Abstract
The earliest stage in cell division in bacteria is the assembly of a Z ring at the division site at midcell. Other division proteins are also recruited to this site to orchestrate the septation process. FtsA is a cytosolic division protein that interacts directly with FtsZ. Its function remains unknown. It is generally believed that FtsA localization to the division site occurs immediately after Z-ring formation or concomitantly with it and that FtsA is responsible for recruiting the later-assembling membrane-bound division proteins to the division site. Here, we report the development of an in vivo chemical cross-linking assay to examine the association between FtsZ and FtsA in Bacillus subtilis cells. We subsequently use this assay in a synchronous cell cycle to show that these two proteins can interact prior to Z-ring formation. We further show that in a B. subtilis strain containing an ftsA deletion, FtsZ localized at regular intervals along the filament but the majority of Z rings were abnormal. FtsA in this organism is therefore critical for the efficient formation of functional Z rings. This is the first report of abnormal Z-ring formation resulting from the loss of a single septation protein. These results suggest that in this organism, and perhaps others, FtsA ensures recruitment of the membrane-bound division proteins by ensuring correct formation of the Z ring.
In Bacillus subtilis, the division septum is formed at the cell center during vegetative growth and involves the assembly of a complex of several proteins at the midcell site. The first protein known to assemble is FtsZ, which forms a ring structure (Z ring), via GTP-dependent polymerization, on the inside of the cytoplasmic membrane that subsequently contracts during septation (39). Z-ring assembly is essential for the localization of all other division proteins to the division site and is therefore a key step in the division process in bacteria (see Errington et al. [16]).
FtsA is another division protein present in many eubacteria, but its function remains elusive. It is a member of the actin family of ATP-binding proteins (5, 57) and has been shown to bind ATP (18, 34, 55, 57, 62), although ATPase activity has been demonstrated only with purified protein from B. subtilis (18). It is not known what function this ATP hydrolysis serves. Although in Escherichia coli and B. subtilis, FtsA appears to form dimers (18, 62), FtsA from Streptococcus pneumoniae has recently been shown to form long helix-like polymers in vivo (34). In E. coli, FtsA is essential for cell division. In contrast, a viable B. subtilis ftsA-null strain has previously been constructed (3), although it grew slowly, was extremely filamentous during vegetative growth, and had reduced sporulation efficiency. More recently, a B. subtilis mutant of ftsA has been isolated that shows normal vegetative division but is defective in sporulation septation, engulfment, and activation of the forespore transcription factor σF (32).
In E. coli, the pattern and timing of FtsA localization to the midcell division site is similar to that for FtsZ (11, 40, 54), suggesting that both proteins localize to this site coincidently or that FtsA is recruited immediately after FtsZ. Localization of FtsA to the division site in both organisms requires localization of FtsZ (18, 23). However, midcell Z-ring formation in E. coli does not require localization of FtsA to the division site (1, 23). More recently, it has been shown that in E. coli, in the absence of both functional FtsA and ZipA (an essential membrane-bound division protein) or in the absence of the ability of FtsZ to interact with both of these proteins, new Z rings were unable to form and preformed Z rings were destabilized (48). In other words, either FtsA or ZipA is required for the formation and stabilization of Z rings in E. coli. Interestingly, a gain of function mutation in ftsA can bypass the requirement for the essential ZipA in E. coli (19), indicating similar functions for both FtsA and ZipA. In contrast to the situation in E. coli, B. subtilis has no ZipA homolog and the requirement of FtsA for Z-ring formation in this organism has not been tested.
FtsA is probably localized to the division site in bacteria via a direct interaction with FtsZ, as this has been demonstrated in several yeast two-hybrid studies (13, 24, 34, 41, 42, 58, 61). Furthermore, the intracellular ratio of FtsZ to FtsA in both E. coli and B. subtilis is 5:1 (18, 54). It has been demonstrated for E. coli that this ratio must be maintained for correct division; overexpression of either ftsZ or ftsA leads to a division block that can be reversed by simultaneous overexpression of the other gene (4, 9, 12, 40). The number of FtsA molecules per cell in B. subtilis (and probably E. coli [54]) is sufficient to form a complete circumferential ring at the division site (18). Although it has been shown in E. coli that FtsA is not required for Z-ring formation at the division site, the demonstration that these two proteins interact in a yeast two-hybrid system suggests that this interaction can occur independently of Z-ring formation and supports the idea that FtsZ and FtsA localize coincidently to the division site (48, 54).
Recruitment of the various cell division proteins in E. coli occurs largely through the sequential addition of individual proteins to this site (see Weiss [59]). In this organism, localization of FtsA to the division site is required for recruitment of the later-assembling membrane-bound division proteins. In contrast, the B. subtilis division proteins localize in a more concerted manner (see Errington et al. [16]). In other words, if one of the membrane-bound proteins (DivIB, DivIC, FtsL, PBP 2B) is depleted or absent, none of the other three proteins are recruited to the division site (see Errington et al. [16]). As with E. coli, localization of FtsA to the division site in B. subtilis does not appear to depend on localization of the membrane-bound division proteins (18). But it is not known if FtsA is required for the recruitment of the later-assembling membrane-bound division proteins in B. subtilis.
FtsZ can polymerize in vitro into protofilaments and higher-order structures (6, 15, 37, 47). However, how FtsZ polymerizes in vivo to form a Z ring is unknown. The ZipA (present only in E. coli) and ZapA (present in both E. coli and B. subtilis) proteins appear to have a role in stabilizing Z rings by promoting the bundling of FtsZ protofilaments (20, 22, 36, 50). Since ZipA and FtsA of E. coli seem to have overlapping roles in forming and stabilizing the Z ring and both bind to a similar interaction site at the extreme C terminus of FtsZ from this organism (41), it is possible that FtsA is also involved in promoting the bundling of FtsZ protofilaments.
In this study, we have used chemical cross-linking to investigate the interaction between FtsZ and FtsA in B. subtilis cells and have consequently developed the first in vivo assay for their association. We subsequently used this assay during the first cell cycle following spore germination and outgrowth to show that FtsZ and FtsA associate prior to Z-ring assembly at the midcell site. This raised the possibility that in B. subtilis, FtsA is actually required for Z-ring formation in vivo. Consequently, we found that in the absence of FtsA, FtsZ localized at regular intervals in these filaments, but in the majority of cases, abnormal Z rings were observed. Therefore, unlike the situation in E. coli, there is an absolute requirement for FtsA in B. subtilis for efficient Z-ring assembly.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and spore preparation/germination.
Bacterial strains used in this study are listed in Table 1. All B. subtilis strains were grown on tryptose blood agar base plates supplemented with thymine (20 μg ml−1) or in antibiotic medium 3 (Penassay broth [PAB]) at 37°C unless otherwise stated. These media were supplemented with chloramphenicol (5 μg ml−1), phleomycin (2 μg ml−1), spectinomycin (80 μg ml−1), isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM), and xylose (0.2%, wt/vol) when required. B. subtilis-competent cells were prepared according to the method of Anagnostopoulos and Spizizen (2), including the modification suggested by Wilson and Bott (60). In addition, the G-medium contained 1× trace metals (29).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 168 (SU5) | trpC2 | E. Nester |
| BB11 | trpC2 ftsZ::pJSIZΔpble (Pspac-ftsZ ble) | 3 |
| 804 | trpC2 ftsL::pSG441 (Pspac-pbpB aphA-3) φ105J106 (Pxyl-ftsL cat) | 10 |
| SU434 | thyA thyB trpC2 amyE::(Pxyl-ftsZ-yfp spc) | 46 |
| SU456 | trpC2 ftsZ::pJSIZΔpble (Pspac-ftsZ ble) | This study (strain 168 transformed with BB11 DNA) |
| SU457 | trpC2 ftsZ::pJSIZΔpble (Pspac-ftsZ ble) ftsA::cat | This study (see Materials and Methods) |
Strain SU434 (ftsZ-yfp) spores were prepared by the method described by McGinness and Wake (45), except 2% Difco potato dextrose agar supplemented with an additional 1.25% Difco Bacto agar was used instead of Difco potato extract. Germination (2 × 108 spores ml−1) and outgrowth of spores was performed at 34°C in GMD medium (26) supplemented with thymine (20 μg ml−1) and xylose (0.2%, wt/vol).
Cross-linking and immunoprecipitation.
Formaldehyde cross-linking was performed as described by Skare et al. (56) with the following modifications. B. subtilis cells were grown in PAB at 37°C to an optical density at 600 nm (OD600) of 0.6 and pelleted by centrifugation. The cell pellet was washed in an equal volume of 0.1 M NaPO4 buffer, pH 6.8, and resuspended in the same buffer, and formaldehyde was added to final concentrations of either 0.1 or 1% (wt/wt). Cells were incubated at room temperature for various times, pelleted, and washed in 0.1 M NaPO4 buffer, pH 6.8, and then solubilized in lysis buffer as for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (27). By using 1% formaldehyde, the majority (60 to 75%) of outgrown cells are dead within 30 s to 1 min, indicating rapid cell death under these conditions. The washing and centrifugation step had no effect on the appearance or frequency of Z rings. Cross-linked protein samples were heated either at 37°C for 10 min to maintain the formaldehyde cross-links or at 95°C for 30 min to break them prior to electrophoresis on 10% polyacrylamide gels.
Immunoprecipitation was performed using protein G-agarose beads (Roche). Sixty microliters of cross-linked whole-cell extract (lysis buffer suspension) was precleared with 50 μl of protein G-agarose bead suspension in 1 ml of wash buffer 1 (50 mM Tris buffer, 500 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100) at 4°C for 4 h on a rotator. The agarose beads were pelleted by centrifugation and the supernatant added to 15 μl of polyclonal anti-FtsZ sheep serum and incubated at 4°C for 1 h on a rotator. 50 microliters of protein G-agarose bead suspension was added to the sample and incubation continued overnight at 4°C. Antibody-protein complexes bound to the agarose beads were pelleted by centrifugation and washed twice with 1 ml of wash buffer 1, twice with wash buffer 2 (wash buffer 1 containing 0.1% Triton X-100), and once with wash buffer 3 (wash buffer 2 containing 150 mM NaCl). Forty microliters of SDS-PAGE sample loading buffer was added to the agarose beads, and the immunoprecipitates were removed by heating at 100°C for 3 min. The agarose beads were then pelleted and the supernatant used for SDS-PAGE and Western blot analysis.
Affinity purification of anti-FtsA antibodies.
Polyclonal anti-FtsA rabbit serum was affinity purified by passaging 50 μl (5×) over Affigel-15 (Bio-Rad) MicroSpin columns (Amersham Pharmacia Biotech) to which FtsA had been coupled as per the manufacturer's instructions. The column was washed with 5 column volumes of 10 mM Tris-HCl, pH 7.5, and then 10 mM Tris-HCl-0.5 M NaCl, pH 7.5. Anti-FtsA antibodies were eluted in 50 μl of 4.5 M MgCl2-Tris-HCl, pH 7.5. This was added to an equal volume of a solution containing 2× phosphate-buffered saline, 80% glycerol, 1% bovine serum albumin, and 0.04% sodium azide and stored at 4°C.
SDS-PAGE and Western blotting.
Solubilization of B. subtilis cells in lysis buffer, SDS-PAGE and Western transfers were performed as described previously (27). Primary antibodies used were either affinity-purified rabbit anti-FtsA antibodies at a 1:200 dilution or rabbit anti-FtsZ serum at a 1:10,000 dilution. Detection of primary antibodies was performed using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Promega) at a 1:2,500 dilution and the enhanced chemiluminescence Western blotting detection system (Amersham Pharmacia Biotech). Protein levels were quantified using ImageJ software, version 1.32 (Wayne Rasband, National Institutes of Health, Bethesda, Md.). For each sample, an appropriate dilution in the linear range for immunoblotting, with either FtsZ or FtsA antibodies, was chosen for densitometric analysis.
Microscopy.
Cells were ethanol-fixed as described by Hauser and Errington (30). Detection of FtsZ-yellow fluorescent protein (YFP) in live cells of outgrown SU434 spores incubated with thymine (20 μg ml−1) and xylose (0.2%, wt/vol) was performed using 2% agarose pads prepared with the same medium. Immunofluorescence microscopy for the detection of FtsZ was performed as described by Harry and Wake (28) with the following modifications. Lysozyme (10 mg ml−1) treatment was performed for 3 min after poly-l-lysine treatment of microscope slides. Secondary antibody incubation was performed using fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch) at a 1:100 dilution and 4′,6′-diamidino-2-phenylindole (DAPI; 0.2 μg ml−1).
All phase contrast and fluorescence images were obtained using a Zeiss Axioplan 2 fluorescence microscope equipped with a 100× phase objective and an AxioCam MRm cooled charge-coupled device camera controlled through AxioVision software, version 4.2 (Carl Zeiss). YFP, FITC, and DAPI fluorescence were visualized with filter sets 41029 (490- to 510-nm excitation filter, 515-nm beamsplitter, 520-nm long-pass (LP) barrier filter; Chroma Technology), 09 (450- to 490-nm band-pass excitation filter, 510-nm beamsplitter, 515-nm LP barrier filter; Zeiss), and 02 (365-nm excitation filter, 395-nm beamsplitter, 420-nm LP barrier filter; Zeiss), respectively. Image analysis and processing were performed using AxioVision 4.2, and images were prepared for publication using Adobe Photoshop 7.
Construction of the ftsA-null strain.
The ftsA gene was replaced with a drug cassette based on the PCR joining method described by Fabret et al. (17). The chloramphenicol resistance gene and DNA fragments flanking the ftsA gene were amplified using Pfu DNA polymerase (Stratagene). The 1.4-kb upstream ftsA fragment was amplified using oligonucleotides ftsAp1 (5′-GGAATGACTGAAAAAGCG-3′) and ftsAp2 (5′-GGATTTGAGCGTAGCGAAAATCACTTTGGTATTGGACG-3′) and the 1.4-kb downstream ftsA fragment using oligonucleotides ftsAp3 (5′-CCCGTTAGTTGAAGAAGGTTCCGAACATAATAAACAGAGC-3′) and ftsAp4 (5′-TGTCCTTTACATTAGCCG-3′). The 800-bp chloramphenicol resistance gene was amplified from the plasmid pPY18 (14) using oligonucleotides cmlp1 (5′-TTTTCGCTACGCTCAAATCC-3′) and cmlp2 (5′-AACCTTCTTCAACTAACGGG-3′). The 5′ end of ftsAp2 contained the reverse complement sequence of cmlp1 (20 bp), and the 5′ end of ftsAp3 contained the reverse complement of cmlp2 (20 bp). Equal amounts (1 μg) of the ftsA and chloramphenicol fragments were mixed and subjected to a PCR joining reaction (5 min at 94°C [10 s at 94°C, 10 s at 55°C, 10 min at 68°C] for 36 cycles; 10 min at 68°C) using Platinum Pfx DNA polymerase (Invitrogen). The joining reaction mix was then used to directly transform strain SU456-competent cells (Table 1). Transformants were selected on tryptose blood agar base plates containing chloramphenicol (5 μg ml−1), phleomycin (2 μg ml−1), and IPTG (1 mM). DNA extracted from several transformants was checked for inactivation of the ftsA gene by PCR using ftsAp1 and cmlp2. The integrated region upstream of ftsA from one transformant was then sequenced to check that no base errors had been incorporated. This transformant was subsequently chosen as the ftsA-null strain (SU457).
RESULTS
In vivo cross-linking of FtsZ and FtsA using formaldehyde.
Many of the known bacterial division proteins are either membrane associated (FtsZ and FtsA [33, 49, 51, 55]) or membrane bound, and little is known about the in vivo interactions which comprise the division complex. As a means of studying these interactions in B. subtilis, we used in vivo chemical cross-linking. We chose formaldehyde as an in vivo cross-linker, as it is a fairly nonspecific cross-linker and has previously been used to study membrane protein complexes (35, 43, 56). We initially focused on FtsZ and FtsA, as these proteins are known to interact in a yeast two-hybrid system and therefore provide a positive control for testing our in vivo approach. In addition, this would enable us to establish an in vivo assay that would allow examination of FtsZ and FtsA interaction under different conditions and at different stages in the cell cycle.
Vegetatively growing wild-type B. subtilis cells (strain 168) were collected and treated with 0.1 and 1% formaldehyde. Whole-cell extracts were then subjected to SDS-PAGE and cross-linked complexes detected by Western blotting with either FtsZ or FtsA polyclonal antibodies. In the case of FtsA, a higher-molecular-mass complex of ∼150 kDa was readily formed using both treatments (0.1 and 1% formaldehyde) and an additional complex of ∼235 kDa could be visualized using 1% formaldehyde (Fig. 1A, lanes 3 and 4). Extended cross-linking (≥20 min) resulted in almost no detectable FtsA protein at the apparent molecular mass of monomer ∼53 kDa (theoretical value, 48.1 kDa) (Fig. 1A, lanes 3 and 4).
FIG. 1.
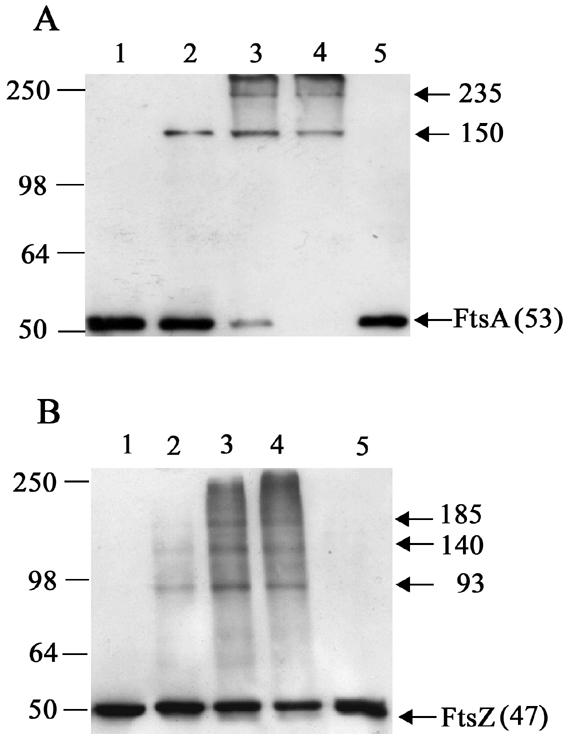
Western blot analysis of FtsA (A) and FtsZ (B) in B. subtilis strain 168 cells cross-linked in vivo with formaldehyde. B. subtilis strain 168 cells were treated with formaldehyde as follows: lane 1, no formaldehyde; lane 2, 0.1% for 10 min; lane 3, 1% for 10 min; lanes 4 and 5, 1% for 20 min. Samples in lane 5 have been incubated at 95°C for 30 min to break the cross-links. All other samples were heated at 37°C for 10 min to maintain the formaldehyde cross-links. Arrows indicate FtsZ and the FtsA monomer or specific complexes and their apparent molecular masses in kDa. Positions and sizes of the protein standards (SeeBlue; Invitrogen) are shown on the left.
In contrast to FtsA, several higher-molecular-mass FtsZ complexes could be visualized among a smeared background by using 1% formaldehyde for 10 or 20 min (Fig. 1B, lanes 3 and 4). The most dominant of these complexes was present using both treatments and had molecular masses of ∼93, ∼140, and ∼85 kDa, which possibly represent FtsZ oligomerization (dimer, trimer, and tetramer, respectively). The smeared background is most likely caused by FtsZ intramolecular cross-links and/or FtsZ oligomers interacting with a number of different proteins, identified by in vitro studies (20, 21, 31). Note that both of the FtsA and FtsZ cross-linked complexes were absent from cells not treated with formaldehyde and could be destroyed by heating samples at 95°C for 30 min (Fig. 1A and 1B, lanes 1 and 5, respectively).
FtsZ and FtsA are present in the same cross-linked complexes (150 and 235 kDa).
Formaldehyde cross-linking allows the reproducible visualization of two distinct FtsA-containing complexes (150 and 235 kDa). Since there is good evidence that FtsZ and FtsA interact directly in B. subtilis (13, 24, 41, 42, 58, 61), it is likely that these complexes contain FtsZ; however, they are not present in levels high enough to allow detection among the more abundant FtsZ-containing complexes using the anti-FtsZ serum. To determine if FtsZ is present in these complexes, we performed two experiments. Firstly, we depleted FtsZ in strain SU456. If FtsZ is present in the 150- and 235-kDa complexes, then depletion of this protein should result in a significant decrease in their level. In this strain, ftsZ expression is controlled by the IPTG-inducible Pspac promoter. Strain SU456 was grown in the presence of IPTG and then resuspended in PAB without IPTG for 120 min. During this time period, the cellular level of FtsZ decreased to ∼15% of that present in cells grown with IPTG while FtsA levels remained constant (Fig. 2A). Cells were then treated with both 0.1% and 1% formaldehyde for 10 min. In the absence of IPTG, the 150- and 235-kDa FtsA-containing complexes detected with the FtsA polyclonal antibodies decreased in intensity (Fig. 2B, lanes 2 and 4). This indicates that the cellular level of these complexes depends on a normal level of FtsZ and is consistent with FtsZ being present in these complexes. The decreased level of the complexes appears to be specific to the depletion of FtsZ and not a general block in cell division, as the levels of these complexes remain constant in other situations that block division, such as in a divIB-null mutant strain (28) at the nonpermissive temperature or in an FtsL/PBP2B depletion strain (10) when either FtsL or PBP2B is depleted (data not shown).
FIG. 2.
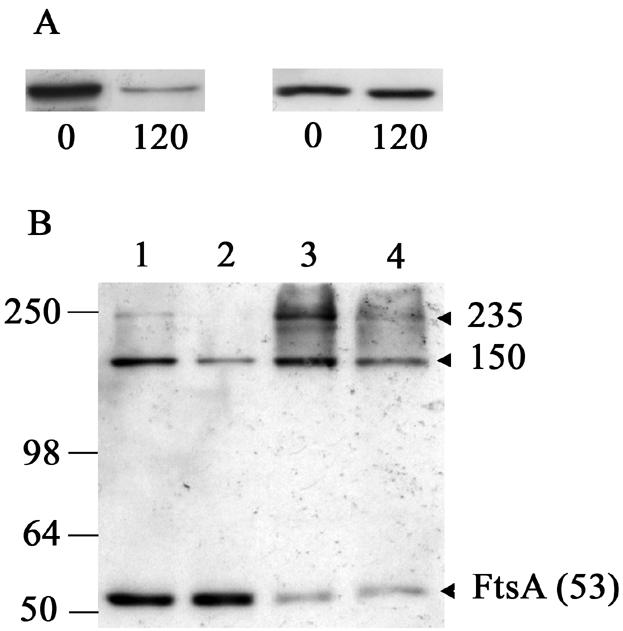
Western blot analysis of FtsZ and FtsA in B. subtilis strain SU456 (Pspac-ftsZ) in the presence and absence of IPTG. (A) SU456 cells were grown at 37°C in PAB in the presence of IPTG to mid-exponential phase (0, left lane) and resuspended in PAB in the absence of IPTG for 120 min (120, right lane). FtsZ (left) and FtsA (right) were detected with antiserum and purified polyclonal antibodies, respectively. (B) SU456 cells grown in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of IPTG for 120 min were cross-linked with 0.1% (lanes 1 and 2) and 1% (lanes 3 and 4) formaldehyde for 10 min. Arrows indicate the FtsA monomer or specific FtsA-containing complexes and their apparent molecular masses in kDa. Positions and sizes of the protein standards (SeeBlue; Invitrogen) are shown on the left.
To definitively establish the presence of FtsZ in these complexes, we performed immunoprecipitation experiments using anti-FtsZ sheep serum. Vegetatively growing cells of the wild-type strain (strain 168) and strain SU456 (Pspac-ftsZ, with or without IPTG) were treated with 1% formaldehyde for 10 min. These whole-cell extracts were then subjected to immunoprecipitation using anti-FtsZ sheep serum, and precipitated complexes were immunodetected using FtsA polyclonal antibodies (from rabbit). The anti-FtsZ sheep serum coimmunoprecipitated the 150- and 235-kDa FtsA-containing complexes from the wild-type strain (Fig. 3, lane 2). No FtsA monomer was detected with the FtsA antibodies following immunoprecipitation, demonstrating that FtsA alone does not precipitate with the anti-FtsZ serum (Fig. 3, lanes 2 through 4). When FtsZ was depleted using strain SU456 (Pspac-ftsZ) as described above, a decreased amount of the 150-kDa and 235-kDa complexes could be coimmunoprecipitated (Fig. 3, lane 3), reflecting diminished association between FtsZ and FtsA, which is again consistent with these complexes containing both FtsZ and FtsA. These results establish that FtsZ and FtsA are associated in the 150- and 235-kDa complexes. We have therefore successfully developed an in vivo assay for FtsZ and FtsA association in B. subtilis.
FIG. 3.
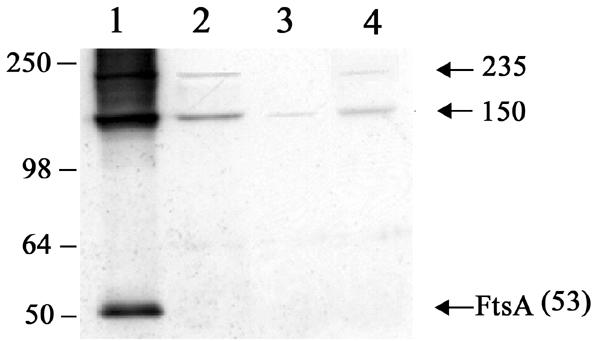
Western blot analysis of FtsA-containing cross-linked complexes immunoprecipitated with anti-FtsZ serum. B. subtilis strains 168 (without IPTG; lane 2) and SU456 (Pspac-ftsZ; without or with IPTG in lanes 3 and 4, respectively) were cross-linked with 1% formaldehyde for 10 min and whole-cell extracts immunoprecipitated with anti-FtsZ sheep serum. FtsA polyclonal antibodies (rabbit) were then used to detect FtsA-containing complexes. Lane 1 contained a cross-linked lysate of B. subtilis strain 168 (without IPTG) that had not been immunoprecipitated. Arrows indicate the FtsA monomer or specific FtsA-containing complexes and their apparent molecular masses in kDa. Positions and sizes of the protein standards (SeeBlue; Invitrogen) are shown on the left.
In vivo association of FtsZ and FtsA in the absence of Z rings. At present, it is generally believed that FtsA is recruited to the midcell site immediately after FtsZ. However, our observation that the FtsA monomer can almost be completely cross-linked into higher-molecular-mass complexes in vegetatively growing cells (Fig. 1A) raises the possibility that FtsA is always associated with FtsZ and therefore colocalizes as such to the midcell division site (although this is not the only interpretation of this observation). We utilized our in vivo assay to determine whether this is in fact the case. We took advantage of the single synchronous cell cycle that occurs following spore germination to determine if we could detect an association between FtsZ and FtsA prior to Z-ring assembly.
We used strain SU434 to visualize Z rings in live cells during spore outgrowth. This strain contains a xylose-inducible ftsZ-yfp construct in addition to a wild-type copy of ftsZ under the control of its endogenous promoter (46). It has been shown previously that the FtsZ-YFP fusion product alone cannot form Z rings (46); therefore, wild-type FtsZ is required to detect Z rings in this way. Strain SU434 spores were germinated in GMD medium (26) containing 0.2% xylose and 20 μg ml−1 thymine at 34°C. Samples were collected at 30-min intervals between 1 and 5 h after germination, and Z-ring formation was monitored using fluorescence microscopy (100 outgrown cells were scored). A number of different localization patterns were apparent during the time course, and these were similar to those previously described for DivIB (28). Representative fields of the 1.5-, 2-, and 4-h time points are shown in Fig. 4. At 1.5 h following the addition of spores to GMD medium, we could not detect any FtsZ localization (Fig. 4a). At 2 h, cells started displaying possible early FtsZ localization as a dot(s) on the side of the membrane in 2.7% of cells (Fig. 4b). By 4 h, 85% of cells contained a sharp Z ring (Fig. 4c). Various levels of xylose were tested in these experiments and at the lowest concentration used (0.02%) that allowed detection of FtsZ-YFP in cells, essentially the same results were obtained.
FIG. 4.
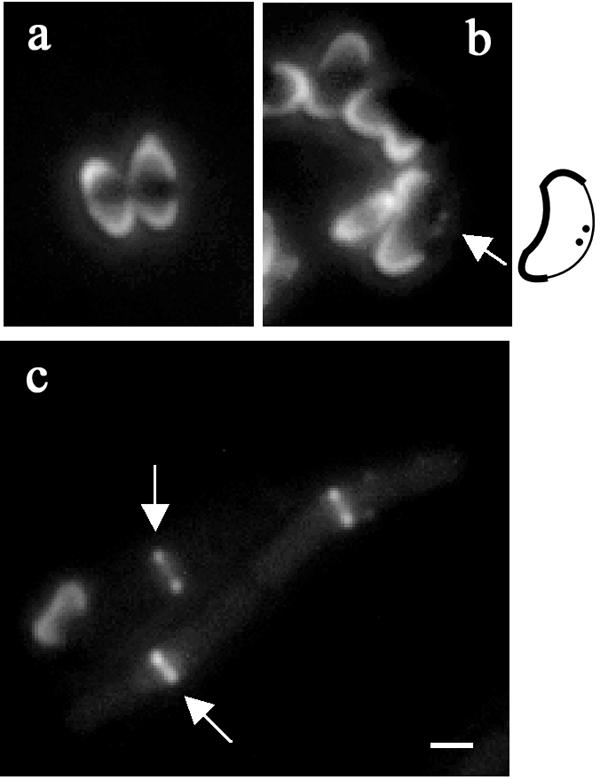
Localization of FtsZ-YFP in B. subtilis strain SU434 (Pxyl-ftsZ-yfp) during spore outgrowth. The 1.5-, 2-, and 4-h time points (after germination) are represented in panels a, b, and c, respectively. The fluorescent images are shown. Arrows indicate FtsZ localization [dot(s) on the side of the membrane or medial band]. Note that the attached spore coats in the 1.5- and 2-h time points are autofluorescent. A schematic is included next to panel b to show the outgrowing spore boundaries and position of the FtsZ localizations. Scale bar represents 1 μm.
The 1.5-h (no FtsZ localization detected) and 4-h (sharp Z rings) time points were chosen as representing the absence and presence of FtsZ localization, respectively, and outgrown cells were subjected to formaldehyde cross-linking and immunodetection using the FtsA polyclonal antibodies. Prior to conducting the cross-linking experiments, cells were lysed as per vegetative cells and the approximate protein concentration of the soluble fraction was determined from a fixed volume of cells using the UV A260 method (25). In an individual cell, the total protein concentration will change during spore outgrowth; therefore, based on these concentrations (which were reproducible), cell densities in subsequent experiments were adjusted so that an equal amount of total protein was cross-linked at each time point. A protein sample of wild-type vegetatively growing cells, treated with formaldehyde, was included as a control. Interestingly, the 150-kDa complex was present at both 1.5 and 4 h after the beginning of germination (Fig. 5, lanes 1 and 2). The presence of this complex at the 1.5-h time point, when no FtsZ localization had occurred, indicates that FtsZ and FtsA associate prior to any observable localization of FtsZ to the division site. It is likely that the 235-kDa complex was present in these samples but present at insufficient levels to allow consistent detection.
FIG. 5.
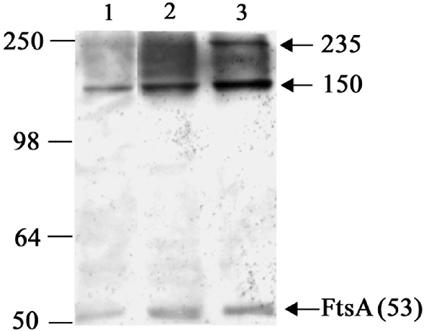
Western blot analysis of FtsA cross-linked in vivo with formaldehyde during the first cell cycle (that is, prior to cell division) following spore germination. B. subtilis strain SU434 (Pxyl-ftsZ-yfp) spores were incubated in GMD medium (supplemented with 0.2% xylose, 20 μg ml−1 thymine). After 1.5 (lane 1) and 4 (lane 2) h of incubation in GMD medium, proteins were cross-linked with 1% formaldehyde for 1 min. FtsA-containing complexes were detected with FtsA polyclonal antibodies. An equal amount of total protein was cross-linked at each time point. Arrows indicate the FtsA monomer or specific FtsA-containing complexes and their apparent molecular masses in kDa. A whole-cell extract of vegetatively grown B. subtilis strain 168 cells treated with 1% formaldehyde for 10 min (lane 3) was included as a control to compare the electrophoretic migration of the 150- and 235-kDa complexes in the different cell lysates. Positions and sizes of the protein standards (SeeBlue; Invitrogen) are shown on the left.
It is interesting to note that FtsZ-YFP does not appear to interact significantly with FtsA, as there were no additional high-moleclular-mass complexes that were at least 28 kDa (mass of YFP) larger than 150 kDa or 235 kDa. This is consistent with our previous observation that FtsZ-YFP cannot form Z rings or support growth and division in the absence of wild-type FtsZ (46). It is possible that this is a consequence of the fusion protein not being able to interact with FtsA.
Z-ring localization in an ftsA-null strain.
Our discovery that FtsZ and FtsA associate in the complete absence of any FtsZ localization to the division site raises the interesting possibility that FtsA in B. subtilis may actually be required for Z-ring formation. To investigate this, we created an ftsA-null strain by replacing the ftsA gene with a chloramphenicol resistance gene in strain SU456 (Pspac-ftsZ). In SU456, the expression of the downstream ftsZ gene is controlled by the IPTG-inducible Pspac promoter (see Materials and Methods). We obtained 10 Cmr transformants in duplicate transformation experiments. The low number of transformants was probably due to a low frequency of homologous recombination between the short region of DNA downstream of ftsA in the PCR construct and the chromosome (see below and Materials and Methods). This transformation efficiency is normal under our conditions. Two transformants from each duplicate experiment were grown in PAB. All grew at similar rates and formed filaments (see below). One of each of these was used in Western analysis and immunofluorescence microscopy to confirm the absence of FtsA and determine the FtsZ localization pattern, respectively. The same results were obtained in each case and are discussed below. The ftsA deletion was confirmed for one of these transformants, named SU457, by sequencing. Chromosomal DNA from this strain was used to transform SU456 again. Several hundred Cmr transformants were obtained, indicating the ease with which this null mutation can be obtained and tolerated. The ftsA-null strain (SU457; ΔftsA Pspac-ftsZ) and the parent strain (SU456; Pspac-ftsZ) were grown in PAB supplemented with 1 mM IPTG at 37°C, and samples were collected during vegetative growth at an OD600 of 0.3 for analysis. It took twice as long for the OD600 of strain SU457 cells to double compared to strain SU456 (42 min in comparison to 24 min), and SU457 cells typically reached a lower stationary-phase OD600 (∼1.2 for the mutant compared to 2 for the parent strain). This difference in the rate of increase in OD600 is at least partly due to lysis of the ftsA-null cells (see below). Viability assays were performed on the ftsA-null strain at various times during early-, mid-, and late-exponential-phase growth and compared with the isogenic parent. In the ftsA-null strain at any given OD600, CFU counts were lower than its isogenic strain. This was expected, as the ftsA-null cells are approximately four times longer, so the maximum frequency of division is 25%. However, since it is clear that there is a significant lysis of cells in the ftsA-null strain, the frequency of division cannot be determined. Significantly, CFU per OD600 is constant throughout the growth curve, indicating that ftsA is not undergoing a major lytic event at a particular stage of growth but rather appears to be susceptible to cell lysis over the entire growth curve.
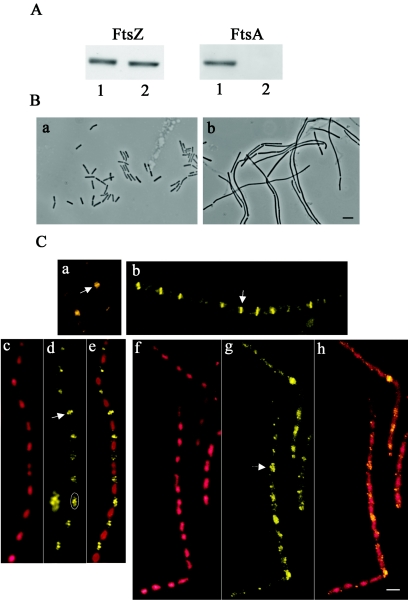
FtsZ levels were essentially the same in strain SU457 (ftsA null) and the parent strain, SU456, and, as expected, FtsA was not detectable in SU457 (Fig. 6A). Interestingly, in contrast to the previously constructed ftsA-null strain (3), which was reported as being extremely filamentous, SU457 produced filaments of varying length (Fig. 6B, panel b). The average cell lengths of SU456 (Pspac-ftsZ) and SU457 (ΔftsA Pspac-ftsZ) were 5.9 ± 1.0 and 22.6 ± 11.9 μm (± standard deviation of the mean), respectively. However we did observe cell lysis (Fig. 6B, panel b), suggesting that our mutant also has a severe phenotype. These differences may be explained by the different genetic backgrounds, the different media used, or the different null alleles constructed.
FIG. 6.
Analysis of the B. subtilis ftsA-null strain SU457 (ΔftsA Pspac-ftsZ) during vegetative growth. SU456 (Pspac-ftsZ) and SU457 were grown in PAB supplemented with 1 mM IPTG at 37°C and collected at an OD600 of 0.3 for analysis. (A) Western blot analysis of FtsZ and FtsA levels in SU456 (lane 1) and SU457 (lane 2) using anti-FtsZ serum and FtsA polyclonal antibodies. (B) Phase-contrast images of ethanol-fixed cells. SU456 and SU457 are represented by panels a and b, respectively. Scale bar represents 6 μm. (C) Immunolocalization of FtsZ in vegetatively growing cells of strains SU456 (a), 804 depleted of FtsL (Pxyl-ftsL, Pspac-pbpB (10) (b), and SU457 (two sets of images [c through e and f through h] of the different FtsZ localization patterns observed are shown). FITC immunofluorescence (a and b, yellow) is shown for strains SU456 and 804. DAPI fluorescence (c and f, red), FITC immunofluorescence (d and g, yellow), and an overlay of these images (e and h) are shown for the two SU457 images. Typical (sharp band) and atypical (diffuse band) FtsZ localizations are indicated by solid and dotted lined arrows, respectively. An example of a spiral-like structure (enlargement shown in inset) has been circled in panel d. Scale bar represents 1.8 μm.
Analysis of vegetatively growing cells using immunofluorescence microscopy (Fig. 6C) revealed normal formation of Z rings in SU456 (Pspac-ftsZ) (Fig. 6C, panel a). Approximately 90% of these cells had a Z ring. In contrast, FtsZ localization in SU457 (ΔftsA Pspac-ftsZ) was abnormal (Fig. 6C, panels c through h). In this strain, FtsZ mainly localized as regular, diffuse bands of fluorescence that appeared to occupy the nucleoid-free regions of a filament. These FtsZ localizations occurred at a frequency of 0.17 localizations per μm, which is very similar to the frequency of Z rings at midcell in SU456 (0.18 Z rings per μm; 50 filaments scored in each case). In some cases, FtsZ localizations in the absence of ftsA resembled a spiral-like structure (Fig. 6C, panel d, circle). Although the occasional typical Z ring (sharp band) could be observed, it occurred at a frequency of only 10% when normalized to the number of Z rings per μm for SU456. To check that these abnormal FtsZ localizations were not due to any effect that filament formation may have on Z-ring assembly, we also examined the appearance of Z rings in the filaments of strain 804 (Pxyl-ftsL, Pspac-pbpB) (10) under identical conditions. As previously reported, when FtsL was depleted in strain 804, FtsZ localized as typical Z rings at regular intervals along the length of a filament (Fig. 6C, panel b). Therefore, the atypical FtsZ localizations observed in SU457 are a direct result of the absence of FtsA in the cell. This demonstrates that FtsA is required for the efficient formation of Z rings in the cell. In its absence, FtsZ is able to localize to approximately the division site at the same frequency but is rarely able to form normal-looking Z rings. This is likely to be a result of a requirement for FtsA in Z-ring formation and/or Z-ring stability.
DISCUSSION
Several previous yeast two-hybrid analyses have shown that FtsZ and FtsA interact directly (13, 24, 34, 41, 42, 58, 61). We have now examined the in vivo association between FtsZ and FtsA in B. subtilis using chemical cross-linking experiments. This is the first time this approach has been used to monitor the association of these two proteins in bacterial cells. Using formaldehyde as the cross-linker, we have identified two higher-molecular-mass protein complexes, of 150 kDa and 235 kDa, that contain both FtsZ and FtsA. These were visualized in wild-type cells by using FtsA polyclonal antibodies. We demonstrated that these complexes do indeed contain both FtsZ and FtsA by showing that (i) the level of these complexes decreases when the cellular level of FtsZ decreases and (ii) anti-FtsZ serum could precipitate these FtsA-containing complexes. Our in vivo assay for FtsZ and FtsA association will enable an examination of this association under different conditions and at different stages in the cell cycle.
What is the full composition of the 150-kDa and 235-kDa complexes? It is most likely that the FtsZ and FtsA proteins are directly interacting in both complexes. The 150-kDa complex is seen more readily in our cross-linking experiments, with only 0.1% formaldehyde and 10-min incubation. In both E. coli and B. subtilis, there is evidence that FtsA dimerizes in vivo (18, 52, 62). Therefore, the 150-kDa complex may contain a dimer of FtsA and a monomer of FtsZ. The 235-kDa complex could also contain only FtsZ and FtsA in a specific stoichiometry. Alternatively, both complexes may contain other division proteins or new, uncharacterized proteins of B. subtilis. It will be interesting to determine whether other proteins are present in these complexes and, if so, to identify them.
Although it is possible that FtsA was present in very large cross-linked complexes not observed here, our observation that most or all of the monomeric FtsA in the cell could be cross-linked raised the possibility that this protein is always associated with FtsZ in B. subtilis, even when FtsZ is not polymerized to form a ring structure. Consistent with this suggestion, previous studies have shown that in E. coli, the timing of FtsA localization to the midcell division site is similar to that of FtsZ (11, 40, 54). However, since FtsZ can form a ring at the division site independently of FtsA (1, 23, 48) in E. coli, it is not clear whether FtsA colocalizes with FtsZ to form a Z ring or whether it is recruited immediately after Z-ring formation. It has been shown that in B. subtilis, localization of FtsA to the division site requires FtsZ (18), but it is not known if midcell Z-ring formation actually requires FtsA in this organism. We examined the first cell cycle following spore germination and outgrowth in B. subtilis to determine if the association between FtsZ and FtsA could occur prior to midcell Z-ring assembly. We found that FtsZ and FtsA do indeed associate prior to any FtsZ localization or Z-ring formation at the division site. This suggests that these proteins are always complexed together, possibly as a subunit of an assembled ring structure, and localize as such to the division site. This is consistent with previous yeast two-hybrid results that show a direct interaction between these two proteins in the nuclei of nonnative organisms (yeast), where they would not be expected to form normal Z rings (13, 24, 34, 41, 42, 58, 61). The finding that FtsZ and FtsA associate prior to Z-ring formation at midcell tempted us to speculate that FtsA is actually required for Z-ring assembly in B. subtilis. We therefore constructed an ftsA-null strain and tested its ability to form Z rings. An ftsA-null strain has been constructed previously (3), but the ability of FtsZ to localize to the division site was not tested. Similar to previous results (3), our ftsA-null strain was viable. It grew slowly and produced filaments of varying length during vegetative growth. Most intriguingly, although FtsZ localizations occurred at regular intervals, very few normal-looking Z rings were observed. Since the ftsA-null strain can divide, albeit inefficiently, it is likely that the few normal Z rings observed (10%) actually constrict to result in division. This is the first situation identified where loss of a single septation protein has led to such a drastic reduction in the level of apparently normal Z rings. This result suggests a critical role for FtsA in Z-ring formation and/or Z-ring stability in B. subtilis. Although the possibility that a secondary (suppressor) mutation has occurred in the vicinity of the ftsA gene in our strain cannot be ruled out, the ease with which the ftsA-null mutation was obtained in B. subtilis in two independent experiments here, and by other researchers previously (3), makes this highly unlikely.
The dependency of Z-ring formation on FtsA in B. subtilis is entirely consistent with the recent discovery that FtsA contains a membrane-targeting sequence that anchors the Z ring to the membrane in E. coli (49). FtsA from B. subtilis also contains this membrane-targeting sequence (49). Presumably, ZipA can perform a role similar to FtsA in E. coli because either ZipA or FtsA, but not both, is required for normal Z-ring formation (48). Since there is no ZipA homolog in B. subtilis, FtsA is likely to be absolutely required for anchoring the Z ring to the membrane. ZipA has been identified in only a subset of gram-negative bacteria (50), whereas FtsA is more widely conserved, being present in over 20 bacterial genera (44). Our findings raise the possibility that FtsA plays a more critical role in Z-ring assembly in other organisms that don't have a ZipA homolog.
The ability of the ftsA-null strain of B. subtilis to divide at all is interesting, given that in E. coli, depletion of the FtsA protein or ftsA mutation blocks septation (7, 16, 59). In E. coli, these situations also block the recruitment of membrane-bound division proteins and it has been suggested that FtsA plays a role in this recruitment. This could be via its ability to bind these proteins directly or by ensuring that the Z ring is formed correctly so that it can recruit them. Recent evidence supports the former possibility for E. coli, as it was demonstrated that the FtsA protein can recruit the two membrane-bound division proteins FtsI and FtsN independently of FtsZ in this organism (8). In contrast, our results suggest that in B. subtilis, there is no absolute requirement for FtsA in the recruitment of the membrane-bound division proteins. The ability of the ftsA-null strain of B. subtilis to divide is most likely to be due to a low level of normal Z-ring formation observed in this strain that subsequently recruits the membrane-bound division proteins. So the role of FtsA in B. subtilis in the recruitment of other division proteins to the division site appears to be an indirect one, through ensuring that FtsZ forms a proper ring. Perhaps this apparent difference in the role of FtsA in these organisms reflects an important difference in the way in which the division complex is assembled.
What is the role of FtsA in Z-ring formation? The ease with which FtsZ can polymerize on its own in vitro indicates that no other protein is required for its self association. However, several positive and negative regulators of FtsZ polymerization exist that influence the ability of FtsZ to form a ring structure in vivo (38, 53). In E. coli, both FtsA and ZipA appear to positively regulate Z-ring formation (1, 23, 48). Either one is required for the formation and stabilization of Z rings (48). Our work here suggests that FtsA plays a similar but more critical role in B. subtilis, which does not have a ZipA homolog. A more highly conserved protein than ZipA that enhances FtsZ polymerization in B. subtilis is ZapA (20). This protein interacts directly with FtsZ and induces the bundling of FtsZ protofilaments in vitro (20). Our results are consistent with the proposal that FtsA plays a similar role to ZapA in B. subtilis (20). Like ZapA, FtsA does not appear to be required for the recruitment of essential division proteins in this organism. Unlike zapA, however, the deletion of ftsA does result in an observable effect on Z-ring formation and cell division. zapA becomes essential for cell division only when ezrA, a negative regulator of FtsZ ring assembly, is also deleted (20). Our data indicate that FtsA is required for efficient Z-ring formation in B. subtilis, and it will be important to examine this role and that of other FtsZ modulators at the molecular level in order to understand how these accessory proteins coordinate Z-ring assembly.
In bacteria, septation occurs between two replicated nucleoids precisely at midcell. The spatial precision of cell division relies on the prior precise placement of the Z ring at the division site. The ability of FtsZ to localize at regular intervals between nucleoids in ftsA-null filaments of B. subtilis suggests that FtsA is not required for the localization of FtsZ to regions close to the precise division site. This appears to be the case for ZipA and FtsA in E. coli (48). How do our observations with B. subtilis help us understand the tight spatial regulation of Z-ring assembly? They raise the possibility that FtsZ can localize approximately to division sites (between nucleoids) without these proteins, but its precise positioning in the form of a ring requires the modulation of FtsZ polymerization by these proteins. In this way, the kinetics of Z-ring formation may be intimately connected to the precision of Z-ring placement at the cell center.
Acknowledgments
We thank Shigeki Moriya for the gift of anti-FtsZ rabbit serum and Peter Lewis for the gift of FtsA protein and anti-FtsA rabbit serum.
This work was supported by a grant and a QEII Research Fellowship from the Australian Research Council (ARC) to E.J.H.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., and J. Lutkenhaus. 1992. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J. Bacteriol. 174:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg, K., Y. Nikolaichik, N. Crossland, and W. D. Donachie. 1998. Roles of FtsA and FtsZ in activation of division sites. J. Bacteriol. 180:881-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91:5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the Escherichia coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 8.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29:593-604. [DOI] [PubMed] [Google Scholar]
- 11.den Blaauwen, T., N. Buddelmeijer, M. E. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewar, S. J., K. J. Begg, and W. D. Donachie. 1992. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol. 174:6314-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Din, N., E. M. Quardokus, M. J. Sackett, and Y. V. Brun. 1998. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27:1051-1063. [DOI] [PubMed] [Google Scholar]
- 14.Duggin, I. G., P. A. Andersen, M. T. Smith, J. A. Wilce, G. F. King, and R. G. Wake. 1999. Site-directed mutants of RTP of Bacillus subtilis and the mechanism of replication fork arrest. J. Mol. Biol. 286:1325-1335. [DOI] [PubMed] [Google Scholar]
- 15.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 18.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 19.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haeusser, D. P., R. L. Schwartz, A. M. Smith, M. E. Oates, and P. A. Levin. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52:801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale, C., A. Rhee, and P. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. de Boer. 2001. Genetic analysis of the Escherichia coli FtsZ:ZipA interaction in the yeast two-hybrid system characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 25.Harris, E. L. V., and S. Angal (ed.). 1989. Determination of total protein concentration, p. 10-40. In Protein purification methods: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 26.Harry, E. J., J. Rodwell, and R. G. Wake. 1999. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 33:33-40. [DOI] [PubMed] [Google Scholar]
- 27.Harry, E. J., B. J. Stewart, and R. G. Wake. 1993. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol. Microbiol. 7:611-621. [DOI] [PubMed] [Google Scholar]
- 28.Harry, E. J., and R. G. Wake. 1997. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol. Microbiol. 25:275-283. [DOI] [PubMed] [Google Scholar]
- 29.Harwood, C. R., and S. M. Cutting (ed.). 1990. Sporulation, germination and outgrowth, p. 391-450. In Molecular biological methods for Bacillus. John Wiley and Sons Ltd., Chichester, Surrey, United Kingdom.
- 30.Hauser, P. M., and J. Errington. 1995. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J. Bacteriol. 177:3923-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kemp, J. T., A. Driks, and R. Losick. 2002. FtsA mutants of Bacillus subtilis impaired in sporulation. J. Bacteriol. 184:3856-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koppelman, C. M., M. E. Aarsman, J. Postmus, E. Pas, A. O. Muijsers, D. J. Scheffers, N. Nanninga, and T. den Blaauwen. 2004. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 51:645-657. [DOI] [PubMed] [Google Scholar]
- 34.Lara, B., A. I. Rico, S. Petruzzelli, A. Santona, J. Dumas, J. Biton, M. Vicente, J. Mingorance, and O. Massidda. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55:699-711. [DOI] [PubMed] [Google Scholar]
- 35.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 36.Low, H. H., M. C. Moncrieffe, and J. Lowe. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839-852. [DOI] [PubMed] [Google Scholar]
- 37.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Löwe, J., F. van den Ent, and L. A. Amos. 2004. Molecules of the bacterial cytoskeleton. Annu. Rev. Biophys. Biomol. Struct. 33:177-198. [DOI] [PubMed] [Google Scholar]
- 39.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 40.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, X., Q. Sun, R. Wang, G. Singh, E. L. Jonietz, and W. Margolin. 1997. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol. 179:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manting, E. H., C. van der Does, and A. J. Driessen. 1997. In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J. Bacteriol. 179:5699-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 45.McGinness, T., and R. G. Wake. 1979. Division septation in the absence of chromosome termination in Bacillus subtilis. J. Mol. Biol. 134:251-264. [DOI] [PubMed] [Google Scholar]
- 46.Migocki, M. D., M. K. Freeman, R. G. Wake, and E. J. Harry. 2002. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee, A., and J. Lutkenhaus. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 50.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.RayChaudhuri, D., and J. T. Park. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251-254. [DOI] [PubMed] [Google Scholar]
- 52.Rico, A. I., M. Garcia-Ovalle, J. Mingorance, and M. Vicente. 2004. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 53:1359-1371. [DOI] [PubMed] [Google Scholar]
- 53.Romberg, L., and P. A. Levin. 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez, M., A. Valencia, M. J. Ferrandiz, C. Sander, and M. Vicente. 1994. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 13:4919-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skare, J. T., B. M. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 57.van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 60.Wilson, G. A., and K. F. Bott. 1968. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 95:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270:387-392. [DOI] [PubMed] [Google Scholar]
- 62.Yim, L., G. Vandenbussche, J. Mingorance, S. Rueda, M. Casanova, J. M. Ruysschaert, and M. Vicente. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182:6366-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]