein (forward) 5′-CCTATGATGAAGCGGTGC-3′, link protein (reverse) 5′-TTGTGCTTGTGGAACCTG-3′ (618 bp); SOX4 (forward) 5′-CAAACCAACAATGCCGAGAAC-3′, SOX4 (reverse) 5′-CTCTTTTTCTGCGCCGGTTTG-3′ (584 bp); SOX5 (forward) 5′-AGCCAGAGTTAGCACAATAGG-3′, SOX5 (reverse) 5′-CATGATTGCCTTGTATTC-3′ (619 bp); SOX6 (forward) 5′-ACTGTGGCTGAAGCACGAGTC-3′, SOX6 (reverse) 5′-TCCGCCATCTGTCTTCATACC-3′ (562 bp); SOX9 (forward) 5′-GAACGCACATCAAGACGGAG-3′, SOX9 (reverse) 5′-TCTCGTTGATTTCGCTGCTC-3′ (631 bp); parathyroid hormone-related peptide (PTHrP) (forward) 5′-CTCGGAGCGTGTGAACATTCC-3′, PTHrP (reverse) 5′-CTTCCGGAAAGTTGATTCCAC-3′ (216 bp); PTHrP-R (forward) 5′-AGGAACAGATCTTCCTGCTGCA-3′, PTHrP-R (reverse) 5′-TGCATGTGGATGTAGTTGCGCGT-3′ (571 bp); BMP-2 (forward) 5′-CAGAGACCCACCCCCAGCA-3′, BMP-2 (reverse) 5′-CTGTTTGTGTTTGGCTTGAC-3′ (688 bp); BMP-6 (forward) 5′-CTCGGGGTTCATAAGGTGAA-3′, BMP-6 (reverse) 5′-ACAGCATAACATGGGGCTTC-3′ (412 bp); and β-ACTIN (forward) 5′-CCAAGGCCAACCGCGAGAAGATGAC-3′, β-ACTIN (reverse) 5′-AGGGTACATGGTGGTGCCGCCAGAC-3′ (587 bp).
DNA Assays.
To prelabel MSCs, they were plated at 50 cells/cm2 in 175-cm2 dishes and incubated in complete medium. After 5 days, [3H]thymidine (1 μCi (1 Ci = 37 GBq)/ml; NEN) was added to the medium, and the incubation was continued for 2 more days. The medium was removed, the cells were washed 3 times with PBS, and the cells were used to prepare micromass cultures. DNA was prepared from 4 million undifferentiated MSCs as day 0, and from 40 pellets each at 7, 14, and 21 days. To digest the matrix, pellets differentiated 7 days or more were incubated with 3 mg/ml collagenase, 1 mg/ml hyaluronidase, and 0.25% trypsin for about 3 h at 37°C. To isolate DNA, the cells were digested in 100 mM NaCl, 10 mM Tris⋅HCl (pH 8), 25 mM EDTA (pH 8), 0.5% SDS, and 0.1 mg/ml protease (Qiagen) at 50°C for 24 h. DNA was extracted with phenol/chloroform/isoamyl alcohol. The concentrations of DNA were measured by using diphenylamine reaction (12). Forty microliters of diphenylamine reagent (100 mg of diphenylamine and 275 μl of H2SO4 in 10 ml of glacial acetic acid) and 20 μl of DNA solution were boiled at 100°C for 20 min, and the absorbance at 595 nm was measured. To measure the [3H]thymidine, 100 ng of DNA was assayed in a scintillation counter.
Results
Time Course for Synthesis of Cartilage by MSCs.
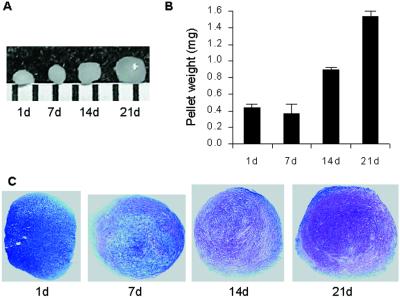
MSCs were pelleted into micromasses and then differentiated in serum-free medium in the presence of TGF-β3, dexamethasone (8), and BMP-6 (9). Immediately after centrifugation, the cells appeared as a flattened pellet at the bottom of the tube. One day later, the pellet had a thickened lip. Between day 2 and day 7, the pellet became spherical without any increase in size. As the pellet grew in size (Fig. 1A), it changed from white and opaque to a glistening transparent structure. Also, the weight of the pellets increased about 4-fold (Fig. 1B). In addition, a cartilage matrix was synthesized, as demonstrated by the appearance of proteoglycans stained with toluidine blue sodium borate (Fig. 1C) or safranin O (9), and the cell density decreased.
Figure 1.
Time course for synthesis of cartilage by MSCs. (A) Macro picture of differentiating MSC pellet at days 7, 14, and 21. A 1-mm scaled ruler is shown. (B) Wet weight of pellets at days 1, 7, 14, and 21. Data are expressed as mean ± SD (n = 3). (C) Histology of paraffin sections stained with toluidine blue sodium borate. The purple color is indicative of proteoglycans, and the blue color is background. Proteoglycan content increased and cell density decreased throughout the course of differentiation.
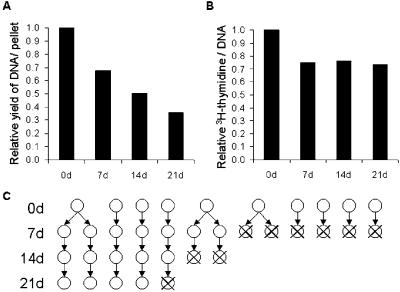
Assays of total DNA indicated a progressive loss of cells as MSCs differentiated into chondrocytes, so that the total DNA content on day 21 was only about 40% of the initial content (Fig. 2A). A pulse-labeled and chase experiment with [3H]thymidine indicated that there was a 25% decrease in the specific activity of cellular DNA between day 0 and day 7, but thereafter the values remained constant (Fig. 2B). The results were consistent with about a 30% increase in cell number between day 0 and day 7, possibly explained by the remaining effects of fetal bovine serum in the early cultures. Thereafter, there was a continued loss of cells (Fig. 2C), apparently through apoptosis (1).
Figure 2.
Cell proliferation and death during chondrocyte differentiation. (A) DNA per pellet; prepared from 20 pellets at day 0, and from 40 pellets each at 7, 14, and 21 days. The amount of DNA decreased throughout the course of differentiation. (B) Specific activity of [3H]DNA per 100 ng DNA in cells prelabeled with [3H]thymidine. This result suggests MSCs proliferate between day 0 and day 7 but not thereafter. (C) Representative model of MSCs proliferation and death during chondrocyte differentiation in pellet culture system.
Time Sequence of Gene Expression Assayed by Microarray.
To define the time sequence in gene expression, RNA was extracted from the pellets on days 0, 1, 7, 14, and 21, and assayed with a microarray containing probes for 12,000 human genes. The intensity differences observed were used to calculate fold changes as recommended by the manufacturer (Affymetrix). All values are presented here as fold changes relative to the data for day 0. The mRNAs on day 21 for 10 genes increased over 100-fold, about 100 genes increased over 10-fold, and about 1,000 genes increased over 2-fold. The 10 genes that increased over 100-fold or more were all genes that coded for known constituents of the cartilage matrix. There was about a 500-fold increase in expression of mRNA from the COL2A1 gene for type II collagen, the major protein component of cartilage. There were smaller increases in mRNAs for COL9A3, -10A1, -11A1, and -11A2, collagens that are less abundant in cartilage. There were also large increases in the mRNAs for aggrecan, which is abundant in cartilage and for cartilage oligomeric protein (COMP) and Matrilin-3 (Table 1).
Table 1.
Time sequence of gene expression assayed by microarray
| GenBank | Gene | Fold
changes vs. day 0
|
|||
|---|---|---|---|---|---|
| 1 day | 7 day | 14 day | 21 day | ||
| Collagens | |||||
| L10347 | COL2A1 | 1.0 | 49.0 | 358.8 | 530.7 |
| X14420 | COL3A1 | 2.0 | 11.0 | 12.1 | 9.2 |
| X54412 | COL9A1 | 2.6 | 2.4 | 6.1 | 12.7 |
| M95610 | COL9A2 | 1.0 | 27.6 | 63.4 | 68.0 |
| L41162 | COL9A3 | 1.0 | 25.5 | 104.4 | 172.7 |
| X60382 | COL10A1 | 1.4 | 17.6 | 66.8 | 115.8 |
| J04177 | COL11A1 | 5.8 | 122.0 | 217.0 | 243.5 |
| AL031228 | COL11A2 | 1.0 | 6.0 | 47.7 | 119.1 |
| M92642 | COL16A1 | 1.1 | 4.1 | 5.6 | 4.0 |
| Proteoglycans and other matrix components | |||||
| L32137 | COMP | 2.0 | 103.7 | 233.9 | 234.9 |
| Z22865 | Dermatopontin | 1.3 | 88.8 | 159.4 | 193.0 |
| AJ001047 | Matrilin-3 | 1.0 | 4.8 | 34.3 | 140.9 |
| X17406 | Aggrecan | 1.0 | 30.3 | 89.8 | 115.8 |
| U05291 | Fibromodulin | 1.0 | 7.6 | 47.6 | 100.0 |
| AB000114 | Osteomodulin | 1.0 | 12.5 | 38.4 | 58.6 |
| U21128 | Lumican | 1.4 | 20.5 | 55.8 | 55.4 |
| U96769 | Chondroadherin | 1.0 | 1.0 | 10.7 | 47.5 |
| J05213 | Sialoprotein precursor | 1.0 | 1.0 | 3.8 | 45.8 |
| M28439 | Keratin type 16 | 1.0 | 30.5 | 24.9 | 26.3 |
| U41344 | PRELP | 2.4 | 8.0 | 16.6 | 24.4 |
| AB011792 | ECM2 | 1.2 | 18.6 | 36.1 | 23.6 |
| M55172 | Aggrecan core protein | −2.1 | 8.7 | 17.2 | 19.1 |
| AB006000 | Chondromodulin I precursor | 1.0 | 1.0 | 3.3 | 16.9 |
| AB011792 | ECM1 | 1.0 | 13.9 | 19.8 | 16.8 |
| U59111 | Dermatan sulfate proteoglycan 3 | 1.0 | 1.3 | 4.6 | 13.1 |
| U90441 | Prolyl 4-hydroxylase alpha 2 | 2.8 | 3.7 | 5.9 | 8.1 |
| U69263 | Matrilin-2 precursor | 1.0 | 3.1 | 3.8 | 5.4 |
| SOX genes | |||||
| X70683 | SOX4 | 7.9 | 5.3 | 3.3 | 1.7 |
| S83308 | SOX5 | 1.0 | 1.7 | 3.4 | 5.0 |
| Z46629 | SOX9 | 2.9 | 11.9 | 9.9 | 14.3 |
| HOX genes | |||||
| M97676 | HOXA7 | −1.7 | 2.7 | 5.1 | 3.1 |
| Hedgehog-related | |||||
| L38517 | IHH | 2.9 | 1.0 | 1.0 | 5.9 |
| U43148 | Patched | 1.9 | 1.5 | 2.6 | 6.4 |
| U17418 | PTHrP-R | 1.5 | 21.2 | 70.4 | 127.1 |
| WNT-related | |||||
| Y12692 | WNT11 | 1.0 | 1.3 | 4.4 | 8.7 |
| L37882 | Frizzled-2 | −2.5 | −3.1 | −7.3 | −16.0 |
| Other transcription factors | |||||
| AF032885 | Forkhead | 25.4 | 76.1 | 58.5 | 62.6 |
| Z50781 | hDIP | 18.6 | 16.3 | 13.1 | 29.2 |
| M98539 | Prostaglandin D2 | 8.3 | 43.5 | 21.3 | 14.1 |
| L13463 | G0S8 | 3.9 | 14.1 | 10.3 | 13.5 |
| AF009801 | BAPX1 | 1.4 | 1.0 | 2.0 | 5.0 |
| Cadherins & CAMs | |||||
| L34059 | Cadherin 4 | −2.4 | −2.5 | −5.9 | −7.7 |
| U59289 | Cadherin 13 | 1.1 | −1.5 | −6.3 | −7.6 |
| CDs | |||||
| J03779 | CD10 | 3.8 | 7.6 | 2.8 | 2.0 |
| L33930 | CD24 | 2.4 | 17.5 | 27.8 | 22.8 |
| AF039917 | CD39L3 | 1.0 | 1.0 | 2.5 | 7.2 |
| L05424 | CD44 | −1.6 | −2.7 | −7.1 | −5.7 |
| Integrins | |||||
| M59911 | Integrin alpha 3 | −1.8 | −7.4 | −10.4 | −6.6 |
| IGFs | |||||
| X57025 | IGF1 | 1.0 | 2.9 | 5.4 | 4.2 |
| M65062 | IGFBP5 | 10.6 | 19.7 | 29.8 | 26.1 |
| M62402 | IGFBP6 | 1.6 | −1.4 | −4.2 | −6.1 |
| ILs | |||||
| M27492 | IL1R1 | 7.7 | 8.6 | 9.3 | 8.4 |
| M83667 | NF-IL6beta | 5.7 | 9.2 | 7.8 | 5.8 |
| TGF-β superfamily | |||||
| M22489 | BMP2 | 4.3 | 11.6 | 5.6 | 5.4 |
| X14885 | TGF-β3 | 5.0 | 5.4 | 5.2 | 3.4 |
| EGFs and related | |||||
| AF024710 | VEGF | 8.3 | 3.2 | 2.3 | 3.7 |
| X94216 | VEGF-C | −3.2 | −5.6 | −9.7 | −11.2 |
| X00588 | EGFR-precursor | 1.2 | −2.0 | −6.4 | −10.2 |
| FGFs | |||||
| M27968 | FGF2 | −8.7 | −8.7 | −8.4 | −5.2 |
| M37825 | FGF5 | −11.5 | −11.5 | −11.5 | −11.5 |
| Z71929 | FGFR2 | 1.1 | 3.0 | 14.7 | 14.9 |
| Retinols | |||||
| X00129 | RBP | 1.0 | 5.1 | 23.1 | 44.6 |
| SMAD | |||||
| AB004922 | SMAD3 | −2.8 | −5.5 | −5.5 | −5.5 |
| MMPs and related | |||||
| X75308 | MMP13 | 1.0 | 6.3 | 1.6 | 1.1 |
| X83535 | MMP14 | 6.2 | 2.2 | 1.7 | 1.3 |
| D83646 | MMP16 | 1.0 | 4.6 | 6.4 | 9.3 |
Among the genes that increased significantly were three SOX genes that had previously been implicated in cartilage differentiation. SOX9 increased about 12-fold by day 7, decreased slightly on day 14, and increased again on day 21. The results were consistent with the previous observations that expression of SOX9 is an early event in cartilage differentiation (13–15). SOX5 is thought to cooperate with SOX9 for cartilage differentiation (16) and, in our results, SOX5 increased continuously. However, the data also revealed that SOX4 increased about 8-fold on day 1 and then decreased progressively on days 7, 14, and 21 (Table 1), indicating the increase in expression of SOX4 preceded that of SOX5 and SOX9.
The gene for prolyl-4-hydroxylase alpha 2 increased progressively (see Proteoglycans and other matrix components in Table 1), an observation consistent with requirement of this subunit for the hydroxylation of prolyl residues during the synthesis of collagens (17). The gene for BMP-2 peaked with a 10-fold increase on day 7 (see TGF-beta superfamily in Table 1), an observation consistent with the role of BMPs in cartilage differentiation. Expression of the gene for PTH/PTHrP receptor (PTHrP-R) also increased progressively so that, by day 21, it was about 130-fold higher than its initial value (see Hedgehog-related in Table 1).
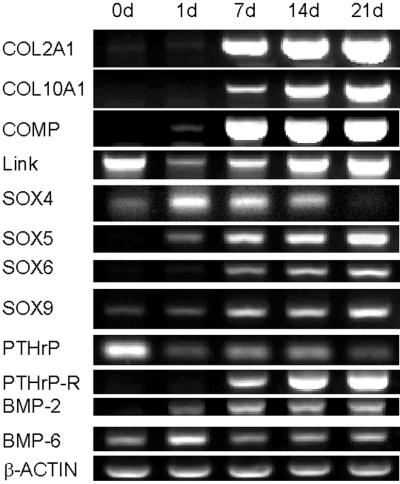
Confirmations by RT-PCR Assays.
RT-PCR assays (Fig. 3) of parallel samples were consistent with the microarray data. The expression of COL2A1, COL10A1, and COMP increased progressively beginning on about day 7. The mRNA for the link protein of large proteoglycans, an mRNA that was not present on the microarray, was expressed on day 0 and decreased on day 1, and then it increased progressively. As indicated by the microarray data, SOX4 increased markedly on day 1 and then decreased progressively. SOX5, SOX6, and SOX9 were expressed at later stages. The data were also in agreement with the microarray assays in that there was a decrease in the level of SOX9 between day 7 and day 14. The gene for PTHrP was expressed at high levels on day 0 and then decreased progressively as the gene for the receptor increased. Also, consistent with the microarray data, the gene for BMP-2 increased to day 7 and then decreased slightly on days 14 and 21. Expression of BMP-6, a gene not assayed on the microarray, peaked on day 1 and then was expressed at a lower level throughout the time course.
Figure 3.
Time sequence of gene expression assayed by RT-PCR. RT-PCR was used to further verify the data from the microarray (COL2A1, COL10A1, COMP, SOX4, SOX5, SOX9, PTHrP-R, BMP-2) and to examine the expression of genes that were not on microarray chip (link protein, SOX6, PTHrP, and BMP-6).
Time Course for Expression of Other Genes of Interest.
Further examination of the microarray data are shown in Table 1, which presents genes whose expression altered 5-fold or greater. COL3A1, a fibril-forming collagen, and COL16A1, a FACIT collagen, peaked on day 14. The data for proteoglycans and related matrix components indicated that, in addition to the marked increases in the expression of genes for aggrecan, COMP, and matrilin-3 (Fig. 3), there were large increases in dermatopontin, fibromodulin, osteomodulin, lumican, chondroadherin, sialoprotein precursor, keratin type 16, proline arginine-rich end leucine-rich repeat protein (PRELP), extracellular matrix protein 2 (ECM2), aggrecan core protein, chondromodulin-1 precursor, ECM1, dermatan sulfate proteoglycan 3, and matrilin-2 precursor.
Among HOX genes, HOXA7, which is expressed in prevertebrae from T3 to T13 at 12.5 p.c (18), increased 5-fold on day 14. Among hedgehog and related genes, Indian hedgehog (IHH) and its receptor, patched, peaked on day 21. Among WNT related genes, there was a progressive increase in WNT11, which is expressed in perichondrium of the developing skeleton (19), a progressive decrease in a frizzled-2, which had been reported to be expressed in fetal kidney and lung, and in adult colon and ovary (20). Among other transcription factors, there were marked increases in forkhead, which is required for skeletal tissue development and regulated by BMPs (21); hDIP, which is a potential transcriptional regulator related to murine TSC-22 and Drosophila shortsighted (22); prostaglandin D2, which functions as a neuromodulator and/or trophic factor in the central nervous system (23); G0S8, which is a basic helix-loop-helix phosphoprotein gene and represents a common consequence of dysregulated growth (24); and BAPX1, which is a homeobox gene and plays a critical role in embryonic development of the axial skeleton and spleen (25).
Among cell adhesion genes, there were progressive decreases in cadherin4, which had been reported to be expressed in the brain, and cadherin13, which also is called H-cadherin for its high expression in the heart (26). Among other surface proteins, CD10, which is also known as common acute lymphocytic leukemia antigen (CALLA), or enkephalinase, which is particularly abundant in the kidney (27), peaked on day 7. In addition, CD24, which has been implicated in both activation and differentiation of B lymphocytes (28), peaked on day 14. CD39L3, which is also known as eNTPDase-3 and is one of the extracellular enzymes that hydrolyze nucleotides (29), peaked on day 21. There was a marked decrease in CD44 and integrin alpha 3. In the family of IGF genes, IGF1 and IGFBP5 peaked on day 14, and IGFBP6 decreased on day 21. Among IL genes, IL1R1 peaked on day 14 and NF-IL6β, which is also known as CCAAT/enhancer-binding protein beta (CEBPB) or transcription factor 5 (TCF5) for transcriptional activator in the immune and inflammatory responses (30), peaked on day 7. In the TGF-β superfamily, TGF-β3, one of the components of the differentiating medium, increased.
Among EGFs and related proteins, vascular endothelial growth factor (VEGF) increased beginning on day 1 accompanied by decrease in VEGF-C, a related ligand of the FLT4 receptor that does not bind VEGF (31), and a decrease in EGFR-precursor. Among FGFs, there were decreases in FGF2 and FGF5 (32) and an increase in FGFR2 on days 14 and 21. Among retinols, retinol binding protein (RBP) increased markedly during differentiation, but there were no significant changes in a number of other related genes. Among the SMAD genes, expression of SMAD3, which transduces TGF-β signaling (33), decreased on day 7. Among the MMPs and related proteins, MMP-13, which is also known as collagenase3, peaked on day 7, and MMP14, which is also called MT-MMP1, peaked on day 1. There was a progressive increase of MMP16, which is also called MT-MMP3 and which induces the activation of pro-gelatinase A (34).
Discussion
Understanding chondrogenesis will obviously require examining the process in several different experimental systems, any one of which will have some limitations. The system used here has the advantage that it uses a relatively homogeneous population of undifferentiated cells (6, 7) that can be extensively differentiated to chondrocytes under defined conditions (8, 9). Therefore, the system is free of the important but complex influences that ectodermal and mesodermal cells, such as those in the apical epidermal ridge (AER) or zone of polarizing activity (ZPA), have on chondrogenesis during limb development. Also, because the initial MSCs are early progenitors that can differentiate into multiple cell lineages, the cells may not be significantly affected by any preprogramming that occurs in the lateral plate mesoderm and that may influence chondrogenesis during limb development or in aggregates of mesenchymal cells isolated from limb buds.
The robustness of chondrogenesis in the system was reflected by the progressive and marked increases in mRNAs for the known constituents of cartilage matrix. The microarray technology used here does not allow quantitative comparisons between the levels of expression of different genes, but the data does make it possible to compare the fold changes with time and differentiation in the expression of multiple genes. With chondrogenesis, it is clear that the largest increases in gene expression must occur with genes that code for the macromolecules of the cartilage matrix and that account for a major part of the bulk of the tissue. In the data obtained here, the time sequences for expression of the genes for the cartilage matrix were in general consistent with previous analyses of developing cartilage. One of the few exceptions was the appearance of mRNA for COL10A1 before the chondrocytes demonstrated significant hypertrophy.
Chondrogenesis by the MSCs was induced by artificially condensing the cells by centrifugation and the addition of TGF-β3, dexamethasone (8), and BMP-6 (9) to the medium. The medium requirements are consistent with previous observations on the effects of TGF-βs and BMPs on chondrogenesis in other systems (35). The condensation phase was accompanied by a decrease in expression of genes for cadherin 4 and 13. At the same time, there was a large increase in expression of CD24, a member of a large family of ATP-binding cassette genes encoding a family of transport proteins not previously linked to chondrogenesis (36).
The expression of SOX genes was shown to occur early in chondrogenesis (14, 37–39). Surprisingly, the SOX4 gene (40), which has received little attention, was expressed much earlier than the SOX5, SOX6, and SOX9 genes, which have been extensively studied (16).
One of the most unexpected observations was a lack of marked changes in a long list of developmental genes that have been assigned important roles in chondrogenesis in other systems. The genes on the list include a series of HOX genes; the hedgehog genes Shh; the Wnt genes, except for a late increase in WNT11; the frizzled genes, except for a decrease in the frizzled-2; and the SMAD genes, except for a decrease in SMAD3. One major exception was a very large and progressive increase in expression of the PTHrP-R that is part of the cycle in which Ihh synthesized by prehypertrophic chondrocytes increases synthesis of PTHrP by endochondral cells, and then PTHrP binds to its receptor on the prehypertrophic chondrocytes to inhibit their hypertrophy. The lack of impressive changes in a variety of developmental genes may be explained by the fact that small changes in the mRNA levels from these development genes may have major effects. An alternate explanation is that impressive changes in the genes were not seen because of the simplicity of the system, in which there is limited regulation of the morphology of the structure formed. The simplicity of the system, however, is part of its value as an experimental tool in relating the time sequence of cellular and molecular events to the differentiation of the cells and the assembly of the cartilage matrix.
Acknowledgments
This work was supported in part by National Institutes of Health Grants AR47796 and AR44210, the Oberkotter Foundation, HCA–the Health Care Company, and the Louisiana Gene Therapy Research Consortium.
Abbreviations
- MSC
marrow stromal cell
- BMP
bone morphogenetic protein
- TGF
transforming growth factor
- FC
fold change
- DSI
difference in signal intensity
- RT
reverse transcription
- PTHrP
parathyroid hormone-related peptide
- COMP
cartilage oligomeric protein
References
- 1.DeLise A M, Stringa E, Woodward W A, Mello M A, Tuan R S. In: Methods in Molecular Biology: Developmental Biology Protocols. Tuan R S, Lo C W, editors. Totowa, NJ: Humana; 2000. pp. 359–376. [Google Scholar]
- 2.Irvine K D, Vogt T F. Curr Opin Cell Biol. 1997;9:867–876. doi: 10.1016/s0955-0674(97)80090-7. [DOI] [PubMed] [Google Scholar]
- 3.Niswander L. Curr Opin Genet Dev. 1997;7:530–536. doi: 10.1016/s0959-437x(97)80082-2. [DOI] [PubMed] [Google Scholar]
- 4.Ahrens P B, Solursh M, Reiter R S. Dev Biol. 1977;60:69–82. doi: 10.1016/0012-1606(77)90110-5. [DOI] [PubMed] [Google Scholar]
- 5.Oster G F, Murray J D, Maini P K. J Embryol Exp Morphol. 1985;89:93–112. [PubMed] [Google Scholar]
- 6.Prockop D J. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 7.Pittenger M F, Mackay A M, Beck S C, Jaiswal R K, Douglas R, Mosca J D, Moorman M A, Simonetti D W, Craig S, Marshak D R. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 8.Johnstone B, Hering T M, Caplan A I, Goldberg V M, Yoo J U. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 9.Sekiya I, Colter D C, Prockop D J. Biochem Biophys Res Commun. 2001;284:411–418. doi: 10.1006/bbrc.2001.4898. [DOI] [PubMed] [Google Scholar]
- 10.Lipshutz R J, Fodor S P, Gingeras T R, Lockhart D J. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 11.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 12.Burton K. Biochem J. 1956;62:315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 14.Wright E, Hargrave M R, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre V, de Crombrugghe B. Matrix Biol. 1998;16:529–540. doi: 10.1016/s0945-053x(98)90065-8. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre V, Li P, de Crombrugghe B. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko K I. J Biol Chem. 1997;272:17342–17348. doi: 10.1074/jbc.272.28.17342. [DOI] [PubMed] [Google Scholar]
- 18.Knittel T, Kessel M, Kim M H, Gruss P. Development (Cambridge, UK) 1995;121:1077–1088. doi: 10.1242/dev.121.4.1077. [DOI] [PubMed] [Google Scholar]
- 19.Lako M, Strachan T, Bullen P, Wilson D I, Robson S C, Lindsay S. Gene. 1998;219:101–110. doi: 10.1016/s0378-1119(98)00393-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z, Lee C C, Baldini A, Caskey C T. Genomics. 1995;27:370–373. doi: 10.1006/geno.1995.1060. [DOI] [PubMed] [Google Scholar]
- 21.Nifuji A, Miura N, Kato N, Kellermann O, Noda M. J Bone Miner Res. 2001;16:1765–1771. doi: 10.1359/jbmr.2001.16.10.1765. [DOI] [PubMed] [Google Scholar]
- 22.Vogel P, Magert H J, Cieslak A, Adermann K, Forssmann W G. Biochim Biophys Acta. 1996;1309:200–204. doi: 10.1016/s0167-4781(96)00177-7. [DOI] [PubMed] [Google Scholar]
- 23.Shibanuma M, Kuroki T, Nose K. J Biol Chem. 1992;267:10219–10224. [PubMed] [Google Scholar]
- 24.Wu H K, Heng H H, Shi X M, Forsdyke D R, Tsui L C, Mak T W, Minden M D, Siderovski D P. Leukemia. 1995;9:1291–1298. [PubMed] [Google Scholar]
- 25.Tribioli C, Lufkin T. Development (Cambridge, UK) 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 26.Lee S W. Nat Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 27.Letarte M, Vera S, Tran R, Addis J B, Onizuka R J, Quackenbush E J, Jongeneel C V, McInnes R R. J Exp Med. 1988;168:1247–1253. doi: 10.1084/jem.168.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duperray C, Boiron J M, Boucheix C, Cantaloube J F, Lavabre-Bertrand T, Attal M, Brochier J, Maraninchi D, Bataille R, Klein B. J Immunol. 1990;145:3678–3683. [PubMed] [Google Scholar]
- 29.Chadwick B P, Frischauf A M. Genomics. 1998;50:357–367. doi: 10.1006/geno.1998.5317. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita S, Akira S, Kishimoto T. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Gray A, Yuan J, Luoh S M, Avraham H, Wood W I. Proc Natl Acad Sci USA. 1996;93:1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hebert J M, Rosenquist T, Gotz J, Martin G R. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 33.Sakou T, Onishi T, Yamamoto T, Nagamine T, Sampath T, Ten Dijke P. J Bone Miner Res. 1999;14:1145–1152. doi: 10.1359/jbmr.1999.14.7.1145. [DOI] [PubMed] [Google Scholar]
- 34.Takino T, Sato H, Shinagawa A, Seiki M. J Biol Chem. 1995;270:23013–23020. doi: 10.1074/jbc.270.39.23013. [DOI] [PubMed] [Google Scholar]
- 35.Hanada K, Solchaga L A, Caplan A I, Hering T M, Goldberg V M, Yoo J U, Johnstone B. J Cell Biochem. 2001;81:284–294. doi: 10.1002/1097-4644(20010501)81:2<284::aid-jcb1043>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 36.Schriml L M, Dean M. Genomics. 2000;64:24–31. doi: 10.1006/geno.1999.6102. [DOI] [PubMed] [Google Scholar]
- 37.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 38.Sekiya I, Koopman P, Tsuji K, Mertin S, Harley V, Yamada Y, Shinomiya K, Nifuji A, Noda M. J Cell Biochem. 2001;81,Suppl.:71–78. doi: 10.1002/jcb.1077. [DOI] [PubMed] [Google Scholar]
- 39.Sekiya I, Koopman P, Tsuji K, Mertin S, Harley V, Yamada Y, Shinomiya K, Nifuji A, Noda M. J Endocrinol. 2001;169:573–579. doi: 10.1677/joe.0.1690573. [DOI] [PubMed] [Google Scholar]
- 40.Reppe S, Rian E, Jemtland R, Olstad O K, Gautvik V T, Gautvik K M. J Bone Miner Res. 2000;15:2402–2412. doi: 10.1359/jbmr.2000.15.12.2402. [DOI] [PubMed] [Google Scholar]