Abstract
We describe a protein family in Drosophila containing six adenosine deaminase-related growth factors (ADGFs), which are homologous to a mitogenic growth factor discovered in conditioned medium from cells of a different fly species, Sarcophaga. Closely related proteins have been identified in other animals, and a human homolog is implicated in the genetic disease Cat-Eye Syndrome. The two most abundantly expressed ADGFs in Drosophila larvae are ADGF-A, which is strongly expressed in the gut and lymph glands, and ADGF-D, which is mainly expressed in the fat body and brain. Recombinant ADGF-A and ADGF-D are active adenosine deaminases (ADAs), and they cause polarization and serum-independent proliferation of imaginal disk and embryonic cells in vitro. The enzymatic activity of these proteins is required for their mitogenic function, making them unique among growth factors. A culture medium prepared without adenosine, or depleted of adenosine by using bovine ADA, also stimulates proliferation of imaginal disk cells, and addition of adenosine to this medium inhibits proliferation. Thus ADGFs secreted in vivo may control tissue growth by modulating the level of extracellular adenosine.
During animal development the growth of different tissues and organs is finely controlled by signals originating both within the developing structure and outside it. The intrinsic controls of tissue growth are often intimately connected with the mechanisms controlling the development of a covert or overt pattern of differentiation within the cell population (1). For example, the Dpp and Wg signal transduction pathways in imaginal discs are required for normal cell proliferation as well as for the control of the developing pattern (2). Extrinsic factors controlling tissue growth include hormones and polypeptide growth factors that are released in a regulated fashion either from central endocrine organs or from diffuse sources, and these often have tissue-specific functions. Extrinsic control of tissue growth is a familiar concept in mammals, where the main signals are growth hormone, insulin-like growth factors, and other polypeptide growth factors (3). However, it is a relatively unexplored area of investigation in Drosophila and other insects. We are developing approaches to the investigation of extrinsic controls on tissue growth by using the imaginal discs of Drosophila as a model system. This work has already led to the identification of two families of secreted growth factors: the imaginal disk growth factors (IDGFs) (4), which are related to chitinases, and the adenosine deaminase-related growth factors (ADGFs), which are the subject of this report.
Most known growth factors function by binding to cell surface receptors and activating signal transduction pathways. However, growth could also be regulated indirectly by proteins that control the level of low molecular weight substances such as adenosine, which is an important signaling molecule in mammals (5). Here, we show that adenosine has a negative effect on growth of several Drosophila cell types, and that ADGFs stimulate cell growth by an indirect mechanism, depleting the levels of extracellular adenosine.
Materials and Methods
cDNA Clones.
cDNAs encoding ADGF-A, -C, -D, and -E were obtained from Research Genetics (Huntsville, AL). cDNAs of Drosophila ADGF-B and adenosine deaminase (ADA) were predicted from genomic sequence (Celera Genomics; BDGP-FlyBase Informatics (2000); http://hedgehog.lbl.gov:8000/annot/) and prepared by reverse transcription–PCR using 5 μg of total RNA isolated from adult Drosophila. The PCR primers for ADA were as follows: ada-F, 5′-CAAATGGAGCAGTTTCTTAAAGG-3′; ada-R, 5′-ATTTGACCAATATGTCTGCGTAG-3′. Primers for ADGF-B were as follows: gfb-F, 5′-CTAAAAGGGTTTCATTAACAGTAC-3′; gfb-R, 5′-TGGGCAACGAAAAGAGCTCAC-3′.
RNA Isolation and Northern Blots.
Drosophila embryos, first-, second-, and third-instar larvae, pupae, and adult males and females, as well as larval organs and adult body parts, were used for total RNA isolation by using the commercial RNeasy kit from Qiagen (Chatsworth, CA). RNA aliquots (5 μg) were separated by formaldehyde-agarose gel electrophoresis and transferred to Hybond-N nylon membranes (Amersham Pharmacia Biotech). The membranes were stained in 1% methylene blue to verify the integrity of rRNA bands. Hybridization and washing conditions were as recommended by the manufacturer of the nylon membrane. Hybridization probes were prepared by random priming of gel-purified cDNA inserts with [α-32P]dATP (Amersham Pharmacia Biotech).
In Situ Hybridization.
Sense and antisense digoxigenin-labeled RNA probes were prepared from linearized vectors by using SP6 and T7 polymerases (Takara Shuzo, Kyoto) and digoxigenin labeling mixture (Roche Diagnostics; ref. 4). Dechorionated embryos and partially dissected larvae were fixed and hybridized with RNA probes in 50% formamide, 5× SSC, 250 μg/ml salmon sperm DNA, 50 μg/ml heparin, and 0.1% Tween 20 at 55°C (6). Signals were detected by anti-digoxigenin-AB Fab fragments (Roche Diagnostics). There were no detectable signals from sense RNA probes except from the proventriculus.
Protein Expression Using Baculovirus.
Baculovirus expression constructs were His-tagged at the C termini by using PCR from tailed primers, including sequences encoding six histidines. The constructs were sequenced and subcloned into the shuttle vector pVL1393 or pBlueBac4.5 (Invitrogen). Constructs were cotransfected with BaculoGold DNA into Sf9 cells, and recombinant baculoviruses were purified by plaque assay as described in the supplier's instructions (Invitrogen). Recombinant proteins were purified either from “Ultimate serum-free medium” (ADGF-A, ADGF-D) or from a cell lysate (ADGF-E and ADA) by using Ni-nitrilotriacetic acid (NTA) column and instructions from the manufacturer (Qiagen). Lepidopteran cells Sf9 and Hi5 were maintained in Trichoplusia ni medium TNM-FH (Sigma T1032) supplemented with 10% FBS or ultimate serum-free medium (Invitrogen).
Cell Culture and Growth Rate Analysis.
Drosophila Cl.8+ imaginal wing disk cells were maintained in Shields and Sang medium (ref. 7; Sigma, catalog no. S 8398) supplemented with 2% FBS, 2.5% fly extract, and 0.125 international units/ml insulin. Embryonic S2 and Kc167 cells, as well as the Mbn-2 blood cell line (8), were maintained in Shields and Sang medium supplemented with 10% FBS. BG2-c6 cells were maintained in the same medium with 10% FBS and 0.125 international units/ml insulin. NIH-Sape-4 cells were cultured in Mitsuhashi and Maramorosh insect medium (Sigma, catalog no. M 9257) supplemented with 10% FBS. “Supplement-free medium” was Shields and Sang medium (catalog no. S 8398) containing yeast extract. “Minimal” medium (MM) was Shields and Sang medium (Sigma, catalog no. S 8523) lacking yeast extract, supplemented with l-leucine, -l-methionine, fumaric acid, folic acid, myo-inositol, pyridoxine, riboflavin, and thiamine. Adenosine and inosine (Sigma) were used at final concentrations of 10–200 μM and 125 μM-1.25 mM, respectively. ADGF-A was used at a concentration of 12.5–50 ng/ml; bovine ADA (Roche Diagnostics) was used at concentration of 50 ng/ml or 4 ng/ml. For growth assays, the cells were plated on six-well plates at 5 × 105/ml in appropriate media. Media and supplements were changed every day. Proliferation of Cl.8+ cells and S2 cells was measured by direct counting of cells by using digital photographs of identical areas (0.8 × 0.8 mm) taken every 24 h. Proliferation of BG2-c6 cells was measured by using a haemocytometer. Each value represents an average of three experiments.
ADA Assays.
Assays were performed by using continuous spectrophotometric measurement according to the published protocol (9) with modifications suggested in the instructions by Sigma. Recombinant proteins were incubated with 45 μM adenosine, and the decrease of absorbance in 265 nm was measured for 5 min. For ADA activity of cell extracts, about 1 × 107 cells were homogenized in 500 μl of lysis buffer (50 mM Tris, pH 8.5/5 mM β-mercaptoethanol/100 mM KCl/1 mM PMSF/1% Nonidet P-40) and sonicated by 10 strikes of 2 s each. Extracts were centrifuged at maximum speed for 1 min in a microcentrifuge, and 10 μl of extract was directly used for the assay. Activity measurements were corrected for protein concentration as measured by the Coomassie Protein Assay Reagent and protocol by Pierce.
Results
ADGFs.
By searching databases for sequences encoding proteins related to the insect-derived growth factor from Sarcophaga cells (10), we identified six Drosophila genes encoding a family of ADGFs. The genomic organization of these genes is shown at our web site, http://mamba.bio.uci.edu/∼pjbryant/lab/ADGFs/index.htm, and has been independently described (11). The genes include one triplet in tandem at 3L;74E2, encoding ADGF-A and ADGF-B, as well as ADGF-A2, previously reported as male-specific insect-derived growth factor, or MSI (12). The ADGF-A2 gene is included within the 5′ intron of the ADGF-A gene. Two genes in reversed tandem at 3R;87F12 encode ADGF-C and ADGF-D, and a single gene at 2R;51C1 encodes ADGF-E. Database searching also revealed expressed sequence tag (EST) clones corresponding to full-length cDNAs for Drosophila ADGF-A, -C, -D, and -E. We have obtained these clones and completed their sequencing. cDNA clones encoding ADGF-B and a homolog (CG11994) of authentic ADA from other organisms, which were predicted only from the genomic DNA sequence, were obtained by reverse transcription–PCR.
Partial sequence alignments of the predicted ADGF proteins are shown in Fig. 1a. They show 30–56% identity among themselves, and also show significant similarity to the sequence of ADA, especially in the conserved amino acid residues necessary for ADA catalytic activity (13). All ADGFs have longer N termini than classical ADA enzymes. In ADGF-E, the glutamic acid residue involved in catalysis is replaced by glutamine (Gln-377), and two otherwise highly conserved aspartates are replaced by a glycine-serine pair (Gly-459, Ser-460). ADGF-A, -C, and -D have strongly predicted signal peptides (13) with putative cleavage sites within the N-terminal 32 aa, so these are very likely to be secreted proteins. For ADGF-B and -E, the predictions are weaker. Note, however, that mammalian ADA has been shown to be partly on the cell surface, but it has no predicted signal peptide so the mechanism of its transport to the surface is unclear (14). Database searches showed genes encoding ADGF-related proteins in several other species, and the phylogenetic tree shows that ADGFs and classical ADA enzymes form distinct branches (Fig. 1b).
Figure 1.
Similarity of ADGFs and ADAs. (a) Section of the sequence alignment of Drosophila ADGFs with ADA generated by megalign and decorated with boxshade. Triangles indicate several of the residues required for catalytic activity in ADA. Asterisks above the ADGF-A sequence indicate the two amino acids (His,Ala) replaced in the mutagenesis experiment. ADGF-A (CG5992), ADGF-B (CG5998), ADGF-C (CG9345), ADGF-D (CG9621), ADGF-E (CG10143), and putative Drosophila ADA (CG11994) were predicted from genomic sequence and confirmed by cDNA sequencing. ADGF-A2 is a proposed new name for male-specific insect-derived growth factor (which equals MSI; FBgn0043025; ref. 12), which is needed because MSI is already in use for an unrelated gene. CECR1 is the predicted gene product from the human cat eye syndrome critical region (35), which is more closely related to ADGFs than to ADA. Dark shading indicates agreement with a >50% identity consensus, and gray shading indicates agreement with a >50% similarity consensus. Complete alignment at http://mamba.bio.uci.edu/∼pjbryant/lab/ADGFs/index.htm. (b) Phylogenetic tree of the ADGF family. Drosophila ADGFs are compared with Sarcophaga (GenBank accession no. D83125), Glossina (accession nos. AAD52850 and AAD52851), and Lutzomyia (accession no. AF234182) homologs, as well as with several authentic ADAs (human, accession no. NM_000022; mouse, no. NM_007398; Caenorhabditis elegans, no. U61947; Streptomyces, no. AL589164; yeast Saccharomyces cerevisiae, no. NC_001146; Arabidopsis, no. AL161501) and the Drosophila homolog (DrosADA) of authentic ADA. The length of each pair of branches represents the distance between sequence pairs, and the units at the bottom of the tree indicate the number of substitution events. Human CECR1 (accession no. 190746) is clearly a member of the ADGF rather than the ADA family. DrosADA (CG11994) falls within the ADA branch and is therefore considered a homolog of authentic ADAs rather than an ADGF, despite its lack of ADA catalytic activity.
Spatial and Temporal Pattern of ADGF Expression.
Developmental Northern blots show that ADGF-A is expressed at all tested stages from embryo to adult (Fig. 2a). ADGF-D is expressed only in postembryonic stages, with a lower level of expression in larvae and pupae and a higher level in adult males. ADGF-B and -E are expressed mainly in adult males, but weak expression is also detected in pupae. Northern blots using RNA from dissected larval tissues show that ADGF-A is expressed mainly in the gut, whereas ADGF-D is expressed mainly in the fat body (Fig. 2b). In adults, both genes are expressed in both male and female (both in a head + thorax extract as well as in the abdomen). In contrast, ADGF-B and -E are expressed in the adult only in the male abdomen, possibly reflecting testis-specific expression as described for ADGF-A2 (= MSI).
Figure 2.
Temporal and spatial pattern of ADGF expression. Northern blots of 5 μg of total RNA were examined by hybridization with cDNA probes for specific ADGF family members. (a) Whole-body samples from early embryos (E1), late embryos (E2), first-instar larvae (L1), second-instar larvae (L2), third-instar larvae (L3), pupae (P), adult males (♂), and adult females (♀). Ribosomal RNA stained with methylene blue (bottom row) is shown as a loading control. (b) Tissue specificity of ADGF expression based on Northern blots (probed with α-32P-labeled ADGF-A, -B, -D, and -E cDNAs). Third-instar larval samples were from integument (I), fat body (F), gut (G), and brain + salivary glands + imaginal discs (BS); and adult samples were from female heads + thoraces (♀H), female abdomens (♀A), male heads + thoraces (♂H), and male abdomens (♂A). Ribosomal RNA stained with methylene blue is shown as a loading control (bottom row).
ADGF expression was also analyzed by in situ hybridization to whole mounts using digoxigenin-labeled RNA probes. ADGF-A is first strongly expressed at the blastoderm stage at the anterior pole (Fig. 3a), and, at stages 12–15, it is expressed at high levels in muscle precursor cells and at lower levels in the anterior region (Fig. 3b). It is expressed in third-instar larval anterior and posterior midgut and lymph glands (Fig. 3 d and c, respectively) where the signal is perinuclear, as expected for a functional transcript. ADGF-D is expressed strongly in the fat body, and in specific areas of the larval brain (Fig. 3 e and f). We did not detect any specific staining of larval tissues with ADGF-C or -E probes.
Figure 3.
Tissue specificity of ADGF expression based on in situ hybridization of RNA probes to whole tissues. a–d, Probed with ADGF-A antisense RNA; e–f, probed with ADGF-D antisense RNA. ADGF-A shows strong expression at the anterior pole at the cellular blastoderm stage (a), and in the mesoderm at stage 14 (b). In third-instar larvae ADGF-A shows expression in the midgut (d) and lymph glands (c). ADGF-D is expressed in fat body (e) and ventral ganglion of the brain (f). Am, anterior midgut; An, anterior; As, amnioserosa; Pv, proventriculus; OL, optic lobe; DV, dorsal vessel; FB, fat body; Ms, mesoderm; MT, Malpighian tubules; LG, lymph glands; Pm, posterior midgut; VG, ventral ganglion. Staining of proventriculus with ADGF-A was not specific because it also occurred with the sense control.
Recombinant ADGFs Are Mitogenic.
To test whether ADGFs are mitogenic and to examine their possible ADA activity, we expressed recombinant ADGF-A, -D, and -E proteins in a baculovirus expression system. As expected from signal peptide predictions, ADGF-A and -D were secreted into the medium whereas ADGF-E was contained within the cell fraction. When separated by SDS/PAGE, recombinant ADGF-A, -D, and -E ran as protein bands of 57–59 kDa, as predicted from the ORFs (data not shown).
The Drosophila imaginal wing disk Cl.8+ cell line was chosen for the in vitro experiments because these cells were previously shown to be very sensitive to growth factors (4). These cells undergo morphological changes and die within 24 h in supplement-free medium (SFM; Fig. 4A), whereas, in medium supplemented with fly extract, insulin, and 2% FBS (Fig. 4C) or growth factors (e.g., imaginal disk growth factors or IDGFs), they survive well. On the addition of ADGF-A or ADGF-D to SFM, the cells display their characteristic elongated cell shape and pseudopodia (Fig. 4B). The addition of ADGF-E does not have this effect and does not prevent cell death within 24 h. Varying concentrations of ADGF-A were added to the Cl.8+ cells in SFM, and growth rates were measured. The results (data not shown) show that ADGF-A and ADGF-D are mitogenic at concentrations as low as 12.5 ng/ml (20 μM).
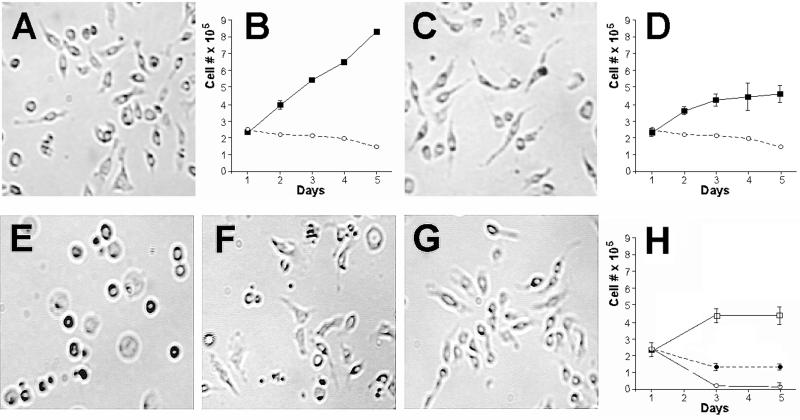
Figure 4.
Effects of recombinant ADGF-A on morphology and growth of Drosophila Cl.8+ cells (A–D), S2 cells (E–H), and BG2-c6 cells (I–L). Cells were grown at low density in SFM (A, E, and I); in SFM plus 50 ng/ml (80 μM) recombinant ADGF-A (SFM + ADGF-A; B, F, and J), and in complete medium (C, G, and K), and are shown 48 h after plating. Complete medium for Cl.8+ cells included 2% serum, insulin (0.125 international units/ml) and 2.5% fly extract, complete medium for S2 cells contained 10% fetal serum, and complete medium for BG2-c6 cells contained 10% fetal serum and 0.125 international units/ml of insulin. Charts show the growth of the three cell types in SFM (broken lines with open circles) and media supplemented with 50 ng/ml of ADGF-A (solid lines with filled squares). In SFM, most Cl.8+ cells are flat and round, and slowly die (A) whereas, in complete medium or SFM supplemented with 50 ng/ml ADGF-A, they are elongated and develop pseudopodia (C and B). In SFM, S2 and BG2-c6 cells become flattened and survive with occasional division (E and I, respectively), whereas, in medium supplemented by 10% serum or in SFM supplemented with 50 ng/ml ADGF-A, S2 cells grow rapidly and are only weakly attached to the surface. Addition of ADGF-A to SFM does not produce any morphological effect on BG2-c6 cells, but in the presence of 10% serum these cells grow very rapidly and form large clusters.
ADGF-A and ADGF-D have different effects on different cell types. They induce morphological changes and/or serum-independent growth of S2 cells (ref. 15; derived from Drosophila whole embryos; Fig. 4F), flesh fly (Sarcophaga) embryonic cells NIH-Sape-4 (ref. 10; data not shown), as well as Drosophila Cl.8+ imaginal disk cells, but show no significant effect on Drosophila BG2-c6 neuroblasts (ref. 16; Fig. 4J), Kc167 embryonic cells (17), or Mbn-2 haematopoietic cells (ref. 8; Table 1). ADGF-E does not show these effects on any of the tested cell types.
Table 1.
Adenosine and ADGF-A sensitivity of Drosophila cell lines
| Cells | Toxicity of 100 μM adenosine | Growth-promoting activity of ADGF-A (50 μg/ml) | Endogenous ADA activity in cell extract, units/mg |
|---|---|---|---|
| C1.8+ | + | + | 0.011 ± 0.003 |
| S2 | + | + | 0.006 ± 0.003 |
| BG2-c6 | − | − | 0.065 ± 0.009 |
| Mbn-2 | − | − | 0.035 ± 0.004 |
| Kc167 | − | − | 0.020 ± 0.011 |
Survival in the presence of 100 μM adenosine, growth promoting activity of ADGF-A, and endogenous ADA activity were examined for five Drosophila cell lines. Note: fresh cultures of C1.8+ cells and Kc167 cells have higher ADA activity than older overgrown cultures (data not shown).
Some ADGFs Have ADA Activity, Which Is Necessary for Their Mitogenic Function.
The similarity of ADGFs to ADA, especially the conservation of the amino acids necessary for catalytic activity, suggested that these proteins might be active ADAs. Using a spectrophotometric assay (9), we found that recombinant ADGF-A and ADGF-D are very active ADAs (ADGF-A showing 157 units/mg, and ADGF-D showing 145 units/mg) whereas ADGF-E does not show detectable activity. Because ADGF-A and -D have both mitogenic and enzymatic activities, whereas ADGF-E lacks both activities, the mitogenic activities of ADGFs may depend on their enzymatic activity. To test this hypothesis, we produced an enzymatically inactive ADGF-A by site-directed mutagenesis. Based on the similarity of ADGF-A and the known structure of mouse ADA (13), we substituted two amino acids (His-386,Ala-387 for Glu,Leu) in the catalytic domain (see Fig. 1a) and expressed the mutant protein in a baculovirus expression system. This mutant ADGF-A lacks both enzymatic and mitogenic activities (data not shown), strongly suggesting that the mitogenic activity of ADGFs depends on their ADA activity.
As a further test of the activity hypothesis, we expressed in baculovirus a Drosophila homolog (gene CG11994) of authentic ADA from other organisms. Surprisingly, as with the mutated ADGF-A and ADGF-E, this protein showed no enzymatic activity or effect on growth of Cl.8+ or S2 cells (data not shown). The lack of catalytic activity may be caused by substitutions (Fig. 1a) in the conserved catalytic sites in this Drosophila ADA as compared with authentic ADAs, especially the presence of Cys-199, Ala-200, where other homologs contain Ala,Gly. In contrast, commercial bovine ADA (catalytically active) strongly promotes growth of Drosophila Cl.8+ cells in vitro (Fig. 5 A and B).
Figure 5.
Effect of bovine ADA, adenosine-free conditions, and adenosine on Cl.8+ cells (shown 48 h after plating). Cells grown in SFM + 4 ng/ml bovine ADA (A) and in minimal medium (MM) (C), which lacks known sources of adenosine, are elongated and develop pseudopodia characteristic of normal Cl.8+ cells (see Fig. 4 B and C). Counts of cell numbers show that treatment of cells with bovine ADA (B, solid line, filled squares) is as effective as ADGF-A in SFM (see Fig. 4D, solid line), whereas cells grown in MM grow better than cells in SFM with added yeast extract (D, solid line vs. broken line). The growth of Cl.8+ cells in MM is slower than in SFM with ADGF-A (Fig. 4D, solid line) probably because of the lack of nutrients that are otherwise supplied in yeast extract. (E) Cells in MM with adenosine 500 μM; (F) cells in MM with adenosine 500 μM and ADGF-A 100 ng; (G) cells in MM with ADGF-A 100 ng. (H) Cell counts show no effect of ADGF-A on cell growth in the absence of adenosine in MM (open squares on H, compared with filled squares on D), that a high concentration of adenosine kills the cells (open circles, H), and that ADGF-A partially protects the cells from adenosine (filled circles, H).
Adenosine Inhibits Growth of Cl.8+ Cells, and ADGFs Might Act by Depleting Extracellular Adenosine.
ADAs catalyze the deamination of adenosine to inosine and of deoxyadenosine to deoxyinosine. If the conversion of adenosine to inosine were involved in growth control, we would expect that changes in the adenosine or inosine concentration in the medium would have an effect on cell growth and/or on ADGF mitogenic activity. Adenosine at concentrations higher than 50 μM is toxic for Cl.8+ cells in SFM (Table 1), but even higher concentrations of inosine have no detectable effect on growth. Other cell types show differences in sensitivity to adenosine, with S2 cells being very sensitive and failing to proliferate in a concentration as low as 10 μM, whereas BG2-c6, Kc167 cells, and Mbn-2 cells are resistant to concentrations higher than 100 μM (Table 1). The adenosine resistance of these cell types is correlated with their endogenous ADA activity (Table 1).
The positive effect of several active ADAs, including ADGF-A, ADGF-D, and bovine ADA, on the growth of Cl.8+ and S2 cells (Figs. 4 D and H and 5B) suggested that these proteins might act simply by depleting extracellular adenosine. To test this possibility, we produced a “minimal” medium (MM; see Materials and Methods) that lacks the usual yeast supplement, and therefore has no known source of adenosine. This change in the medium dramatically increased survival, polarization, and proliferation of Cl.8+ cells (Fig. 5C), to a degree comparable to that seen with the ADA and ADGF treatments (Fig. 5B). Addition of ADGF-A under adenosine-free conditions had no effect on growth (Fig. 5 G and H), but, when adenosine was added to this medium, the cells rounded up and failed to proliferate (Fig. 5E), and this effect was to some extent prevented when ADGF was added along with adenosine (Fig. 5 F and H). The results show that extracellular adenosine blocks cell polarization and proliferation, and that these effects can be prevented by the addition of active ADA or ADGF.
Discussion
The six ADGFs constitute a family of proteins that may play important roles in the extrinsic control of tissue growth in Drosophila. These proteins are not related to known growth factors but instead show sequence and functional similarity to the enzyme ADA (Fig. 1a). Like classical ADA, ADGF-A and ADGF-D catalyze the hydrolytic deamination of adenosine or 2′-deoxyadenosine to inosine or 2′-deoxyinosine, respectively, and thus could regulate adenosine and deoxyadenosine levels (14). Recombinant forms of ADGF-A and ADGF-D promote survival, polarization, and proliferation of an imaginal disk cell line, and they are also mitogenic on at least one other Drosophila cell line derived from embryos and a Sarcophaga embryonic cell line NIH-Sape-4 (data not shown).
ADGFs Stimulate Cell Growth by a Novel Mechanism.
Within the ADA/ADGF family, mitogenic activity is correlated with ADA activity. Thus ADGF-A, ADGF-D, and bovine ADA are active ADAs and are mitogenic, whereas ADGF-E, Drosophila ADA, and a mutant form of ADGF-A lack enzymatic activity and are not mitogenic. Thus, these proteins apparently require catalytic activity for their mitogenic activity, making them unique among growth factors. Removing sources of extracellular adenosine can mimic their action on imaginal disk cells, and addition of adenosine inhibits growth, providing a clear indication that these proteins may promote growth by depleting extracellular adenosine.
Responses of Mammalian and Drosophila Cells to Adenosine.
Extracellular adenosine and ADA affect various mammalian cell types in different ways. In severe combined immune deficiency, lack of ADA and the consequent accumulation of adenosine is associated with absence of T cells, decrease in B cells, and some hepatotoxicity (18), but other organ systems are not significantly affected (19). ADA knockouts in mouse also show widespread accumulation of adenosine associated with severe T and B cell loss as well as pulmonary insufficiency, and bone and kidney abnormalities (20). Some of the abnormalities can be alleviated by administering ADA in both human (21) and mouse (22) disorders. Thus T and B cells may be negatively affected by extracellular adenosine in the same way as imaginal disk cells are. Adenosine, in the presence of ADA inhibitors, is also toxic to many human carcinoma cell lines (23). In contrast, human endothelial cells show a different response: they are stimulated to proliferate by physiological concentrations (10 μM) of adenosine and are inhibited by ADA (24, 25). Adenosine at 10 μM also promotes mesangial cell proliferation (26). However, at the higher concentration of 100 μM, adenosine causes apoptosis of endothelial cells (27). These and other results suggest that extracellular adenosine levels must be finely regulated and that different cell types may have different adenosine optima. Different Drosophila cell types also respond differently to extracellular adenosine; in contrast with Cl.8+ or S2 cells, the tested BG2-c6, Mbn-2, and Kc167 cells appear resistant to adenosine, and their proliferation is not significantly affected by either recombinant ADGF-A or ADGF-D. Measurements of ADA activity suggest that these cells may acquire resistance to adenosine by producing enough endogenous proteins, presumably ADGFs, with ADA activity (Table 1). Thus, the different and highly tissue-specific expression patterns of the ADGFs in Drosophila may reflect a mechanism that has evolved to regulate extracellular adenosine levels locally, to provide appropriate environments for different cell types.
Tissue Distribution of ADA in Drosophila and Mammals.
The tissue distribution of ADA activity in mammals shows an interesting parallel to the expression pattern of ADGF-A in Drosophila. Although human ADA is expressed in all cell types, the enzyme activity is highest in gastrointestinal tract and lymphoid tissues (28), analogous to the highest expression of ADGF-A in gut and lymph glands. It has been suggested that human lymphocytes are rich in ADA owing to the need for destruction of excess of adenosine (5), whereas the gut expression might be a barrier against exogenous adenosine (29). A similar distinction could be made in Drosophila. Because Drosophila ADA appears to have accumulated mutations blocking its catalytic activity, it seems likely that the ADGF family members have replaced this function. Thus, ADGFs could have evolved an expression pattern that best replaces the required ADA functions.
ADGF Subfamily Members in Other Species.
ADGFs have homologs in other species that represent a subfamily distinct from classical ADAs. The first member of the family was identified from the culture medium of an embryonic cell line from the flesh fly Sarcophaga. Because it has a specific activity similar to that of mammalian growth factors, it was named insect-derived growth factor; it is expressed in embryos, larvae, and adult females. To avoid confusion with the unrelated proteins that have been published under the name IDGF in Drosophila (4), we refer to the Sarcophaga insect-derived growth factor as Sarcophaga ADGF-A (named after its Drosophila ortholog). Another family member [AGSA (atrial gland specific antigen)] was reported from the exocrine atrial gland of the mollusc Aplysia californica (30) and has also been reported as mollusc-derived growth factor (MDGF; ref. 31). Homologous genes are expressed in the salivary gland of the tsetse fly Glossina (32) and the sand fly Lutzomyia (33), and the Lutzomyia protein has been shown to be an ADA (34). Closely related sequences are present in the silkworm Bombyx database SilkBase (http://www.ab.a.u-tokyo.ac.jp/silkbase/) and in a cDNA (AU037658) from the slime mold Dictyostelium.
The ADGF genes have a close human homolog, CECR1, strongly implicated in cat eye syndrome (35), a disorder characterized by hypoplastic kidneys, congenital heart malformation, and anomalous pulmonary venous connections. The predicted gene product is 32–36% identical with the ADGFs, but only 17% identical to human ADA. The cat eye syndrome is associated with the duplication of a 2-Mb region of chromosome 22q11.2 (36) containing CECR1, and overexpression of the gene may play an important role in the disorder. The CECR1 region is deleted in DiGeorge syndrome, which is characterized by hypoplasia of the parathyroid glands and a deficit of T cells (37). The abnormalities in both syndromes could be due to detrimental effects of extracellular adenosine that are not adequately regulated in these tissues by classical ADA.
Acknowledgments
We thank Dr. Martin Milner (Fife, United Kingdom) for Cl.8+, Dr. Dan Hulmark (Umea, Sweden) for Mbn-2, Dr. Yasumitsu Takagi (Fukuoka, Japan) for BG2-c6, and Dr. Peter Cherbas (Bloomington, IN) for Kc167 cells. This work was supported by grants from the U.S. National Science Foundation (440860-21565), the Grant Agency of the Czech Republic (204/01/1022), and the Grant Agency of the Czech Academy of Sciences (A5007107).
Abbreviations
- ADGF
adenosine deaminase-related growth factor
- ADA
adenosine deaminase
- MM
minimal medium
- MSI
male-specific insect-derived growth factor
- SFM
supplement-free medium
References
- 1.Bryant P J, Simpson P. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Benjumea F J, Cohen B, Cohen S M. Nature (London) 1994;372:175–179. doi: 10.1038/372175a0. [DOI] [PubMed] [Google Scholar]
- 3.Cross M, Dexter T M. Cell. 1991;64:271–280. doi: 10.1016/0092-8674(91)90638-f. [DOI] [PubMed] [Google Scholar]
- 4.Kawamura K, Shibata T, Saget O, Peel D, Bryant P J. Development. 1999;126:211–219. doi: 10.1242/dev.126.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Wintrobe M M. Clinical Hematology. Philadelphia: Lea & Febiger; 1956. pp. 1–1185. [Google Scholar]
- 6.Theisen H, Haerry T E, O'Connor M B, Marsh J L. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- 7.Shields G, Sang J H. J Embryol Exp Morphol. 1970;23:53–69. [PubMed] [Google Scholar]
- 8.Samakovlis C, Åsling B, Boman H G, Gateff E, Hultmark D. Biochem Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- 9.Murphy J, Baker D C, Behling C, Turner R A. Anal Biochem. 1982;122:328–337. doi: 10.1016/0003-2697(82)90291-3. [DOI] [PubMed] [Google Scholar]
- 10.Homma K, Matsushita T, Natori S. J Biol Chem. 1996;271:13770–13775. doi: 10.1074/jbc.271.23.13770. [DOI] [PubMed] [Google Scholar]
- 11.Maier S A, Podemski L, Graham S W, McDermid H E, Locke J. Gene. 2001;280:27–36. doi: 10.1016/s0378-1119(01)00762-4. [DOI] [PubMed] [Google Scholar]
- 12.Matsushita T, Fujii-Taira I, Tanaka Y, Homma K J, Natori S. J Biol Chem. 2000;275:36934–36941. doi: 10.1074/jbc.M003455200. [DOI] [PubMed] [Google Scholar]
- 13.Wilson D K, Rudolph F B, Quiocho F A. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 14.Franco R, Casado V, Ciruela F, Saura C, Mallol J, Canela E I, Lluis C. Prog Neurobiol. 1997;52:283–294. doi: 10.1016/s0301-0082(97)00013-0. [DOI] [PubMed] [Google Scholar]
- 15.Schneider I. Drosoph Inform Serv. 1971;46:111. [Google Scholar]
- 16.Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, Miyake T. In Vitro Cell Dev Biol. 1994;30A:209–216. doi: 10.1007/BF02632042. [DOI] [PubMed] [Google Scholar]
- 17.Echalier G, Ohanessian A. C R Acad Sci Hebd Seances Acad Sci D. 1969;268:1771–1773. [PubMed] [Google Scholar]
- 18.Hershfield M S, Arredondo-Vega F X, Santisteban I. J Inherit Metab Dis. 1997;20:179–185. doi: 10.1023/a:1005300621350. [DOI] [PubMed] [Google Scholar]
- 19.Resta R, Thompson L F. Immunol Today. 1997;18:371–374. doi: 10.1016/s0167-5699(97)01047-5. [DOI] [PubMed] [Google Scholar]
- 20.Blackburn M R, Datta S K, Kellems R E. J Biol Chem. 1998;273:5093–5100. doi: 10.1074/jbc.273.9.5093. [DOI] [PubMed] [Google Scholar]
- 21.Hershfield M S, Chaffee S, Sorensen R U. Pediatr Res. 1993;33:S42–S47. doi: 10.1203/00006450-199305001-00236. [DOI] [PubMed] [Google Scholar]
- 22.Blackburn M R, Aldrich M, Volmer J B, Chen W, Zhong H, Kelly S, Hershfield M S, Datta S K, Kellems R E. J Biol Chem. 2000;275:32114–32121. doi: 10.1074/jbc.M005153200. [DOI] [PubMed] [Google Scholar]
- 23.Barry C P, Lind S E. Cancer Res. 2000;60:1887–1894. [PubMed] [Google Scholar]
- 24.Ethier M F, Chander V, Dobson J G., Jr Am J Physiol. 1993;265:H131–H138. doi: 10.1152/ajpheart.1993.265.1.H131. [DOI] [PubMed] [Google Scholar]
- 25.Meininger C J, Granger H J. Am J Physiol. 1990;258:H198–H206. doi: 10.1152/ajpheart.1990.258.1.H198. [DOI] [PubMed] [Google Scholar]
- 26.MacLaughlin M, Martinez-Salgado C, Eleno N, Olivera A, Lopez-Novoa J M. Cell Signal. 1997;9:59–63. doi: 10.1016/s0898-6568(96)00091-5. [DOI] [PubMed] [Google Scholar]
- 27.Rounds S, Yee W L, Dawicki D D, Harrington E, Parks N, Cutaia M V. Am J Physiol. 1998;275:L379–L388. doi: 10.1152/ajplung.1998.275.2.L379. [DOI] [PubMed] [Google Scholar]
- 28.Hirschhorn R, Martiniuk F, Rosen F S. Clin Immunol Immunopathol. 1978;9:287–292. doi: 10.1016/0090-1229(78)90100-9. [DOI] [PubMed] [Google Scholar]
- 29.Hershfield M, Mitchel B S. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C L, Beaudet A L, Sly W S, Valle D, editors. Vol. 2. New York: McGraw–Hill; 1995. pp. 1725–1768. [Google Scholar]
- 30.Sossin W S, Kreiner T, Barinaga M, Schilling J, Scheller R H. J Biol Chem. 1989;264:16933–16940. [PubMed] [Google Scholar]
- 31.Akalal D B, Nagle G T. Brain Res Mol Brain Res. 2001;91:163–168. doi: 10.1016/s0169-328x(01)00127-9. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Aksoy S. Gene. 2000;252:83–93. doi: 10.1016/s0378-1119(00)00226-2. [DOI] [PubMed] [Google Scholar]
- 33.Charlab R, Rowton E D, Ribeiro J M. Exp Parasitol. 2000;95:45–53. doi: 10.1006/expr.2000.4503. [DOI] [PubMed] [Google Scholar]
- 34.Charlab R, Valenzuela J G, Andersen J, Ribeiro J M. Gene. 2001;267:13–22. doi: 10.1016/s0378-1119(01)00393-6. [DOI] [PubMed] [Google Scholar]
- 35.Riazi M A, Brinkman-Mills P, Nguyen T, Pan H, Phan S, Ying F, Roe B A, Tochigi J, Shimizu Y, Minoshima S, et al. Genomics. 2000;64:277–285. doi: 10.1006/geno.1999.6099. [DOI] [PubMed] [Google Scholar]
- 36.Mears A J, el Shanti H, Murray J C, McDermid H E, Patil S R. Am J Hum Genet. 1995;57:667–673. [PMC free article] [PubMed] [Google Scholar]
- 37.Carey A H, Kelly D, Halford S, Wadey R, Wilson D, Goodship J, Burn J, Paul T, Sharkey A, Dumanski J. Am J Hum Genet. 1992;51:964–970. [PMC free article] [PubMed] [Google Scholar]