Abstract
Moraxella catarrhalis is a gram-negative bacterium that is mainly responsible for respiratory tract infections. In this study we report a novel outer membrane protein (OMP), designated M35, with a molecular mass of 36.1 kDa. This protein was structurally homologous to classic gram-negative porins, such as OMP C from Escherichia coli and OMP K36 from Klebsiella pneumoniae, with a predicted structure of 8 surface loops and 16 antiparallel β-sheets. The DNA sequences of the genes from 18 diverse clinical isolates showed that the gene was highly conserved (99.6 to 100% of nucleotides), with only one isolate (ID78LN266) having base variations that resulted in amino acid substitutions. Electrophoresis and analysis of recognition of the protein using mouse anti-M35 sera showed that M35 was expressed on the bacterial surface and constitutively expressed across M. catarrhalis isolates, with only ID78LN266 showing poor antibody recognition. Our results showed that the single amino acid mutation in loop 3 significantly affected antibody recognition, indicating that loop 3 appeared to contain an immunodominant B-cell epitope. The antibody specificity to loop 3 may be a potential mechanism for evasion of host immune responses targeted to M35, since loop 3 should theoretically orientate into the porin channel. Thus, M35 is a highly conserved, surface-expressed protein that is of significance for its potential functional role as an M. catarrhalis porin and is of interest as a vaccine candidate.
Moraxella catarrhalis, a gram-negative bacterium, is predominantly responsible for respiratory tract infections, such as otitis media, sinusitis, and exacerbations of chronic obstructive pulmonary disease (17, 24, 30). On rare occasions, it can also cause more serious diseases, such as meningitis and septicemia, although these occur mainly in immunocompromised populations and neonates (7, 23). M. catarrhalis pathogenesis and virulence factors are not well understood. For a long time, limited research was undertaken on this bacterium, since it was considered a normal commensal. The acceptance of M. catarrhalis as a pathogen, and its remarkably rapid development of resistance to β-lactam antibiotics (12, 14), have stimulated research into the pathogenesis of infection and characterization of the bacterium.
The majority of research into M. catarrhalis has been focused on the identification and characterization of outer membrane proteins (OMP), with a view to assessing their suitability as vaccine antigens. An ideal vaccine candidate should be highly conserved so that it can elicit an immune response that protects against all strains of the bacterium. A number of potential vaccine candidates have been identified so far, and one particular characteristic of these is conservation across strains. Two proteins that demonstrate potential as vaccine antigens, OMP E and OMP CD, appear to be well conserved (24-26) and demonstrate some similarity with porins from Escherichia coli and Pseudomonas species, respectively (24). This study reports the characterization of a novel OMP from M. catarrhalis, designated M35, that we identified as part of a search for vaccine antigens (unpublished data). The protein has a predicted structural homology with known porins from other species and is constitutively expressed and genetically well conserved. This study is the first to report the identification and characterization of a protein from M. catarrhalis that fits the classic bacterial porin structure.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
The Moraxella catarrhalis bacterial isolates used in this study are listed in Table 1. Isolate K65 was used for all aspects unless stated otherwise. Bacteria were grown overnight at 37°C with 5% CO2 on chocolate blood agar (brain heart infusion agar supplemented with 5% partially lysed horse blood) unless stated otherwise.
TABLE 1.
M. catarrhalis strains used in this study
| Isolate designation | Origin and other notesa |
|---|---|
| ATCC 25240 | Standard laboratory strain; American Type Culture Collection |
| K65 | Sputum; adult; Perth, WA, 1985; penicillin sensitive |
| K114 | Sputum; adult; Perth, WA, 1985; produces β-lactamase; piliate |
| 4223 | Middle ear fluid; child with otitis media; Buffalo, New York |
| K65NCb | Nonclumping variant of K65 |
| K114NC | Nonclumping variant of K114 |
| 4223NC | Nonclumping variant of 4223 |
| 8314 | Nasopharyngeal swab; baby (24 days) with no otitis media evident; NT, 1992 |
| 8354 | Nasopharyngeal swab; baby (30 days) with otitis media with effusion; NT, 1992 |
| 8361 | NT |
| 8402 | NT |
| 8423 | Nasopharyngeal swab; baby (26 days) with otitis media with effusion; NT, 1992 |
| 8596 | Nasopharyngeal swab; baby (20 days) with otitis media with effusion; NT, 1993 |
| 8724 | Nasopharyngeal swab; baby (21 days) with no otitis media evident; NT, 1993 |
| 9170 | NT |
| 9315 | Nasopharyngeal swab; baby (47 days) with otitis media with effusion; NT, 1994 |
| ID 002 LN 010 | Nasopharyngeal swab; infant (<2 years); Kalgoorlie, WA |
| ID 3 LN 259 | Nasopharyngeal swab; infant (<2 years); Kalgoorlie, WA |
| ID 23 LN 175 | Nasopharyngeal swab; infant (<2 years); Kalgoorlie, WA |
| ID 60 LN 148 | Nasopharyngeal swab; infant (<2 years); Kalgoorlie, WA |
| ID 78 LN 266 | Nasopharyngeal swab; infant (<2 years); Kalgoorlie, WA |
WA, Western Australia; NT, Northern Territory.
Further information on the generation of the nonclumping variants can be found in reference 19.
Nucleotide sequencing.
Genomic DNA was extracted from each of the 21 isolates (Table 1) with the QIAGEN Genomic Tip 100/G kit, following the manufacturer's instructions. PCR amplification of the M35 gene was then performed using the following oligonucleotide primers: 5′-GGCGCATGCAAAAAACTTGCTCTAGCAACCGCA-3′ and 5′-GGCCTGCAGGAATTTATATTCTAAACCTGCG-3′. These primers contain restriction enzyme sites that were not necessary for, but did not interfere with, this procedure (underlined). PCR was conducted using the polymerase Pfu Turbo (Stratagene) with an initial denaturing for 2 min at 95°C, 32 cycles of denaturing for 1 min at 95°C, annealing for 1 min at 50°C, and elongation for 1.5 min at 72°C, followed by 10 min at 72°C. The resulting PCR products were purified to remove primers and unincorporated nucleotides using the MinElute gel extraction kit (QIAGEN), following the manufacturer's instructions. The purified PCR products were then used as template DNA in a dye terminator cycle sequencing reaction using the DTCS Quick-Start Mix (Beckman Coulter), following the manufacturer's instructions. A sequence was determined for each isolate with each of the primers shown in Table 2. The nucleotide sequences were then determined using the CEQ 8000 genetic analysis system (Beckman Coulter). The primers were designed to enable sequencing of the gene (1,077 bp including stop codon) in three overlapping sections on both strands. The sequences of each of the overlapping sections were then pieced together manually, and the complementary sequences were compared to check the integrity of the results. The sequences from each of the 21 isolates were then compared by alignment, and any differences from the consensus were noted and confirmed by repeating the above protocol.
TABLE 2.
Primers used for DNA sequencing
| Primer designation | Position and primer sequence |
|---|---|
| SGp35pQF | 1 GGCGCATGCAAAAAACTTGCTCTAGCAACCGCA |
| M35F1 | 298 GATACTTACCTAGGTCTTGC |
| M35F2 | 738 GTCTGAGGGCAAGGTAGGCG |
| M35F3 | 10 CTTGCTCTAGCAACCGCAG |
| SGp35pQR | 1077 GGCCTGCAGGAATTTATATTCTAAACCTGCG |
| M35R1 | 382 ACCTTCCAACATGCTGGC |
| M35R2 | 813 GCCTGACCATAGGCTGTC |
| M35R3 | 298 GCAAGACCTAGGTAAGTATC |
Production of recombinant M35 protein.
A recombinant polyhistidine (six-His)-tagged form of the mature M35 protein (rM35) was created by cloning the amplified gene into a pQE30 plasmid (clones developed previously by Sara Gomez Gallego), expressed in Escherichia coli M15 pREP4 cells (Stratagene), and purified using a nickel-nitrilotriacetic acid immobilized-metal affinity chromatography technique. The first 20 residues of the translated amino acid sequence are predicted to be a leader peptide that is cleaved during the assembly of the mature protein, which is typical of bacterial outer membrane proteins. The cleavage site was predicted to be after the sequence AQA, and N-terminal amino acid sequencing confirmed this. The rM35 protein was therefore designed to not contain this leader peptide.
E. coli clones were grown overnight at 37°C on Luria-Bertani (LB) medium containing 50 μg/ml of both ampicillin and kanamycin (Amresco). Single colonies were then grown in LB broth culture with the same concentration of antibiotics at 37°C in an orbital shaker to a relative optical density of approximately 0.7 at 600 nm. Expression of rM35 was then induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. Expression of the protein was allowed to occur over the next 4 h (under the same culture conditions) before harvesting of the cells by centrifugation at 4,000 × g for 20 min at 4°C. The pellets were stored at −80°C overnight before purification of the six-His-tagged protein using a nickel-nitrilotriacetic acid protein purification kit (QIAGEN), following the manufacturer's instructions, under denaturing conditions (8 M urea).
Production of recombinant M35 protein lacking the third external loop.
A recombinant form of the M35 protein lacking the third external loop (rM35-L3) was constructed. Two PCR products were created using the following primer pairs: 5′-GGCGCATGCAAAAAACTTGCTCTAGCAACCGCA-3′ and 5′-GGCGAATTCACCAGCCAACAATGTGCCGTATT-3′; and 5′-GGCGAATTCAACAATGCCTTTGCTTATGTATC-3′ and 5′-GGCCTGCAGGAATTTATATTCTAAACCTGCG-3′. These products were copies of the M35 gene from nucleotides 4 to 348 and 445 to 1074 (does not include start or stop codon), and EcoRI restriction sites were introduced at the end of the first product and beginning of the second. These two PCR products were then digested with EcoRI and ligated with T4 ligase. The pQE30 plasmid and this product were then digested with PstI and SphI restriction enzymes, and the altered gene was ligated into the plasmid. The plasmid was then heat shock transformed into M15 pREP4 E. coli cells, and the method outlined above was used to express and purify the altered protein.
Polyclonal hyperimmune anti-M35 mouse serum.
Hyperimmune antiserum was raised against the recombinant M35. A solution of the protein was diluted with sterile phosphate-buffered saline (PBS) and emulsified with an equal volume of incomplete Freund's adjuvant (Sigma Chemicals). Adult male BALB/c mice received 10 μg of rM35 in a 100-μl volume by intraperitoneal injection with a 26-gauge needle on days 0, 7, and 14. On day 21, blood was collected for serum. The sera from four mice were pooled, aliquoted, and stored at −20°C until required. Mouse antiserum was generated with the approval of the University of Canberra Animal Ethics Committee.
Flow cytometry.
M. catarrhalis cells (isolates K65 and ID78LN266) were grown overnight, harvested, and washed by centrifugation in sterile PBS. To reduce non-M35-specific binding, the serum from both immunized and nonimmunized mice was adsorbed against E. coli cells for 30 min at 37°C and then overnight at 4°C. M. catarrhalis cells were pelleted by centrifugation at 13,600 × g for 1 min, the supernatant was removed, and the bacteria were resuspended in 1 ml of a 1-in-50 dilution of mouse serum (immune or nonimmune) in sterile PBS and incubated for 1 h at 37°C. The cells were again pelleted by centrifugation at 13,600 × g for 1 min, the supernatant was removed, and the cells were resuspended in 200 μl of Alexa 488 (green) goat antimouse immunoglobulin G (IgG) (Molecular Probes) diluted 1 in 200 in sterile PBS and incubated for 1 h at 37°C. The cells were washed by centrifugation at 13,600 × g for 1 min, the supernatant removed, and the bacteria washed twice more with sterile PBS. The final suspension was diluted to 1 in 20 with PBS for analysis by flow cytometry (Coulter XL-MCL; Beckman Coulter). Approximately 100,000 cells were counted and their relative fluorescence measured. A control for nonspecific binding of the secondary antibody was also included.
Refolding of recombinant M35 protein.
Refolding of the recombinant M35 protein was achieved by a modification of the protocol described by Watanabe (31). Urea was removed from the protein solution by dialysis overnight against 10 mM morpholinepropanesulfonic acid, 0.1 M NaCl, 0.2% (wt/vol) sodium dodecyl sulfate (SDS), pH 6.9. This was followed by dilution with an equal volume of the same buffer including 20-mg/ml octylglucoside, to give a final concentration of 10-mg/ml octylglucoside. The protein was incubated at room temperature in this solution for at least 24 h prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis under nonreducing conditions.
Protein gel electrophoresis and staining.
Whole-cell protein samples from each of the 18 isolates and 3 variants were subjected to one-dimensional SDS-PAGE. Cells were grown overnight, harvested, and washed in PBS, and the concentration was adjusted to 5 × 109 CFU/ml before dilution with an equal volume of SDS reducing buffer (4.0 ml water, 1 ml 0.5 M Tris-HCl, 0.8 ml glycerol, 1.6 ml 10% [wt/vol] SDS, 0.2 ml 0.05% [wt/vol] bromophenol blue, 50 μl β-mercaptoethanol). These samples were then boiled for 10 min before electrophoresis was performed using either the Bio-Rad Mini-Protean II system with 12% polyacrylamide gels (containing 4% polyacrylamide stacking gels) or the Pharmacia PhastSystem with 10 to 15% gradient gels, as indicated. Gels were then stained with Coomassie brilliant blue or silver stained.
Two-dimensional electrophoresis was also performed with whole-cell protein samples from the K65 and ID78LN266 isolates. Cells were harvested and washed in PBS and diluted to approximately 1.25 × 109 CFU/ml before sonication on ice at an output of 40 for six 10-s bursts with 30 s between each burst, using a probe-type sonicator (Sonifier 250; Branson Ultrasonic Corporation). An equal volume of 20% (wt/vol) trichloroacetic acid (Mallinckrodt) was added, and the samples were incubated on ice for 30 min. The precipitate was pelleted by centrifugation (15,900 × g) at 4°C for 15 min, and the supernatant was discarded. The pellet was washed twice by the addition of 300 μl of ice-cold acetone and centrifuged (15,900 × g) for 5 min at 4°C. The supernatant was removed, and the pellets were left to air dry in a fume hood for approximately 30 min before resuspension in 400 μl of rehydration buffer (8.3 M urea, 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 15 mM dithiothreitol, 0.1% Biolytes 3-10 (Bio-Rad), 0.001% bromophenol blue) by sonication in a bath-type sonicator for 1 to 2 min. The proteins were then separated by isoelectric focusing (Bio-Rad IEF Cell system). This process was repeated twice for each sample, with two different pH gradients (3 to 10 and 4 to 6). The second dimension was performed in the same way as the one-dimensional electrophoresis, with the exception that the isoelectric focusing strip was embedded with agarose embedding buffer.
Amino acid sequencing.
A two-dimensional electrophoresis gel was prepared, and the spot corresponding to the M35 protein was excised. The M35 protein was then subjected to N-terminal amino acid sequencing at the Newcastle Protein Facility, NSW, using Edman degradation and high-performance liquid chromatography analysis.
Western blot analysis.
The protein from a one-dimensional electrophoresis gel was transferred to a nitrocellulose membrane (Bio-Rad) using a semidry technique. The gel was soaked in Tris-glycine buffer (200 mM glycine, 25 mM Tris, pH 8.8) for 10 min before transfer to the nitrocellulose membrane between graphite electrodes using a current that was dependent on the size of the membrane (current mA = 0.8 × area [cm2]) for 1 h. Following transfer the nitrocellulose membrane was stained with Ponceau stain and destained in distilled water to confirm the protein transfer. It was then soaked in Tris-buffered saline (TBS) (0.5 M NaCl, 20 mM Tris-Cl, pH 7.5) for 10 min before blocking with 1% (wt/vol) skim milk in TBS for 30 min and washed twice with TBS containing 0.05% Tween 20 (TTBS) for 5 min before the addition of polyclonal antiserum (diluted 1/20 with 1% skim milk powder in TTBS) and left overnight with gentle agitation. The membrane was then washed twice with TTBS for 5 min before addition of peroxidase-conjugated antimouse IgG (Sigma Chemicals) diluted 1/500 with 1% skim milk powder TTBS and incubated for 90 min with gentle agitation. The membrane was washed twice more in TTBS and once in TBS for 5 min each before being exposed to fresh HRP Developer (Bio-Rad) for approximately 1 h. Western blot analysis for nonspecific recognition of M. catarrhalis proteins was also performed using serum from nonimmunized mice, and no protein bands were positive.
Nucleotide sequence accession numbers.
The nucleotide sequences of the M35 genes from isolates ATCC 25240, 4223, K65, and ID78LN266 have been deposited in GenBank with accession numbers AY905610, AY905611, AY905612 and AY905613, respectively.
RESULTS
M35 gene sequence is highly conserved across 18 isolates of M. catarrhalis.
To determine the level of conservation of the M35 gene, DNA for the homologous gene was sequenced from 18 isolates of M. catarrhalis and 3 variants (Table 1). The isolates were chosen for their diversity based on the following criteria: year of isolation, age group of patient, geographical location, anatomical collection site, and the nonclumping variants of three of the clinical isolates. The latter were chosen to ensure that this gene was not lost or varied in these isolates that were produced as a result of passage for selection of the “nonclumping” characteristic (19). Hence, the isolates studied were collected at various times from the 1960s to 2001 and from both adults and children. Our collection contained isolates sent to us from both the United States and different parts of Australia. Some were infection isolates, while others were from commensal flora. In addition, multiple isolates from the same original collections were used in case any variation detected may have been affected by any of the selection parameters.
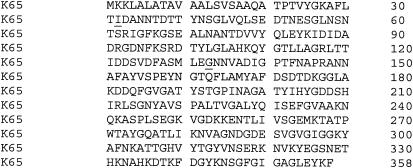
The gene comprised 1,074 bp for the full-length sequence with a translated amino acid sequence of 358 residues (Fig. 1). There is a predicted leader sequence of 20 residues, resulting in a mature protein of 338 residues. The sequencing of the M35 gene demonstrated that it was very highly conserved between these 18 isolates, as well as the 3 variants. Fifteen isolates were 100% homologous, and this sequence was considered the consensus. Several synonymous base changes were found in the other six isolates. Four isolates were found to have one difference from the consensus sequence (99.9% identity), while another two isolates were found to differ by four bases (99.6% identity), and no insertions or deletions were observed.
FIG. 1.
Translated amino acid sequence of the M35 protein from M. catarrhalis. The K65 isolate represents the consensus, and the two substitutions found in isolate ID78LN266 are underlined.
The amino acid translations of all 21 sequences were also compared, and this resulted in the detection of only two nonsynonymous base changes (Fig. 1). Both these amino acid substitutions were present in isolate ID78LN266. At position 32, valine was substituted for isoleucine. Valine and isoleucine are similar amino acids, although valine has a slightly shorter side chain. At position 133, aspartic acid replaced glycine. This is a replacement of a very small and uncharged amino acid with a larger acidic residue. Since the gene was sequenced in both directions and the sequencing process repeated, it is most unlikely that these changes were the results of a reading error.
M35 is putatively an outer membrane porin.
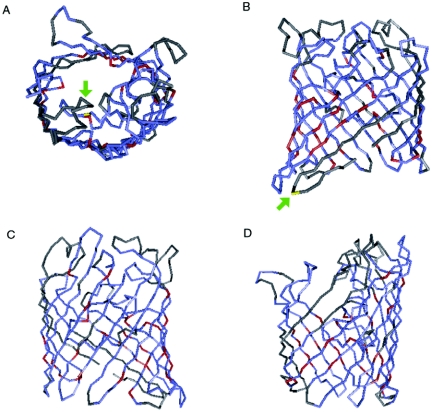
Submission of the DNA sequence of the M35 gene to BLASTN returned no significant alignments (http://www.ncbi.nlm.nih.gov/BLAST/). However, when the translated amino acid sequence was submitted to a BLASTP database search, it revealed that M35 has homology with many other putative porin proteins and some known porins, including PorB from Neisseria meningitidis (3, 6, 29). An RPS-BLAST search of the conserved domain database provided results indicating that M35 is likely to fold into structural domains similar to known gram-negative porins (18). It was found that the amino acid sequence of M35 could be superimposed upon that of the porin OmpC from E. coli when similar residues were aligned. This showed an alignment of 93.8% and an Expect (E) value of 1 × 10−17. The E value is an indication of the statistical significance of the alignment based upon the number of matches that would be expected by chance, and a value less than 1 × 10−2 is generally considered significant. Database searches also showed that the M35 sequence aligns with the consensus theoretical structure for gram-negative porins with an E value of 2 ×10−8 and an alignment of 86.9%. Included within this group of gram-negative porins are OmpF and PhoE (phosphoporin) from E. coli (27) and osmoporin (OMP K36) (2) from Klebsiella pneumoniae, all of which are general outer membrane porin proteins. Figure 2 shows an alignment of the translated amino acid sequence of M35 with OMP K36 using the Cn3D program, including the predicted positions of the two amino acid substitutions found in isolate ID78LN266. The first substitution, valine to isoleucine, appears to be located at the base of the periplasmic region, while the second substitution, glycine to aspartic acid, is predicted to be located on loop 3, which folds into the porin channel. This predicted structure is consistent with the classical β-barrel structure of bacterial outer membrane porins. It has 16 antiparallel β-sheets that would span the outer membrane and are connected by 8 short periplasmic loops and 8 longer external loops. Characteristically, each transmembrane β-sheet is composed of alternating hydrophobic and hydrophilic residues, so that the hydrophobic side chains project out from the barrel into the membrane while the hydrophilic side chains project into the lumen of the channel, providing a hydrophilic environment within the channel allowing the passage of water-soluble molecules. With this prediction model, the third external loop is usually longer and is reported to fold into the interior of the channel, restricting its diameter and thus the size and the preference for anionic or cationic molecules able to pass through it (18).
FIG. 2.
Three-dimensional alignment of the translated M35 amino acid sequence with OMP K36 from Klebsiella pneumoniae. Red indicates residues that are identical between the two proteins, blue represents similar amino acids, and gray represents nonidentity. (A) Location of the substitution of aspartic acid for glycine in ID78LN266 (yellow residue indicated by green arrow), viewed through the channel; (B) location of the substitution of valine for isoleucine in ID78LN266 (yellow residue indicated by green arrow), viewed from the side with the periplasmic region at the bottom and extracellular region at the top; (C and D) other side views of the alignment.
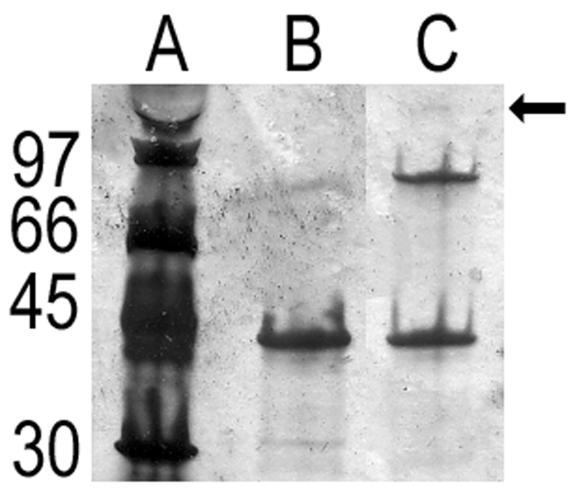
Porins frequently exist as trimers. To determine if M35 would form a trimer, the recombinant M35 protein was refolded and analyzed by SDS-PAGE. Under reducing conditions, a single band was seen, with a very faint band at a higher molecular mass sometimes being evident (Fig. 3). Under nonreducing conditions, an additional band corresponding to the molecular mass expected of a dimer of M35 was observed, along with a very faint band corresponding to the molecular mass expected of the trimer of M35 (Fig. 3). Immunoblotting with anti-M35 confirmed both the monomer and dimer but not the faint trimer band (data not shown).
FIG. 3.
SDS-PAGE analysis of refolded recombinant M35 protein. Lane A: low molecular mass protein standards, molecular masses (kDa) as indicated; lane B: refolded M35 under reducing conditions (with β-mercaptoethanol and heated at 100°C for 5 min); lane C: refolded M35 under nonreducing conditions (no β-mercaptoethanol and incubated at room temperature for 5 min). The arrow indicates the faint band that corresponded to the molecular mass of a trimer of M35.
Expression and antigenic recognition of the M35 protein are well conserved.
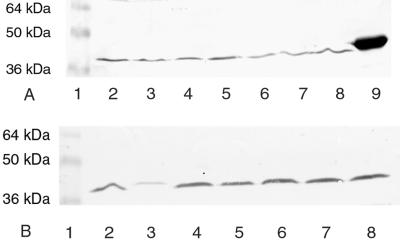
The presence of the M35 gene in each of the isolates evaluated is not an indication that the protein is constitutively expressed by all of the strains. To confirm whether M35 was expressed by all the strains evaluated, Western blots were probed with anti-M35 sera. All strains were positive when probed with anti-recombinant M35 mouse sera (Fig. 4). One isolate (ID78LN266 in lane 3) gave a band of lower density than the others even though the amount of bacterial protein transferred from SDS-PAGE was equivalent. This suggests that M35 in this strain either is expressed in smaller amounts, is not as accessible to antibodies, or is expressed in a form that is not as antigenically well conserved, compared to M35 from the K65 strain and the other isolates. The Western blotting result was confirmed by repeating the procedure on a number of occasions.
FIG. 4.
Western blot analysis. Whole-cell protein preparations of M. catarrhalis from different isolates were separated by SDS-PAGE, and transferred to nitrocellulose, and probed with mouse anti-M35 sera. (A) Lane 1, prestained molecular mass markers; lane 2, ID60LN148; lane 3, ID3LN259; lane 4, 8423; lane 5, 8402; lane 6, 8314; lane 7, 4223; lane 8, 8361; lane 9, rM35; (B) lane 1, prestained molecular mass markers; lane 2, 8724; lane 3, ID78LN266; lane 4, ATCC 25240; lane 5, 8596; lane 6, ID002LN010; lane 7, ID23LN175; lane 8, K65.
The general SDS-PAGE profile for M. catarrhalis strains appears highly conserved, although some minor protein banding pattern differences can be observed. The protein appears to migrate with a molecular mass of approximately 35 to 36 kDa, but the density of bands in the 30 to 40 kDa region was too complex to assess the specific expression of the native M35 protein. Two-dimensional electrophoresis was undertaken to better separate M35 from other proteins of similar molecular masses and to characterize if there were any differences in expression between the K65 and ID78LN266 isolates.
The apparent location of M35 was predicted from both the theoretical isoelectric point and molecular mass, which were found to be 4.94 to 5.1 and 36.1 kDa, respectively, from calculations based on the amino acid sequence using online isoelectric point prediction programs (http://www.embl-heidelberg.de/cgi/pi-wrapper.pl; http://us.expasy.org/tools/pi_tool.html). The predicted position of M35 was then confirmed by Western blotting (data not shown). The location of M35 within the two-dimensional protein profile for K65 is shown in Fig. 5A and B. A protein spot corresponding to M35 was also present for the ID78LN266 profile (Fig. 5C). The identity of the spot deemed to be M35 was confirmed by N-terminal amino acid sequencing, which produced the sequence TPTVYGKAFLT, which corresponds to the first 10 amino acids of the mature M35 protein.
FIG. 5.
Two-dimensional electrophoresis. In the first dimension, isolate K65 was separated across a pH gradient of (A) 3 to 10 or (B) 3 to 6, and isolate ID78LN266 was separated across a pH gradient of 3 to 6 (C). In the second dimension, SDS-PAGE-only lanes are the following: the M. catarrhalis isolates as reference whole-cell protein samples (isolate number) and low molecular mass markers (markers). +ve, positive; −ve, negative.
The isoelectric point of M35 based on mobility on the two-dimensional profile was estimated to be approximately 5.3, which was marginally higher than the theoretical isoelectric point of 4.94. It should be noted that the amino acid sequence used to make this prediction did not include the putative leader peptide, which corresponds to the first 20 amino acid residues of the translated amino acid sequence (Fig. 1).
M35 is expressed on the surface of the bacterium.
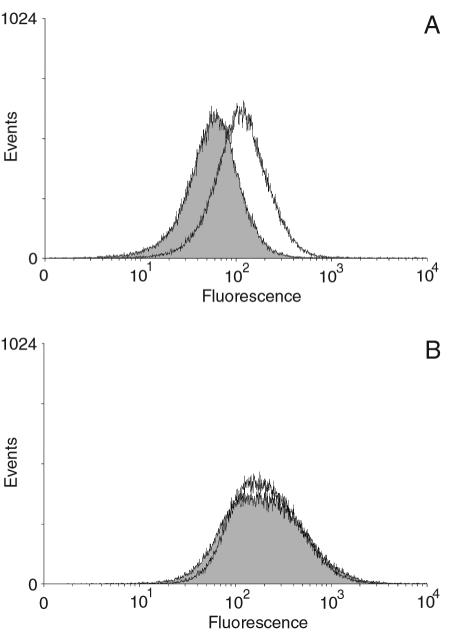
If M35 is a porin, as predicted, then it should have surface-exposed regions. To determine whether there were surface-exposed regions of M35 that were accessible to M35-specific antisera, an indirect immunocytometric assay was conducted. Flow cytometry showed a small but reproducible curve shift between the control cells (exposed to sera from nonimmunized mice) and those exposed to anti-M35 sera (Fig. 6A). This demonstrated that antibodies produced by mice against M35 bound to surface-exposed regions of M. catarrhalis K65. This result shows that antibody generated by immunization with recombinant M35 recognized the conformational form of the native M35 and that M35 was located on the surfaces of the bacteria, as would be expected for a porin. In contrast to the result for K65, no curve shift was observed for strain ID78LN266 (Fig. 6B). Hence, this would suggest that the anti-M35 that was able to recognize M35 on the surface of K65 cells may be specific for an epitope that includes the region where the amino acid variation had occurred on strain ID78LN266. This was predicted to be on the surface-located loop 3.
FIG. 6.
Flow cytometry histograms. Fluorescence curves for (A) isolate K65 and (B) ID78LN266. The bacteria were incubated with anti-M35 sera (unshaded curves) or nonimmune sera (shaded peaks) and binding detected by antimouse IgG-fluorescent isothiocyanate conjugate. The geometric mean (and median) value for fluorescence of K65 cells exposed to anti-M35 sera were 109.12 (112.40), compared with 55.09 (58.82) for nonimmune sera. Isolate ID78LN266 did not demonstrate any significant curve shift, with the cells having a mean fluorescence of 194.56 (194.56) for anti-M35 sera and 189.22 (187.60) for the nonimmune sera.
The third external loop is immunodominant.
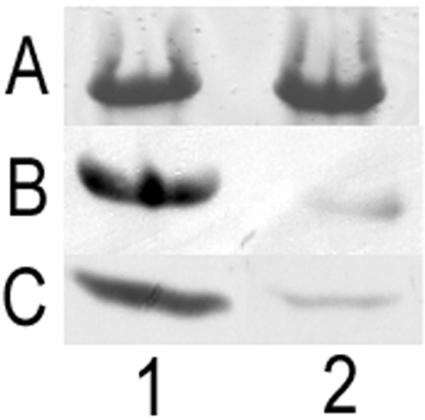
Western blot analysis revealed that the majority of antibodies raised against M35 were apparently specific for loop 3. In the variant isolate, ID78LN266, the density of the resulting band was greatly reduced from that observed for all other isolates (Fig. 4). The specificity of the antibody being directed at the third loop was confirmed by the reduction in antibody binding to a loop 3-deficient recombinant M35 protein (Fig. 7).
FIG. 7.
Effect on antibody recognition of M35 with a loop 3 sequence deletion (rM35loop3−). (A) Coomassie blue-stained SDS-PAGE showing rM35 (lane 1) and rM35loop3− (lane 2). (B) Western blot of rM35 (lane 1) and rM35loop3− (lane 2) probed with mouse anti-M35. (C) Western blot probed with mouse anti-M35 of isolate K65 (lane 1) and isolate ID78LN266 (lane 2). Isolate ID78LN266 shows reduction in antibody recognition similar to observed for rM35loop3−.
DISCUSSION
While little is known about the mechanisms of pathogenesis by M. catarrhalis, the focus on characterizing proteins in the outer membrane has resulted in the elucidation of functional roles for a number of major OMPs. OMPs described in the literature for M. catarrhalis that have been identified as having specific functional and/or pathogenic roles include the following: (i) the ubiquitous surface proteins UspA1 and UspA2 (including the variant UspA2H), which have a role in cellular adhesion and complement sensitivity (20, 22); (ii) several proteins involved in the acquisition of iron, such as transferrin binding proteins, TbpA and TbpB (21), lactoferrin binding proteins, LbpA and LpbB (5), and a protein known as CopB (1); and (iii) major membrane proteins, called OMPE and OMP CD, that are highly conserved across strains (16, 25).
To date, no protein that fits a classic gram-negative porin structure has been identified, although it has been proposed that the OMP CD sequence is related to that of the Pseudomonas aeruginosa porin OprF. This relationship suggests that perhaps OMP CD may be involved in nutrient acquisition (18, 26); however, the only evidence in support of this hypothesis is that Omp CD mutants have lower growth rates (15). The predicted amino acid sequences for OMP CD are highly conserved across clinical isolates (16, 26), and this protein has recently been shown to have adhesin properties for human lung cells (15).
In this study we report the identification of an M. catarrhalis protein that is characteristically typical of an outer membrane porin (18). The protein, named M35, is the first M. catarrhalis protein identified that fits the classic porin structure. M35 has been identified as having potential as a vaccine candidate (13), and this study was undertaken to do the following: (i) investigate the degree of sequence conservation, (ii) determine that the protein was expressed on the bacterial surface, and (iii) predict the structural characteristics and possible functional roles of M35 based on homologies to other proteins. In addition, our study has also identified a possible immunodominant epitope.
Prediction modeling of the DNA and translated sequences from the K65 strain suggested that M35 might be an outer membrane porin, since its sequence shares homology with gram-negative porins such as osmoporin (OMP K36) from Klebsiella pneumoniae (2) and Neisseria meningitidis PorB (3, 6, 29). Typical are the 16 antiparallel membrane-spanning regions comprising β-sheet structures and 8 surface-exposed loops, with loop 3 being longer, orientated into the channel, and thought to influence the size exclusion and ion selectivity of the porin (18). Not only did M35 meet the criteria with the β-sheet and loop regions, the predicted loop 3 section was also a longer sequence and the modeling alignment predicted it would fold into the pore as has been demonstrated for other porins, such as OMP K36 from K. pneumoniae (2). Additionally, in vitro refolding of the recombinant protein indicated that M35 would form a trimer like many other porins, with results seen in this study being consistent with those reported by Watanabe (31) for the E. coli porin OmpF. The flow cytometry results confirmed that M35 was expressed on the bacterial surface.
It was hypothesized that if M35 is a porin, then the pattern of conservation in its β-sheet sequence could be similar to that of porins from other species, which tend to be highly conserved, and the surface-exposed loop regions would be quite variable. For example, P2 from nontypeable Haemophilus influenzae (NTHi), another gram-negative bacterium that occupies the same niche in humans as M. catarrhalis, also fits a classic porin structure and has well-conserved membrane-spanning regions, but many of the surface-exposed loops have variable sequence regions (4, 11). The sequencing results of the M35 gene differ from those for these other porins in that it is highly conserved across the entire gene in all the strains sequenced.
The degree of conservation of M35 is as high as, if not higher than, that of other M. catarrhalis proteins. For example, OMP E and CD (25, 26) are considered well enough conserved to be suitable vaccine candidates. Their degree of conservation has been based on restriction fragment length polymorphism, some sequence comparisons, and the abilities of monoclonal antibodies to bind to different strains (16, 25, 26). These proteins are reported to have homologies of 95% or better between strains. Restriction fragment length polymorphism profiles and antibody recognition techniques can potentially overestimate the degree of conservation due to limitations in their ability to detect minor sequence mutations, and so sequencing the genes from a greater library of isolates may lower these estimates. OMP E and CD genes have been shown to be stable over time within the same host, remaining unchanged over a 3- to 9- or 6-month period, respectively, when potentially under selective pressure from the host's immune system (16, 25). Our study has differed from the approach used by these researchers by investigating the level of genetic conservation of M35 by sequencing the genes from 18 isolates of diverse origin, as well as 3 variants. The high level of sequence conservation of the genes in the isolates used in this study would suggest that the M35 gene is extremely stable and may not be a target of host immunity postinfection.
Anti-M35 produced in mice immunized with M35 strongly recognized a single protein band within whole-cell preparations from 20 of the 21 isolates (including the 3 variants), with the exception being isolate ID78LN266. Since the M35 expression did not appear to be reduced or lost by this strain, the weaker Western blot result and loss of antibody recognition of M35 on the bacterial surfaces were most likely due to a lack of recognition of particular epitopes by the antisera. With only two amino acid differences found between ID78LN266 and the consensus sequence based on that from strain K65, it appeared that the majority of M35-specific antibody may be directed against epitopes containing these amino acids, but in particular, the substitution of aspartic acid for glycine within the third external loop region (Fig. 2A).
While the selection of strain ID78LN266 in this study was serendipitous, the specific amino acid mutations have provided a means of determining that it appears that the majority of the M35-specific antibody may be directed against an epitope on loop 3 and that this loop was important for any surface antibody binding to M35. The results observed for ID78LN266 could be replicated in a loop 3-deficient recombinant M35 protein. These results could also explain why there was only a small fluorescence curve shift when antibody bound to M35 on the surface of M. catarrhalis, which was in contrast to a very strong band on the Western blots. It is conceivable that in its conformational state, loop 3 may be less accessible to antibody if it is orientated into the channel. Support of the hypothesis that M35 is on the bacterial surface and accessible to antibody can be extrapolated from a recent study by Troncoso et al. that demonstrated that M. catarrhalis produces a cross-reactive antibody that recognizes N. meningitidis PorB (28). It is feasible that this uncharacterized M. catarrhalis antigen-specific antibody may have been specific to M35. However, the evidence that loop 3 of M35 is an immunodominant epitope is both consistent with and different from reports on PorB. Studies have reported that the PorB-specific IgG responses in convalescent-phase and postimmunization sera are predominantly to loop 1 and are marginally bactericidal but strongly opsonic (8-10). Hence, such precedence makes it feasible that M35-specific antibody could also be directed against a specific surface loop, but unlike PorB, it appears that it may be directed against loop 3.
Loss of antibody recognition due to a single amino acid change in an antigen-specific loop of a porin has also been previously demonstrated. Murphy and colleagues have shown that point mutations resulting in a single amino acid change in loop 5 of the NTHi porin, P2, resulted in a loss of recognition of the epitope by a monoclonal antibody specific to this region (32). Such changes are believed to be part of the mechanism of microbial evasion of host immune responses to NTHi. However, it is unlikely that similar mechanisms for sequence hypervariability demonstrated by P2 and other porins are responsible for the specific sequence changes found in strain ID78LN266, since the M35 genes are so well conserved in all the strains. It is more likely that this was a random mutation that occurred during the evolution of this strain and happens to be in an immunologically significant region. There was nothing unusual or different in the clinical history of this isolate. Hence, it will be important to investigate if individuals produce antibody to M35 postinfection, and if so, whether this is predominantly directed against loop 3. If loop 3 is the main target, then this may be part of an immune evasion mechanism by M. catarrhalis. If in vivo loop 3 is folded into the porin channel, then loop 3 would be a poor target for antibody binding and M. catarrhalis would not be under pressure to alter the M35 sequence to avoid host M35-specific immune responses.
In summary, this study has identified a highly conserved protein with a sequence that structurally meets the criteria for that of a classic porin and hence is the first protein with a typical porin structure to be identified for M. catarrhalis. The protein was constitutively expressed across strains and was detected on the bacterial surface. Our results also indicate that loop 3 may fold into the channel and appears to be an immunodominant epitope. The high degree of sequence conservation and constitutive expression would suggest that M35 may be an excellent vaccine candidate but that the significance of the loop 3 immunodominance on the efficacy of M35-specific immunity should be further investigated.
Acknowledgments
We acknowledge the technical assistance of Claire Batum, Ray Ellett, Nancy Fisher, Gillian Nolen, and Peter Tyrer.
REFERENCES
- 1.Aebi, C., B. Stone, M. Beucher, L. D. Cope, I. Maciver, S. E. Thomas, G. H. McCracken, Jr., P. F. Sparling, and E. J. Hansen. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 64:2024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertí, S., F. Rodríquez-Quiñones, T. Schirmer, G. Rummel, J. M. Tomás, J. P. Rosenbusch, and V. J. Benedí. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bash, M. C., K. B. Leasiak, S. D. Banks, and C. E. Frasch. 1995. Analysis of Neisseria meningitidis class 3 outer membrane protein gene variability regions and type identification using genetic techniques. Infect. Immun. 63:1484-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, J., S. Grass, D. Jeanteur, and R. S. Munson, Jr. 1994. Diversity of the P2 protein among nontypeable Haemophilus influenzae isolates. Infect. Immun. 62:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnah, R. A., H. Wong, S. M. Loosmore, and A. B. Schryvers. 1999. Characterization of Moraxella (Branhamella) catarrhalis lpbB, lpbA, and lactoferrin receptor orf3 isogenic mutants. Infect. Immun. 67:1517-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conceição, A. S. A., M. Minetti, M. S. Blake, and D. P. Remata. 1998. Characterization of the structure, function, and conformational stability of PorB class 3 protein from Neisseria meningitidis. J. Biol. Chem. 273:25329-25338. [DOI] [PubMed] [Google Scholar]
- 7.Daoud, A., F. Abuekteish, and H. Masaadeh. 1996. Neonatal meningitis due to Moraxella catarrhalis and review of the literature. Ann. Trop. Paediatr. 16:199-201. [DOI] [PubMed] [Google Scholar]
- 8.Delvig, A. A., T. E. Michaelsen, A. Aase, A. Hoiby, and E. Rosenqvist. 1997. Vaccine-induced IgG antibodies to the linear epitope on the PorB outer membrane protein promote opsonophagosis of Neisseria meningitidis by human neutrophils. Clin. Immunol. Immunopathol. 84:27-35. [DOI] [PubMed] [Google Scholar]
- 9.Delvig, A. A., E. Wedege, D. A. Caugant, R. Dalseg, J. Kolberg, M. Achtman, and E. Rosenqvist. 1995. A linear B-cell epitope on the class 3 outer-membrane protein of Neisseria meningitidis recognized after vaccination with the Norwegian group B outer-membrane vesicle vaccine. Microbiology 141:1593-1600. [DOI] [PubMed] [Google Scholar]
- 10.Delvig, A. A., E. Wedege, T. E. Michaelsen, A. Hoiby, P. Brandtzaeg, and E. Rosenqvist. 1996. Immune responses to linear epitopes on the PorB protein of Neisseria meningitidis in patients with systemic meningococcal disease. Microbiology 142:2491-2498. [DOI] [PubMed] [Google Scholar]
- 11.Duim, B., J. Dankert, H. M. Jansen, and L. van Alphen. 1993. Genetic analysis of the diversity in outer membrane P2 of non-encapsulated Haemophilus influenzae. Microb. Pathog. 14:451-462. [DOI] [PubMed] [Google Scholar]
- 12.Flemingham, D., R. N. Grüneberg, and the Alexander Project Group. 2000. The Alexander Project 1996-1997: latest susceptibility data from this international study of bacterial pathogens from community-acquired lower respiratory tract infections. J. Antimicrob. Chemother. 45:191-203. [DOI] [PubMed] [Google Scholar]
- 13.Giebink, G. S., L. O. Bakaletz, S. J. Barenkamp, B. Green, X. X. Gu, T. Heikkinen, M. Hotomi, P. Karma, Y. Kurono, J. Kyd, T. F. Murphy, P. L. Ogra, J. Patel, and S. Pelton. 2005. Vaccine panel. Recent advances in otitis media. Report on the 8th Research Conference. Ann. Otol. Rhinol. Laryngol. 114:86-103. [PubMed] [Google Scholar]
- 14.Hoban, D., and D. Flemingham. 2002. The PROTEKT surveillance study: antimicrobial susceptibility of Haemophilus influenzae and Moraxella catarrhalis from community-acquired respiratory tract infections. J. Antimicrob. Chemother. 50:49-59. [DOI] [PubMed] [Google Scholar]
- 15.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiao, C. B., S. Sethi, and T. F. Murphy. 1995. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from human respiratory tract. Microb. Pathog. 16:215-225. [DOI] [PubMed] [Google Scholar]
- 17.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 18.Koebnik, R., K. P. Locher, and P. van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 19.Kyd, J. M., A. W. Cripps, and T. F. Murphy. 1998. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J. Med. Microbiol. 47:159-168. [DOI] [PubMed] [Google Scholar]
- 20.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 15:1754-1760. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, G. A., T. R. Shope, M. J. Waecker, and F. H. Lanningham. 1995. Moraxella (Branhamella) catarrhalis bacteremia in children—a report of two patients and review of the literature. Clin. Pediatr. 34:146-150. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, T. F. 1996. Branhamella catarrhalis—epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, T. F., A. L. Brauer, N. Yuskiw, E. R. McNamara, and C. Kirkham. 2001. Conservation of outer membrane protein E among strains of Moraxella catarrhalis. Infect. Immun. 69:3576-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, T. M., C. Kirkham, and A. J. Lesse. 1993. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol. Microbiol. 10:87-97. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido, H. 1994. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269:3905-3908. [PubMed] [Google Scholar]
- 28.Troncoso, G., S. Sanchez, M. T. Criado, and C. Ferreiros. 2004. Analysis of Moraxella catarrhalis outer membrane antigens cross-reactive with Neisseria meningitidis and Neisseria lactamica. FEMS Immunol. Med. Microbiol. 40:89-94. [DOI] [PubMed] [Google Scholar]
- 29.van der Ley, P., J. E. Heckels, M. Virji, P. Hoogerhout, and J. T. Poolman. 1991. Topology of outer membrane porins in pathogenic Neisseria spp. Infect. Immun. 59:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, Y. 2002. Characterization of the refolding and reassembly of an integral membrane protein OmpF porin by low-angle laser light scattering photometry coupled with high-performance gel chromatography. J. Chromatogr. A 961:137-146. [DOI] [PubMed] [Google Scholar]
- 32.Yi, K., and T. F. Murphy. 1994. Mapping of a strain-specific bactericidal epitope to the surface-exposed loop 5 on the P2 porin protein of non-typeable Haemophilus influenzae. Microb. Pathog. 17:277-282. [DOI] [PubMed] [Google Scholar]