Myosins comprise a diverse family of molecular motor enzymes that use the energy from cycles of ATP binding, hydrolysis, and product release to perform mechanical work along actin filaments. Although all characterized myosins share a conserved catalytic motor domain, referred to as “head,” variations in enzymatic and structural properties allow different myosins to generate diverse types of motility (1). Muscle myosin is not processive; it tugs intermittently on actin filaments and remains dissociated much of the time so multiple myosins are needed to sustain constant movement. In contrast, myosin V is processive, and an individual two-headed motor molecule takes multiple ≈36-nm steps, each coupled to the consumption of a single ATP molecule, and walks unidirectionally along an actin filament for long distances without detaching (2, 3). Myosin V walks following an asymmetric hand-over-hand mechanism (4, 5), where the heads alternate leading and trailing positions along actin, analogous to the hands of a rope climber. The two heads of myosin V are hypothesized to exert pushing and pulling forces that modulate each other's mechanochemical cycles and coordinate their stepping behavior (1, 6–8). In this issue of PNAS, Purcell et al. (9) examine the effects of external loads on the duration of the actin-attached states of myosin V by using optical tweezers to apply forward (push) or backward (pull) loads on single myosin V heads. They discover that a backward force, thought to mimic the force transmitted to the leading head from the attached trailing head of double-headed myosin V, slows the rate of actin detachment. A pushing force in the direction of motion, which may resemble the force transmitted to the trailing head by an attached leading head, has minimal effects on the lifetime of actin-attached states. The work demonstrates that an asymmetry exists between the myosin V heads when bound to actin, where a head in the leading position is more sensitive to load than one in a trailing position, and advances our understanding of how head–head coordination and intramolecular strain regulate structural and kinetic transitions of myosin V.
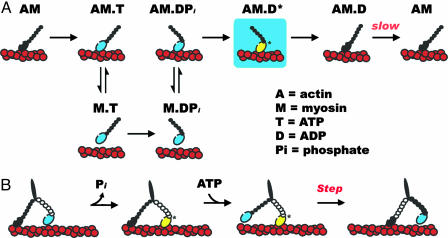
The myosin ATPase cycle has been extensively characterized in solution in the absence of external loads (ref. 1 and Fig. 1A). ATP binding and hydrolysis cycle myosin through a series of conformational states that bind actin filaments strongly (attached) or weakly (detached) depending on the chemical state of the bound nucleotide. In the absence of nucleotide or with bound ADP, myosin V binds actin filaments strongly and dissociates from them very slowly, about once every 20–150 s (10, 11). ATP binding to myosin dissociates it rapidly from actin. Rapid ATP hydrolysis forms ADP and Pi, which remain bound noncovalently to detached myosin, and is associated with a conformational change in detached myosin that rotates the lightchain binding domain, or “lever arm,” to the prepower stroke state. Myosin with bound ADP and Pi is an unstable, high-energy intermediate, but it is kinetically very stable because it releases Pi very slowly. Actin binding accelerates Pi release from myosin–ADP–Pi, resulting in force production and a mechanical displacement as myosin relaxes and rotates the lever arm back to the postpower-stroke position. Subsequent release of bound ADP generates myosin (with no bound nucleotide) attached strongly to actin, ready to repeat the cycle with the binding of another ATP. When bound to actin, the trailing head of myosin V is in a postpower-stroke position, and the leading head is in a prepower-stroke position (12, 13).
Fig. 1.
Effects of backward loads on the ATPase cycle and processive motility of myosin V accounting for the observations of Purcell et al. (9). (A) Strong actin-binding myosin heads are colored black, and weak actin-binding heads are colored blue. Purcell et al. provide evidence for an additional ADP-bound actomyosin state (AM.D*, yellow head) that is populated under backward loads and may represent a transiently populated ATPase cycle biochemical intermediate. (B) Simplified model of coordinated myosin V processive stepping. Purcell et al. demonstrate that a backward load, thought to represent the force on the leading head, slows ADP release from myosin V. A stepping intermediate with the trailing head strongly bound to actin and the leading head, under backward load, bound to actin and ADP is likely to be populated (17).
In the presence of saturating ATP and absence of load, ADP release limits ATP-induced dissociation from actin and dictates the overall myosin V ATPase cycling rate (10). Rate-limiting ADP release causes a single, cycling myosin V head to spend most of its ATPase cycle time strongly bound to actin and ADP (as AM.D shown in Fig. 1 A). This high “duty ratio” is critical for processivity because it enables at least one of the two heads of a myosin V molecule undergoing a processive run to be strongly bound to actin at any time, ensuring that random thermal forces do not cause it to diffuse away from its track. Although a single head of myosin V spends a majority of its cycle time strongly bound to actin, processive motility requires both heads of myosin V, and single-headed myosin V takes only one step per encounter with actin filaments (14).
Most models of myosin V processivity assume that the heads coordinate their ATPase cycles during a processive run (1, 6–8, 15–18), although coordination is not an absolute requirement for processive movement. There are various strategies that myosin V can follow to coordinate the catalytic cycles of its two heads. Although there are significant differences among the plausible models, they fall broadly into three different classes depending on how one head of myosin V affects the actin-binding properties of the other. There is general agreement that during a processive run the trailing head releases ADP, binds ATP, dissociates from actin, and is swung forward to become the leading head. The differences among the models lie in what happens next. In one case, the leading head binds actin strongly and generates (forward) force, which can either introduce strain into the molecule (6) or promote detachment of the trailing head by accelerating ratelimiting ADP release, which allows ATP to bind (8). Alternatively, connection to the attached trailing head may impose a backward stress on the leading head that prevents it from binding actin strongly (1, 7). Analysis of processive runs (15, 16) and myosin V's first step after initial encounter with actin (17) favors combinations of these models and suggests that myosin V can follow multiple kinetic pathways during a processive run. However, direct experimental evidence for head–head-mediated strain in myosin V, strain-accelerated ADP release from the trailing head, or strain inhibition of leading head attachment during a processive run is lacking.
The new work of Purcell et al. (9) provides direct experimental evidence that the effect of external load on the ATPase cycle kinetics of myosin V is asymmetric, where backward loads affect ADP release and actin attachment to a greater extent than forward loads, and permits evaluation of the different proposed models of myosin V processivity. They measured how long myosin V remains bound to actin under forward or backward loads of ≈2 pN, predicted to simulate the forces that each head of a two-headed myosin V molecule would contribute during processive stepping. When they pushed myosin V in the direction of movement there were minimal changes in the lifetimes of the actin-attached states, suggesting that a forward load, such as that exerted on the trailing head by a force-generating leading head, does not accelerate ADP release from myosin V. When they pulled backward, detachment from actin was much slower. The dwell times of the actin-attached states were independent of ATP and ADP concentrations, indicating that actin detachment occurs independent of ATP binding. The most plausible explanation is that myosin V under a backward load does not release bound ADP and dissociates from actin with ADP still bound at its active site. However, the detachment rate constant of myosin V with bound ADP under a backward load is >25 times more rapid than that in the absence of load (11). Therefore, a backward load not only slows ADP release from myosin V but it also accelerates actin detachment and weakens actin binding. Such a biochemical state is likely to represent a prepower-stroke conformation of myosin that is bound to ADP and actin (AM.D* in Fig. 1 A). There is considerable evidence favoring the existence of multiple actomyosin V–ADP states (11, 16, 17, 19, 20) that bind actin with different affinities (11, 17). In the absence of load, the strong actin-binding conformation is favored (11). The backward force on myosin V may shift the equilibrium to favor population of the weak actin-binding state.
Processivity models implicate a backward load in inhibiting strong actin binding by the leading head.
The results of Purcell et al. (9) are consistent with processivity models that implicate a backward load in inhibiting strong actin binding by the leading head (refs. 1, 7, 15, and 17 and Fig. 1B). Inhibition of strong lead-head binding is likely to decrease the average processive run length of myosin V (in fact, compared with highly processive nucleic acid polymerases, helicases, and the microtubule-based motor kinesin, myosin V takes relatively few steps per encounter). The advantage may be to allow myosin with bound cargo to switch actin filament tracks in dense actin networks with reasonable frequency.
Author contributions: A.O.O. and E.M.D.L.C. wrote the paper
See companion article on page 13873.
References
- 1.De La Cruz, E. M. & Ostap, E. M. (2004) Curr. Opin. Cell Biol. 16, 61–67. [DOI] [PubMed] [Google Scholar]
- 2.Mehta, A. D., Rock, R. S., Rief, M., Spudich, J. A., Mooseker, M. S. & Cheney, R. E. (1999) Nature 400, 590–593. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto, T., Amitani, I., Yokota, E. & Ando, T. (2000) Biochem. Biophys. Res. Commun. 272, 586–590. [DOI] [PubMed] [Google Scholar]
- 4.Forkey, J. N., Quinlan, M. E., Shaw, M. A., Corrie, J. E. T. & Goldman, Y. E. (2003) Nature 422, 399–404. [DOI] [PubMed] [Google Scholar]
- 5.Yildiz, A., Forkey, J. N., McKinney, S. A., Ha, T., Goldman, Y. E. & Selvin, P. R. (2003) Science 300, 2061–2065. [DOI] [PubMed] [Google Scholar]
- 6.Rief, M., Rock, R. S., Mehta, A. D., Mooseker, M. S., Cheney, R. E. & Spudich, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De La Cruz, E. M., Ostap, E. M. & Sweeney, H. L. (2001) J. Biol. Chem. 276, 32373–32381. [DOI] [PubMed] [Google Scholar]
- 8.Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. (2002) Nat. Cell Biol. 4, 59–65. [DOI] [PubMed] [Google Scholar]
- 9.Purcell, T. J., Sweeney, H. L. & Spudich, J. A. (2005) Proc. Natl. Acad. Sci. USA 102, 13873–13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De La Cruz, E. M., Wells, A. L., Rosenfeld, S. S., Ostap, E. M. & Sweeney, H. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannemann, D. E., Cao, W., Olivares, A. O., Robblee, J. P. & De La Cruz, E. M. (2005) Biochemistry 44, 8826–8840. [DOI] [PubMed] [Google Scholar]
- 12.Walker, M. L., Burgess, S. A., Sellers, J. R., Wang, F., Hammer, J. A., 3rd, Trinick, J. & Knight, P. J. (2000) Nature 405, 804–807. [DOI] [PubMed] [Google Scholar]
- 13.Burgess, S., Walker, M., Wang, F., Sellers, J. R., White, H. D., Knight, P. J. & Trinick, J. (2002) J. Cell Biol. 159, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell, T. J., Morris, C., Spudich, J. A. & Sweeney, H. L. (2002) Proc. Natl. Acad. Sci. USA 99, 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker, J. E., Krementsova, E. B., Kennedy, G. G., Armstrong, A., Trybus, K. M. & Warshaw, D. M. (2004) Proc. Natl. Acad. Sci. USA 101, 5542–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uemura, S., Higuchi, H., Olivares, A. O., De La Cruz, E. M. & Ishiwata, S. (2004) Nat. Struct. Mol. Biol. 11, 877–883. [DOI] [PubMed] [Google Scholar]
- 17.Rosenfeld, S. S. & Sweeney, H. L. (2004) J. Biol. Chem. 279, 40100–40111. [DOI] [PubMed] [Google Scholar]
- 18.Clemen, A. E., Vilfan, M., Jaud, J., Zhang, J., Barmann, M. & Rief, M. (2005) Biophys. J. 88, 4402–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenfeld, S. S., Houdusse, A. & Sweeney, H. L. (2005) J. Biol. Chem. 280, 6072–6079. [DOI] [PubMed] [Google Scholar]
- 20.Robblee, J. P., Cao, W., Henn, A., Hannemann, D. E. & De La Cruz, E. M. (2005) Biochemistry 44, 10238–10249. [DOI] [PubMed] [Google Scholar]