Abstract
Nitrogenase is a multicomponent metalloenzyme that catalyzes the conversion of atmospheric dinitrogen to ammonia. For decades, it has been generally believed that the [8Fe-7S] P-cluster of nitrogenase component 1 is indispensable for nitrogenase activity. In this study, we identified two catalytically active P-cluster variants by activity assays, metal analysis, and EPR spectroscopic studies. Further, we showed that both P-cluster variants resemble [4Fe-4S]-like centers based on x-ray absorption spectroscopic experiments. We believe that our findings challenge the dogma that the standard P-cluster is the only cluster species capable of supporting substrate reduction at the FeMo cofactor and provide important insights into the general mechanism of nitrogenase catalysis and assembly.
Keywords: MoFe protein, VFe protein
Nitrogenase catalyzes one of the most remarkable chemical transformations in biological systems, the reduction of atmospheric dinitrogen to a bioavailable form, ammonia. Three classes of nitrogenase systems, namely, the Mo-, V-, and Fe-only nitrogenases, have been identified (for recent reviews see refs. 1–11).f Although encoded by different structural genes, all three nitrogenases are comprised of two essential component metalloproteins, component 1 (MoFe, VFe, or FeFe protein) and component 2 (Fe protein).g The homodimeric Fe protein of the Mo-nitrogenase has one [4Fe-4S] cluster bridged between the two subunits, whereas the α2β2-tetrameric MoFe protein contains two unique metal clusters per αβ-subunit: the [8Fe-7S] P-cluster (14), which is located at the αβ-interface, and the [Mo-7Fe-9S-X-homocitrate]h FeMo cofactor (FeMoco), which is situated within the α-subunit. ATP-dependent electron transfer is believed to proceed from the [4Fe-4S] cluster of the Fe protein to the P-cluster of the MoFe protein and finally to FeMoco where substrate reduction takes place. The Fe protein of the V-nitrogenase shares a high degree of homology with the Fe protein of the Mo-nitrogenase and, apart from the presence of an additional γ-subunit, the VFe protein is also believed to be highly homologous to the MoFe protein with respect not only to the structural genes encoding the α- and β-subunits, but also to the redox centers contained within the protein, designated the P-cluster and the FeV cofactor (FeVco), respectively, in this case (4, 10, 16). The structurally homologous heterometal centers, i.e., FeMoco and FeVco, are also categorically termed cofactors.
It is generally believed that the structural integrity of the P-cluster is indispensable for nitrogenase reactivity. However, the exact mechanism by which the P-cluster carries out its function in substrate reduction and, in particular, the oxidation states and structural conformations of the P-cluster involved in this process, remain largely a puzzle (17). Two types of cofactor-deficient MoFe or VFe protein variants, generated by nifH or nifB deletion,i respectively, have been used to study the features of P-clusters without the interference of the cofactor centers (17–25). Recent studies have shown that a MoFe protein variant generated by nifB deletion contains normal P-clusters (22), whereas a MoFe protein variant generated by nifH deletion has catalytically inactive P-cluster precursors comprised of [4Fe-4S]-like fragments (19). These results, apparently, further support the dogma in the field that a fully assembled P-cluster is essential for enzyme activity. Here, we report the observation of catalytic activity in two cofactor-deficient, P-cluster variant-containing species of component 1 upon FeMoco insertion. These two species are (i) ΔnifH Av1′, a MoFe protein variant isolated from a nifH-deletion Azotobacter vinelandii strain after preincubation with Fe protein and MgATP and (ii) ΔnifB Av1V, a VFe protein variant isolated from a nifB-deletion A. vinelandii strain (for designation of strains and protein species related to this work see Table 1). We believe that our findings challenge the concept that the standard [8Fe-7S] P-cluster is the only species responsible for nitrogenase reactivity and provide insights into the general mechanisms of nitrogenase assembly and catalysis.
Table 1. Nomenclature of strains and proteins of A. vinelandii related to this work.
| Strain | Strain designation | Source | Genotype | Designation of component 1* | Designation of component 2* |
|---|---|---|---|---|---|
| Mo-nitrogenase | |||||
| WT | 26 | WT | Av1 | Av2 | |
| DJ1143 | AvDJ1143 | 21 | nifB deletion, His-tagged MoFe protein | ΔnifB Av1 | |
| DJ1165 | AvDJ1165 | 17 | nifH deletion, His-tagged MoFe protein | ΔnifH Av1 | |
| DJ1165 | AvDJ1165 | This study | nifH deletion, His-tagged MoFe protein, preincubated with Av2 and MgATP | ΔnifHAv1′ | |
| V-nitrogenase | |||||
| WT | 27-29 | WT | Av1v | Av2v | |
| YM7A | AvYM7A | This study | nifB deletion, His-tagged VFe protein | ΔnifB Av1v |
The component proteins of the Mo-nitrogenase are designated by the initials of the organism from which they are isolated and the Arabic numeral of the component. For example, the MoFe and Fe proteins of the Mo-nitrogenase of A. vinelandii are designated Av1 and Av2, respectively. This nomenclature is also applied to the V-nitrogenase, with the addition of a superscript V. Therefore, the VFe and Fe proteins of the V-nitrogenase of A. vinelandii are abbreviated as Av1v and Av2v, respectively.
Materials and Methods
Unless otherwise noted, all chemicals and reagents were obtained from Fisher Scientific, Baxter Scientific Products (McGaw Park, IL), or Sigma.
Construction of Variant A. vinelandii Strains. Construction of A. vinelandii nifB- and nifH-deletion strains DJ1143 (AvDJ1143) and DJ1165 (AvDJ1165), which produce His-tagged MoFe proteins designated ΔnifB Av1 and ΔnifH Av1, respectively, has been described (17, 21). A. vinelandii strain DJ1258 (AvDJ1258), which produces a His-tagged WT VFe protein, was a generous gift from Dennis Dean (Virginia Polytechnic Institute and State University, Blacksburg). A. vinelandii nifB-deletion strain YM7A (AvYM7A), which produces a His-tagged VFe protein designated ΔnifB Av1V, was constructed by introducing a deletion/kanamycin resistance cassette insertion in the nifB gene loci of AvDJ1258 following procedures as described (30, 31). For A. vinelandii strains related to this work see Table 1.
Cell Growth and Protein Purification. All A. vinelandii strains were grown and harvested as described (17, 30, 31). Published methods were used for the purification of Av2 (32), ΔnifB Av1, and ΔnifB Av1V (17, 30, 31). Preincubated ΔnifH Av1 (designated ΔnifH Av1′) was purified as described (17) except that the crude extract prepared from 500 g of cells of AvDJ1165 was incubated with 100 mg of purified Fe protein (Av2) and 100 ml of reaction mixture (0.8 mM ATP, 1.6 mM MgCl2, 10 mM creatine phosphate and 80 units/ml of creatine phosphokinase) for 30 min at room temperature before being loaded on a Zn(II)-charged immobilized metal affinity chromatography column. For designation of protein species related to this work see Table 1.
EPR Spectroscopy. All EPR samples were prepared in a Vacuum Atmospheres (Hawthorne, CA) dry box with an oxygen level of <2 ppm. Unless noted otherwise, all samples were in 25 mM Tris·HCl (pH 8.0), 10% glycerol, and 2 mM Na2S2O4. Protein samples were oxidized by incubation with excess indigo disulfonate (IDS) for 30 min. Subsequently, IDS was removed by a single passage over an anion-exchange column as described (33). Samples were either used as they were or concentrated in Centricon-30 (Amicon) in anaerobic centrifuge tubes outside of the dry box. All perpendicular- and parallel-mode EPR spectra were recorded with a Bruker ESP 300 Ez spectrophotometer, interfaced with an Oxford Instruments (Pleasanton, CA) ESR-9002 liquid helium continuous flow cryostat. All spectra were recorded at 10 K by using a microwave power of 50 mW, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. The microwave frequencies of 9.62 and 9.39 GHz were used for the perpendicular-mode (10 scans) and parallel-mode (144 scans) EPR spectra, respectively. Spin quantitation of the S = 1/2 perpendicular-mode EPR signal was carried out under nonsaturating conditions with Cu(II)EDTA as the standard, using the procedures described by Aasa and Vänngård (34). This method has been applied to and outlined for quantitation of similar spin systems in earlier nitrogenase studies (17, 20, 31). Quantitation numbers in this study represent the average of three independently purified protein batches.
Activity Assays, Metal Analysis, and Visible Region Spectroscopy. All nitrogenase activity and FeMoco insertion assays were carried out as described (26, 31, 35). The products H2 and C2H4 were analyzed as described (36). Ammonium was determined by a HPLC fluorescence method (37). Molybdenum (38) and iron (39) were determined as described. Vanadium was analyzed by inductively coupled plasma-MS at the chemical analysis facility of the University of Georgia (Athens). Visible region absorption experiments were carried out as described (17). Acid-labile sulfur was analyzed as described (40, 41).
X-Ray Absorption Spectroscopy (XAS) Data Collection and Analysis. Samples for XAS [60 μl of 33 mg/ml of ΔnifB Av1, 19 mg/ml of ΔnifH Av1, 46 mg/ml of ΔnifH Av1′, and 57 mg/ml of ΔnifB Av1V in 25 mM Tris·HCl (pH 8.0), 200 mM imidazole, 500 mM NaCl, 2 mM dithionite, and 50% glycerol] were loaded into 70 μl of Lucite cells with Kapton tape windows and flash-frozen in a pentane/liquid nitrogen slush. XAS data were measured at the Stanford Synchrotron Radiation Laboratory under 3-GeV, 70- to 100-mA beam conditions by using beam line 9-3, with a Si(220) double-crystal monochromator. During data collection, the samples were maintained at 10 K in an Oxford Instruments CF1208 continuous-flow liquid He cryostat. Data were collected as Fe Ka fluorescence by using a Canberra (Meriden, CT) 30-element solid-state Ge array detector. The x-ray energy was calibrated to the inflection point at 7,111.3 eV of a standard Fe foil measured concurrent with the samples. Seventeen (ΔnifH Av1′), 22 (ΔnifB Av1 and ΔnifB Av1V), or 26 (ΔnifH Av1′) scans to k of 16.5 Å–1 were collected. The edge position at ≈7,119 eV was monitored for evidence of photoreduction; no changes were observed in any sample. The XAS data were normalized by using the program xfit (42) by first subtracting a polynomial background absorbance that was fit to the preedge region and extended over the postedge with weighted control points, followed by fitting a three-segment polynomial spline over the extended x-ray absorption fine structure (EXAFS) region, and finally by normalizing the resulting spectra to an edge-jump of 1.0 at 7,130 eV. Fourier transforms of the EXAFS data in the k-range from 2 to 15.5 Å–1 were calculated with the program exafspak (43).
Results
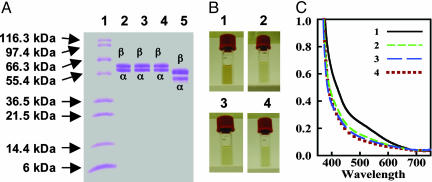
Using the fast one-step purification method described (17, 30, 31), up to ≈750 mg of His-tagged ΔnifH Av1′ and ΔnifB Av1V were purified from 250 g of cells of AvDJ1165 and AvYM7A, respectively. Like ΔnifB Av1 and ΔnifH Av1 (Fig. 1A, lanes 2 and 3), both ΔnifH Av1′ and ΔnifB Av1V (Fig. 1 A, lanes 4 and 5) are composed of α-subunits (≈56 and ≈54 kDa, respectively) and β-subunits (≈59 and ≈55 kDa, respectively).j The molecular masses of ΔnifH Av1′ and ΔnifB Av1V are ≈220 and ≈160 kDa, respectively, based on their elution profiles on a gel filtration Sephacryl S-200 HR column (data not shown). This observation, in combination with results from quantitative analysis of the SDS/PAGE and N-terminal amino acid sequencing (data not shown), indicates a subunit composition of α2β2 for ΔnifH Av1′ and αβ2k for ΔnifB Av1V (Table 2). Additional small subunits, encoded by nafY and vnfG, have been reported to be associated with various species of Av1 and Av1V, respectively (23, 27, 45, 46). However, no additional subunits are detectable in ΔnifH Av1′ or ΔnifB Av1V based on SDS/PAGE (Fig. 1 A, lanes 4 and 5) and quantitative N-terminal amino acid sequencing (data not shown). The absence of additional subunits can also be observed in the cases of ΔnifB Av1 and ΔnifH Av1 (Fig. 1 A, lanes 2 and 3) and may be a common trait of the His-tagged versions of these proteins. Interestingly, the lack of the nafY-encoded γ-subunit from ΔnifB Av1 does not affect the catalytic or structural properties of the protein (21, 22), which strongly suggests that γ is not essential for enzymatic activity or protein assembly in this species.
Fig. 1.
SDS/PAGE, color, and visible region spectra of ΔnifB Av1, ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1V.(A) Coomassie-stained 10–20% gradient SDS/PAGE. Lane 1, 10 μg of protein standard; lane 2, 15 μg of purified ΔnifB Av1; lane 3, 15 μg of purified ΔnifH Av1; lane 4, 15 μg of purified ΔnifH Av1′; and lane 5, 15 μg of purified ΔnifB Av1V.(B) Anaerobic cuvettes filled with dithionite-reduced ΔnifB Av1 (part 1), ΔnifH Av1 (part 2), ΔnifH Av1′ (part 3), and ΔnifB Av1V (part 4). The protein concentrations of all samples were 5 mg/ml. (C) Visible region spectra of samples shown in B. Spectra of dithionite-reduced ΔnifB Av1 (line 1), ΔnifH Av1 (line 2), ΔnifH Av1′ (line 3), and ΔnifB Av1V (line 4) are shown between 350 and 750 nm. The samples were prepared as described in Materials and Methods.
Table 2. Subunit compositions, metal contents, and S = 1/2 EPR signal features.
| Metal content
|
S = 1/2 EPR signal
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Subunit composition | Fe, mol metal/mol protein | Mo,* mol metal/mol protein | V,* mol metal/mol protein | g values | Spin integration, mol spin/mol αβ-dimer | ||
| ΔnifB Av1 | α2β2† | 15.4 ± 0.8 | <0.01 | <0.01 | None | |||
| ΔnifH Av1 | α2β2† | 15.5 ± 0.9 | <0.01 | <0.01 | 2.06 | 1.92 | 1.89 | 0.4 ± 0.1 |
| ΔnifH Av1′ | α2β2† | 15.4 ± 0.4 | <0.01 | <0.01 | 2.06 | 1.92 | 1.89 | 1.1 ± 0.2 |
| ΔnifB Av1v | αβ2‡ | 8.5 ± 0.4 | <0.01 | <0.01 | 2.08 | 1.92 | 1.89 | 0.9 ± 0.1 |
WT Av1 and WT Av1v protein standards show metal contents of 2.0 ± 0.1 Mo and 1.8 ± 0.2 V, respectively.
The α- and β-subunits are encoded by nifD and nifK, respectively. Determination of the subunit composition of the protein is based on quantitative analysis of the SDS/PAGE (Fig. 1A) and the elution profiles on the Sephacryl S-200 HR gel filtration column (data not shown).
The α- and β-subunits are encoded by vnfD and vnfK, respectively. Determination of the subunit composition of the protein is based on quantitative analysis of the SDS/PAGE (Fig. 1A), quantitative analysis of the N-terminal amino acid sequence (data not shown), and the elution profiles on the Sephacryl S-200 HR gel filtration column (data not shown).
Like ΔnifB Av1 and ΔnifH Av1, ΔnifH Av1′ and ΔnifB Av1V do not contain cofactors. This observation is consistent with the absence of molybdenum or vanadium from the proteins (Table 2), as well as the absence of the typical S = 3/2 EPR signals arising from the FeMoco or FeVco centers of Av1 and Av1V, respectively (data not shown). The α2β2-tetrameric ΔnifH Av1′, like ΔnifB Av1 or ΔnifH Av1, contains ≈16 mol Fe/mol protein, whereas the αβ2-trimeric ΔnifB Av1V contains ≈8 mol Fe/mol protein (Table 2). Therefore, it can be assumed that all four protein species have eight Fe per complete αβ-subunit pair.
Despite having presumably the same Fe content per αβ-dimer, the colors of ΔnifH Av1′ and ΔnifB Av1V are similar to that of ΔnifH Av1 and are clearly distinguishable from that of ΔnifB Av1. In contrast to the reddish-brown color of ΔnifB Av1, which is consistent with earlier reports (17, 21), ΔnifH Av1′ or ΔnifB Av1V, like ΔnifH Av1 (17), is light brown (Fig. 1B). The apparent difference in color is further supported by the visible range absorption spectra of these proteins. The spectrum of ΔnifB Av1 shows a broad shoulder in the ≈475- to 575-nm range (17, 21), whereas the spectrum of ΔnifH Av1′ or ΔnifB Av1V, like that of ΔnifH Av1 (17), is essentially featureless throughout the entire visible region (Fig. 1C). These observations indicate that ΔnifH Av1′ and ΔnifB Av1V, like ΔnifH Av1 (19), contain metal centers different from the P-clusters in ΔnifB Av1.
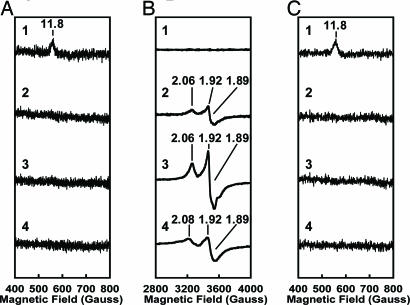
It has been established that the PN state of the P-cluster can be two-electron-oxidized by dyes like IDS to the P2+ state (21), which is then recognized by a g = 11.8 parallel-mode EPR signal (21, 47, 48). Such a signal is observed in the case of the IDS-oxidized ΔnifB Av1 (Fig. 2A, line 1), yet absent in the case of the IDS-oxidized ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1V, even at a protein concentration 10-fold higher than that of ΔnifB Av1 (Fig. 2 A, lines 2–4). Meanwhile, ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1V exhibit an S = 1/2 perpendicular-mode EPR signal in the dithionite-reduced state (Fig. 2B, lines 2–4), which integrates to 0.4, 1.1, or 0.9 spin per αβ-dimer, respectively (Table 2). This signal is absent in the case of ΔnifB Av1 because the dithionite-reduced PN state of the P-cluster is EPR silent in this region (Fig. 2B, line 1). These results strongly suggest that ΔnifH Av1′ and ΔnifB Av1V do not contain normal P-clusters.
Fig. 2.
EPR spectroscopy of ΔnifB Av1, ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1V.(A) Parallel-mode EPR spectra of IDS-oxidized ΔnifB Av1 (line 1), ΔnifH Av1 (line 2), ΔnifH Av1′ (line 3), and ΔnifB Av1V (line 4). (B) Perpendicular-mode EPR spectra of dithionite-reduced ΔnifB Av1 (line 1), ΔnifH Av1 (line 2), ΔnifH Av1′ (line 3), and ΔnifB Av1V (line 4). (C) Parallel-mode EPR spectra of IDS-oxidized, FeMoco-reconstituted ΔnifB Av1 (line 1), ΔnifH Av1 (line 2), ΔnifH Av1′ (line 3), and ΔnifB Av1V (line 4). The protein concentrations of parallel-mode EPR samples were 5, 50, 50, and 50 mg/ml for ΔnifB Av1, ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1V, respectively. The protein concentrations of all perpendicular-mode EPR samples were 10 mg/ml. All spectra were measured as described in Materials and Methods. The g values are indicated.
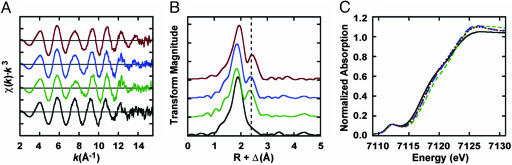
Consistent with the EPR spectroscopic data, XAS provides further evidence of the absence of normal P-clusters in either ΔnifH Av1′ or ΔnifB Av1V. In an earlier study, the absence of a distinct Fe-Fe scattering peak in the Fourier transform of ΔnifB Av1 was identified as being a signature of the all-ferrous P-cluster structure (19). The presence of this peak at ≈2.4 Å in the Fourier transforms of ΔnifH Av1′ and ΔnifB Av1V indicates that these proteins do not contain standard P-clusters (Fig. 3B). Based on the similarity of the EXAFS (Fig. 3A) and Fourier transforms (Fig. 3B) of ΔnifH Av1′ and ΔnifB Av1V to those of ΔnifH Av1, the structures of the clusters in ΔnifH Av1′ and ΔnifB Av1V are expected to be similar to those described previously for ΔnifH Av1, which was shown to contain [4Fe-4S]-like centers (19). The shift in the edge position of the absorption spectra of ΔnifH Av1′ and ΔnifB Av1V, by 0.4 and 0.6 eV, respectively, relative to that of ΔnifH Av1, indicates that, although structurally similar, the clusters of ΔnifH Av1′ and ΔnifB Av1V are in a more reduced state than that of ΔnifH Av1 (Fig. 3C). Despite being shifted relative to ΔnifH Av1, the absorption edges of ΔnifH Av1′ and ΔnifB Av1V are still higher in energy than that of ΔnifB Av1, which contains a standard all-ferrous P-cluster. Taken together, the x-ray absorption edge and EXAFS data provide clear evidence that ΔnifH Av1′ and ΔnifB Av1V contain P-cluster variants, presumably comprising [4Fe-4S]-like fragments.
Fig. 3.
Iron K-edge EXAFS (A), nonphase-shifted EXAFS Fourier transforms (B), and x-ray absorption edge spectra (C) for ΔnifB Av1 (black), ΔnifH Av1 (green), ΔnifH Av1′ (blue), and ΔnifB Av1V (red). The samples were prepared and measured as described in Materials and Methods.
Surprisingly, despite the fact that ΔnifH Av1′ and ΔnifB Av1V do not contain normal P-clusters, these proteins are active in terms of H2 evolution, C2H2 reduction, and N2 fixation upon FeMoco insertion (Table 3).l With all tested substrates, ΔnifH Av1′ shows consistent activities of ≈10% of the ΔnifB Av1 standard, whereas ΔnifB Av1V shows H2 evolution activities of up to ≈79% of the ΔnifB Av1 standard and relatively lower N2 and C2H2 reduction activities, 3% and 6%, respectively (Table 3). Both proteins show significant substrate reducing activities in patterns that closely resemble those of their respective WT counterparts (Av1V proteins show lower activities for all substrates except H2 compared with Av1), strongly suggesting that the metal centers contained within these proteins, which are distinct from P-clusters, are able to function in electron transfer in nitrogenase reactions and that these P-cluster variant-containing species may bear physiological relevance. Note that consistent with earlier reports (17), ΔnifH Av1 cannot be activated upon FeMoco insertion (Table 3), indicating that despite their apparent structural similarity, the cluster species in ΔnifH Av1 is different from those in ΔnifH Av1′ and ΔnifB Av1V.
Table 3. Substrate reducing activities upon FeMoco addition.
| Activities
|
||||||||
|---|---|---|---|---|---|---|---|---|
| C2H4 formation under C2H2/Ar
|
H2 formation under Ar
|
NH3 formation under N2
|
H2 formation under N2
|
|||||
| Protein | Per mg of protein | Per nmol of αβ-dimer | Per mg of protein | Per nmol of αβ-dimer | Per mg of protein | Per nmol of αβ-dimer | Per mg of protein | Per nmol of αβ-dimer |
| ΔnifB Av1* | 1,623 ± 15 | 173 ± 17 (100) | 1,723 ± 82 | 184 ± 9 (100) | 640 ± 23 | 68 ± 2 (100) | 357 ± 43 | 38 ± 2 (100) |
| ΔnifH Av1 | 0 | 0 (0) | 0 | 0 (0) | 0 | 0 (0) | 0 | 0 (0) |
| ΔnifH Av1′ | 148 ± 32 | 16 ± 3 (9) | 172 ± 7 | 18 ± 1 (10) | 85 ± 25 | 9 ± 3 (13) | 26 ± 6 | 3 ± 1 (8) |
| ΔnifB Av1v†‡ | 66 ± 4 | 11 ± 1 (6) | 183 ± 11 | 29 ± 2 (16) | 15 ± 1 | 2 ± 0.2 (3) | 188 ± 23 | 30 ± 2 (79) |
It needs to be noted that IDS-oxidized, FeMoco-reconstituted ΔnifH Av1′ or ΔnifB Av1v does not show the g = 11.8 parallel-mode EPR signal (Fig. 2C), which is typical for the IDS-oxidized P2+ state of the P-cluster. This observation indicates the presence of active P-cluster variants in ΔnifH Av1′ and ΔnifB Av1v after FeMoco reconstitution. Activities are expressed in nmol per min per mg of protein and nmol per min per nmol of complete αβ-dimer. The percentages of activities relative to those per nmol of αβ-dimer of ΔnifB Av1 are given in parentheses. The lower detection limits were 0.01, 0.02. 0.001, and 0.02 nmol per min per mg of protein for C2H4 formation under C2H2/Ar, H2 formation under Ar, NH3 formation under N2, and H2 formation under N2, respectively. ΔnifB Av1, ΔnifH Av1, ΔnifH Av1′, and ΔnifB Av1v do not show any substrate reducing activities without the addition of isolated FeMoco.
Specific activities of FeMoco-reconstituted ΔnifB Av1 are not appreciably different from those of WT Av1 expressed by AvDJ1141 (data not shown).
Isolation of FeVco from WT Av1v on a scale that is required for our experiments has not been reported so far. Therefore, it is not possible at this point to provide any data of substrate reducing activities of FeVco-reconstituted ΔnifB Av1v. Nonetheless, a FeMoco-containing ΔnifB Av1v “hybrid” could be formed, and its specific activities were compared with the activities of fully FeMoco-reconstituted ΔnifB Av1. In this context it should be noted that the FeMoco-containing Av1 shows considerably higher substrate reducing activities than FeVco-containing Av1v (26, 27). Therefore, the FeMoco-reconstituted ΔnifB Av1v will show higher percentages of activation if Av1v is used as a standard and may represent an almost fully activated FeMoco-Av1v hybrid. However, a conclusive assessment of the percentages of activation of ΔnifB Av1v is only feasible when a sufficient amount of isolated FeVco is available.
The FeMoco reconstitution assay contained only purified αβ2-trimeric ΔnifB Av1v and isolated FeMoco. An αβ2-trimeric subunit composition of FeMoco-reconstituted ΔnifB Av1v was confirmed by molecular weight determination of the protein on the Sephacryl S-200 HR gel filtration column (data not shown).
Discussion
For decades it has been assumed that a fully assembled P-cluster is necessary to support nitrogenase reactivity (1–11). Here, we show that certain P-cluster variants can also carry out the enzymatic activity. Both ΔnifH Av1′ and ΔnifB Av1V contain catalytically active P-cluster variants presumably composed of [4Fe-4S]-like centers that are clearly distinct from the normal P-clusters. Interestingly, the S = 1/2 EPR signal exhibited by the P-cluster variant in ΔnifB Av1V is also observed in WT Av1V and thought to be associated with the activity of the protein, indicating that the same P-cluster variant is present in WT Av1V and may represent a physiologically relevant species during the process of nitrogenase assembly and/or turnover (4, 27). The S = 1/2 EPR signal of ΔnifH Av1′ is also observed in ΔnifH Av1, the protein species isolated from the same nifH-deletion A. vinelandii strain without preincubation with Fe protein and MgATP (17). The P-cluster species in ΔnifH Av1, although structurally similar to that in ΔnifH Av1′, is catalytically inactive and cannot be activated by FeMoco insertion (17, 19, 21). Combined EPR and EXAFS data show that the P-cluster variant in ΔnifH Av1′ is more reduced than ΔnifH Av1, indicating that further reduction and/or subtle structural rearrangement of the P-cluster precursor might occur during its preincubation with Fe protein and MgATP.m The association of catalytic activity with this putatively more reduced form of ΔnifH Av1 may indicate that the P-cluster variant in ΔnifH Av1′ represents a physiologically relevant intermediate during the assembly process of MoFe protein.
Conflicting results have been reported earlier on whether a FeMoco-containing Av1V hybrid (in contrast to the FeVco-containing Av1V) is active in terms of N2 reduction (23, 50). Our data clearly show that such a hybrid is indeed able to reduce N2 and other tested substrates (Table 3). More interestingly, our FeMoco-containing Av1V hybrid has a substrate reduction profile similar to that of the WT FeVco-containing Av1V, rather than that of the WT FeMoco-containing Av1, most notably with respect to the following two aspects: (i) C2H2 is a relatively poor substrate and (ii) H2 evolution in the presence of N2 occurs at a high rate. This observation is consistent with the general belief that the enzymology of substrate reduction in A. vinelandii depends on the immediate protein environment surrounding the cofactor.
The incomplete αβ2-trimeric subunit composition has been reported earlier for an Av1V species generated during the time-consuming purification of the highly unstable WT Av1V (27). The same subunit composition of αβ2 is observed here in ΔnifB Av1V, with the notable absence of the small vnfG-encoded γ-subunit. It appears that the lack of cofactor/γ-subunit, which is a direct/indirect result of the deletion of nifB gene, has a significant impact on the structural integrity of the protein. Nevertheless, the intact αβ dimer of ΔnifB Av1V is catalytically active upon insertion of FeMoco (Table 3). This observation is contradictory to some earlier reports (23) and indicates that the vnfG-encoded γ-subunit is not essential for FeMoco insertion and protein activity; rather, it might be required for the overall stability of the protein. Recently, we showed that the two αβ-dimers of Av1 of the Mo-nitrogenase are assembled in a stepwise fashion and that the formation of the second αβ-dimer requires additional factors (31). The incomplete αβ2-trimeric subunit composition of the ΔnifB Av1V, which is a result of the disassembly of the unstable protein, might reflect, in an opposite sense, the fact that the Av1V, like Av1, is assembled stepwise and that the vnfG-encoded γ-subunit might be involved in the assembly of the second αβ-dimer of Av1V and contribute to the stability of the fully assembled protein. It needs to be noted, however, that our study uses FeMoco for the activation experiments and not FeVco, the native cofactor of Av1V. Therefore, the role of vnfG-encoded γ-subunit in the assembly process needs to be addressed further when a sufficient amount of isolated FeVco is available.
In summary, we show that nitrogenase reactivity can be achieved by metalloclusters other than P-clusters. Although the physiological relevance, the detailed structures, and the oxidation states of these species await further investigation, the dogma that “nitrogen is reduced when P-clusters are produced” is shaken, giving way to additional perspectives on how this fascinating enzyme works and what can be learned about it.
Acknowledgments
We thank Professor Agnes Henschen-Edman of the University of California-Irvine for the determination and quantitative analysis of the NH2-terminal polypeptide sequence and Professor Dennis Dean for providing A. vinelandii strains DJ1165 and DJ1258. This work was supported by National Institutes of Health Grant GM-67626 (to M.W.R.) and National Institutes of Health Grant RR-01209 (to K.O.H.). XAS data were measured at the Stanford Synchrotron Radiation Laboratory, which is supported by the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the U.S. Department of Energy, Office of Biological and Environmental Research. M.C.C. was supported by an Evelyn Liang McBain Fellowship.
Author contributions: Y.H., B.H., K.O.H., and M.W.R. designed research; Y.H., M.C.C., A.W.F., J.A.W., and M.W.R. performed research; Y.H., M.C.C., B.H., K.O.H., and M.W.R. analyzed data; and Y.H., M.C.C., B.H., K.O.H., and M.W.R. wrote the paper.
Abbreviations: XAS, x-ray absorption spectroscopy; EXAFS, extended x-ray absorption fine structure; FeMoco, FeMo cofactor; FeVco, FeV cofactor; IDS, indigo disulfonate.
Footnotes
A fourth nitrogenase system has been reported, which is superoxide-dependent and apparently different from the other nitrogenase classes (12). So far, only one organism, Streptomyces thermoautotrophicus (12, 13), is known to contain this nitrogenase.
The Mo-, V-, and Fe-only nitrogenases are encoded by the nif, vnf, and anf genes, respectively (4). It needs to be noted that the Fe proteins of the three nitrogenases are encoded by different structural genes (nifH, vnfH, and anfH, respectively).
The identity of X is unknown but it is considered to be C, O, or N (15).
It should be noted that the biosynthesis of the cofactors apparently starts with the production of a common Fe/S core (designated NifB-co) by NifB, the nifB gene product (18). NifB-co probably contains all of the iron and sulfur that ends up in the cofactors (18). Therefore, deletion of nifB results in expression of a cofactor-free variant of MoFe or VFe protein.
The α-subunit of Av1V is known to migrate faster than the β-subunit on an SDS/PAGE, despite a larger molecular weight of the α-subunit (16, 27). Faster migration of the α-subunit has also been observed in the case of the VFe protein of Azotobacter chroococcum, designated Ac1V (44).
An αβ2-trimeric species of Av1V has also been reported (27).
Nitrogenase combines the reduction of N2 to ammonia with the concomitant production of H2 (49) in a reaction that is commonly referred to as nitrogen fixation. Besides its physiological substrate, N2, nitrogenase is also able to reduce a large variety of small double- and triple-bonded substrates (2, 5, 49), and the reduction of these alternative substrates by nitrogenase can be used to determine the activity of this enzyme. One of the most commonly used alternative substrates in nitrogenase activity assays is acetylene (C2H2), which upon reduction to ethylene (C2H4) is easily detectable by gas chromatography analysis (2). Meanwhile, in the absence of any reducible substrates (e.g., in a gas atmosphere of 100% argon), nitrogenase catalyzes H2 evolution, which results from the reduction of protons (49). This reaction is also commonly included in standard activity assays of nitrogenase.
ΔnifH Av1 and ΔnifH Av1′ contain approximately the same amount of Fe (15.5 ± 0.9 and 15.4 ± 0.4 mol/mol protein, respectively, Table 2) and a similar amount of acid-labile sulfur (15.6 ± 2.4 and 16.2 ± 1.9 mol/mol protein, respectively). These results, in combination with our XAS/EXAFS analysis, indicate the presence of structurally similar [4Fe-4S]-like clusters in both ΔnifH Av1 and ΔnifH Av1′ (for structural models see ref. 19).
References
- 1.Eady, R. R. (1995) Met. Ions Biol. Syst. 31, 363–405. [PubMed] [Google Scholar]
- 2.Burgess, B. K. & Lowe, D. J. (1996) Chem. Rev. 96, 2983–3011. [DOI] [PubMed] [Google Scholar]
- 3.Howard, J. B. & Rees, D. C. (1996) Chem. Rev. 96, 2965–2982. [DOI] [PubMed] [Google Scholar]
- 4.Eady, R. R. (1996) Chem. Rev. 96, 3013–3030. [DOI] [PubMed] [Google Scholar]
- 5.Smith, B. E. (1999) Adv. Inorg. Chem. 47, 159–218. [Google Scholar]
- 6.Rees, D. C. & Howard, J. B. (2000) Curr. Opin. Chem. Biol. 4, 559–566. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen, J., Dean, D. R. & Seefeldt, L. C. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 269–295. [DOI] [PubMed] [Google Scholar]
- 8.Lawson, D. M. & Smith, B. E. (2002) Met. Ions Biol. Syst. 39, 75–119. [PubMed] [Google Scholar]
- 9.Igarashi, R. Y. & Seefeldt, L. C. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 351–384. [DOI] [PubMed] [Google Scholar]
- 10.Eady, R. R. (2003) Coord. Chem. Rev. 237, 23–30. [Google Scholar]
- 11.Seefeldt, L. C., Dance, I. G. & Dean, D. R. (2004) Biochemistry 43, 1401–1409. [DOI] [PubMed] [Google Scholar]
- 12.Ribbe, M., Gadkari, D. & Meyer, O. (1997) J. Biol. Chem. 272, 26627–26633. [DOI] [PubMed] [Google Scholar]
- 13.Gadkari, D., Mörsdorf, G. & Meyer, O. (1992) J. Bacteriol. 174, 6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters, J. W., Stowell, M. H., Soltis, S. M., Finnegan, M. G., Johnson, M. K. & Rees, D. C. (1997) Biochemistry 36, 1181–1187. [DOI] [PubMed] [Google Scholar]
- 15.Einsle, O., Tezcan, F. A., Andrade, S. L. A., Schmid, B., Yoshida, M., Howard, J. B. & Rees, D. C. (2002) Science 297, 1696–1700. [DOI] [PubMed] [Google Scholar]
- 16.Joerger, R. D., Loveless, T. M., Pau, R. N., Mitchenall, L. A., Simon, B. H. & Bishop, P. E. (1990) J. Bacteriol. 172, 3400–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribbe, M. W., Hu, Y., Guo, M., Schmid, B. & Burgess, B. K. (2002) J. Biol. Chem. 277, 23469–23476. [DOI] [PubMed] [Google Scholar]
- 18.Dos-Santos, P. C., Dean, D. R., Hu, Y. & Ribbe, M. W. (2004) Chem. Rev. 104, 1159–1174. [DOI] [PubMed] [Google Scholar]
- 19.Corbett, M. C., Hu, Y., Naderi, F., Ribbe, M. W., Hedman, B. & Hodgson, K. O. (2004) J. Biol. Chem. 279, 28276–28282. [DOI] [PubMed] [Google Scholar]
- 20.Gavini, N., Ma, L., Watt, G. & Burgess, B. K. (1994) Biochemistry 33, 11842–11849. [DOI] [PubMed] [Google Scholar]
- 21.Christiansen, J., Goodwin, P. J., Lanzilotta, W. N., Seefeldt, L. C. & Dean, D. R. (1998) Biochemistry 37, 12611–12623. [DOI] [PubMed] [Google Scholar]
- 22.Schmid, B., Ribbe, M. W., Einsle, O., Yoshida, M., Thomas, L. M., Dean, D. R., Rees, D. C. & Burgess, B. K. (2002) Science 296, 352–356. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee, R., Allen, R. M., Ludden, P. W. & Shah, V. K. (1996) J. Biol. Chem. 271, 6819–6826. [DOI] [PubMed] [Google Scholar]
- 24.Hawkes, T. R. & Smith, B. E. (1983) Biochem. J. 209, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawkes, T. R. & Smith, B. E. (1984) Biochem. J. 223, 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess, B. K., Jacobs, D. B. & Stiefel, E. I. (1980) Biochim. Biophys. Acta 614, 196–209. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard, C. Z. & Hales, B. J. (1996) Biochemistry 35, 472–478. [DOI] [PubMed] [Google Scholar]
- 28.Hales, B. J., Case, E. E., Morningstar, J. E., Dzeda, M. F. & Mauterer, L. A. (1986) Biochemistry 25, 7251–7255. [DOI] [PubMed] [Google Scholar]
- 29.Hales, B. J., Langosch, D. J. & Case, E. E. (1986) J. Biol. Chem. 261, 15301–15306. [PubMed] [Google Scholar]
- 30.Hu, Y., Fay, A. W. & Ribbe, M. W. (2005) Proc. Natl. Acad. Sci. USA 102, 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, Y., Fay, A. W., Dos Santos, P. C., Naderi, F. & Ribbe, M. W. (2004) J. Biol. Chem. 279, 54963–54971. [DOI] [PubMed] [Google Scholar]
- 32.Bursey, E. H. & Burgess, B. K. (1998) J. Biol. Chem. 273, 16927–16934. [DOI] [PubMed] [Google Scholar]
- 33.Bursey, E. H. & Burgess, B. K. (1998) J. Biol. Chem. 273, 29678–29685. [DOI] [PubMed] [Google Scholar]
- 34.Aasa, R. & Vänngård, T. (1975) J. Magn. Reson. 19, 308–315. [Google Scholar]
- 35.Ribbe, M. W. & Burgess, B. K. (2001) Proc. Natl. Acad. Sci. USA 98, 5521–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavini, N. & Burgess, B. K. (1992) J. Biol. Chem. 267, 21179–21186. [PubMed] [Google Scholar]
- 37.Corbin, J. L. (1984) Appl. Environ. Microbiol. 47, 1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark, L. J. & Axley, J. H. (1955) Anal. Biochem. 27, 2000–2003. [Google Scholar]
- 39.Van de Bogart, M. & Beinert, H. (1967) Anal. Biochem. 20, 325–334. [DOI] [PubMed] [Google Scholar]
- 40.Beinert, H. (1983) Anal. Biochem. 131, 373–378. [DOI] [PubMed] [Google Scholar]
- 41.Rabinowitz, J. C. (1978) Methods Enzymol. 53, 275–277. [DOI] [PubMed] [Google Scholar]
- 42.Ellis, P. J. & Freeman, H. C. (1995) J. Synchrotron Rad. 2, 190–195. [DOI] [PubMed] [Google Scholar]
- 43.George, G. N. (1990) exafspak (Stanford Synchrotron Radiation Laboratory, Stanford, CA).
- 44.Robson, R. L., Woodley, P. R., Pau, R. N. & Eady, R. R. (1989) EMBO J. 8, 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Homer, M. J., Dean, D. R. & Roberts, G. P. (1995) J. Biol. Chem. 270, 24745–24752. [DOI] [PubMed] [Google Scholar]
- 46.Rubio, L. M., Rangaraj, P., Homer, M. J., Roberts, G. P. & Ludden, P. W. (2002) J. Biol. Chem. 277, 14299–14305. [DOI] [PubMed] [Google Scholar]
- 47.Pierik, A. J., Wassink, H., Haaker, H. & Hagen, W. R. (1993) Eur. J. Biochem. 212, 51–61. [DOI] [PubMed] [Google Scholar]
- 48.Tittsworth, R. C. & Hales, B. J. (1993) J. Am. Chem. Soc. 115, 9763–9767. [Google Scholar]
- 49.Yates, M. G. (1992) in Biological Nitrogen Fixation, eds. Stacey, G., Burris, R. H. & Evan, H. J. (Chapman & Hall, New York), pp. 685–735.
- 50.Moore, V. G., Tittsworth, R. C. & Hales, B. J. (1994) J. Am. Chem. Soc. 116, 12101–12102. [Google Scholar]