Abstract
The cell-wall-less prokaryote Mycoplasma pneumoniae, long considered among the smallest and simplest cells capable of self-replication, has a distinct cellular polarity characterized by the presence of a differentiated terminal organelle which functions in adherence to human respiratory epithelium, gliding motility, and cell division. Characterization of hemadsorption (HA)-negative mutants has resulted in identification of several terminal organelle proteins, including P30, the loss of which results in developmental defects and decreased adherence to host cells, but their impact on M. pneumoniae gliding has not been investigated. Here we examined the contribution of P30 to gliding motility on the basis of satellite growth and cell gliding velocity and frequency. M. pneumoniae HA mutant II-3 lacking P30 was nonmotile, but HA mutant II-7 producing a truncated P30 was motile, albeit at a velocity 50-fold less than that of the wild type. HA-positive revertant II-3R producing an altered P30 was unexpectedly not fully wild type with respect to gliding. Complementation of mutant II-3 with recombinant wild-type and mutant alleles confirmed the correlation between gliding defect and loss or alteration in P30. Surprisingly, fusion of yellow fluorescent protein to the C terminus of P30 had little impact on cell gliding velocity and significantly enhanced HA. Finally, while quantitative examination of HA revealed clear distinctions among these mutant strains, gliding defects did not correlate strictly with the HA phenotype, and all strains attached to glass at wild-type levels. Taken together, these findings suggest a role for P30 in gliding motility that is distinct from its requirement in adherence.
Mycoplasmas are cell-wall-less prokaryotes with minimal genomes and limited biosynthetic capabilities, dictating a strict dependence on host species for survival in nature (43). Mycoplasma pneumoniae is a human pathogen primarily colonizing the respiratory tract. While the most common clinical manifestations of infection are tracheobronchitis and atypical or “walking” pneumonia (7, 12, 14, 30), recent studies indicate a strong correlation with asthma (5, 24, 38), and extrapulmonary complications are not uncommon (53). Adherence of M. pneumoniae cells to host respiratory epithelium (cytadherence) is required for colonization and pathogenesis (20) and is mediated largely by a differentiated terminal organelle (9, 39). This well-defined apical structure is a membrane-bound extension of the mycoplasma cell distinguished ultrastructurally by an electron-dense core (4), which is a major constituent of the M. pneumoniae cytoskeleton (17, 35).
M. pneumoniae cells exhibit gliding motility, with the terminal organelle always the leading end (6), but details regarding the biological significance and the mechanism of gliding are largely unknown. Although the M. pneumoniae genome has been sequenced and twice annotated (11, 19), close inspection reveals no homology to proteins known to be involved in bacterial motility of any type in walled bacteria. Furthermore, while gliding motility has been described for several mycoplasma species, even within the genus Mycoplasma there appear to be distinct gliding mechanisms, as proteins thought to function in M. mobile gliding are absent from the genomes of the gliding mycoplasmas M. genitalium, M. gallisepticum, and M. pneumoniae (15, 19, 23, 37, 49, 56).
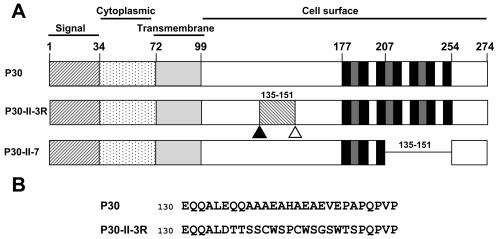
Analysis of M. pneumoniae hemadsorption (HA)-negative mutants has resulted in identification of a number of proteins associated with cytadherence (2, 27, 47, 48), including the putative adhesin P30, a membrane protein which localizes primarily to the terminal organelle (3) and which is predicted to orient with a cytoplasmic N terminus and the C terminus exposed on the cell surface (Fig. 1A) (10, 32). M. pneumoniae HA mutant II-3 lacks detectable P30 due to a frameshift in the corresponding gene (MPN453; Fig. 1A) (44). A second-site mutation in HA revertant II-3R restores the wild-type reading frame for all but 17 residues (Fig. 1A and B) (44). HA mutant II-7 has an in-frame deletion of residues 207 to 254 in a C-terminal Pro-rich repeat region (Fig. 1A) (10), resulting in an internally truncated P30 derivative. Mutants II-3 and II-7 also exhibit reduced levels of the peripheral membrane protein P65, normally found on the mycoplasma cell surface at the attachment organelle (25, 41). The function of P65 is unknown, and it remains undetermined whether the cytadherence defects in strains II-3 and II-7 are due directly to the loss/alteration in P30 or are an indirect result of reduced levels of P65.
FIG. 1.
Cytadherence-associated protein P30 in wild-type M. pneumoniae, mutants II-3 and II-7, and revertant II-3R. (A) Wild-type P30 is predicted to have a single transmembrane domain. Experimental data (10, 32) suggest an orientation with the C terminus on the cell surface. For P30-II-3R, a frameshift (open triangle) resulted in mutant II-3 with no detectable P30. A second site mutation (solid triangle) in revertant II-3R restored P30 except for residues 135 to 151 (see panel B below). P30-II-7 is a P30 derivative resulting from in-frame deletion of residues 207 to 254 in mutant II-7. (B) Amino acid sequence for P30 from wild-type and II-3R M. pneumoniae for the indicated residues.
Possible consequences of the defects in mutants II-3 and II-7 on cell motility have not been investigated. In the current study we assessed gliding motility on the basis of satellite growth around microcolonies and by digital microcinematography for wild-type M. pneumoniae, HA mutants II-3 and II-7, and revertant II-3R, noting a correlation between the rate of satellite growth and cell gliding velocities. Complete loss of P30 was accompanied by failure to glide, while alterations in P30 resulted in reduced gliding velocity and frequency. Complementation studies with recombinant wild-type and mutant P30 alleles confirmed a specific requirement for P30 in gliding motility and established that a gliding defect does not correlate with reduced steady-state levels of P65. Attachment to a substrate is a strict prerequisite for gliding motility, but reduced gliding velocity and frequency for II-7 and II-3R was not a function of substrate-binding capability, suggesting a direct defect in the gliding mechanism resulting from the loss or alteration of P30. Finally, in contrast to the markedly reduced gliding speeds and frequencies that resulted from the changes in P30 in II-7 and II-3R, the fusion of enhanced yellow fluorescent protein (EYFP) at the C terminus of P30, nearly doubling its size, had little effect on P30 function in gliding and HA.
MATERIALS AND METHODS
Mycoplasma strains.
Wild-type M. pneumoniae strain M129 (33) was used at the 18th broth passage. Spontaneously arising HA mutants II-3 and II-7 (29), and HA revertant II-3R (44), all derived from M129, were described previously.
Recombinant P30 derivatives and M. pneumoniae transformation.
Resident alleles for P30 from the wild type, mutant II-7, and revertant II-3R, together with upstream MPN454 (p21) and its corresponding promoter (57), were amplified by PCR, engineering an EcoRI site (underlined) in 5′ primer EcoRIp21pos (5′-GTAGCTTCATGAATTCGGTCT-3′), which was used with the 3′ primer hmw3neg (5′-CAAAGCTAATTGGTTCATCACTGTC-3′). The resulting PCR products were cloned using standard protocols (45) into Tn4001.2062 within plasmid pKV74 (18) by use of the EcoRI site in the upstream primer and a BamHI site downstream of MPN453, then transformed into Escherichia coli, and subsequently purified and sequenced (Integrated Biotechnology Laboratories, University of Georgia, Athens, GA). Mycoplasma transformation was achieved as described previously (18); multiple transformants were isolated from each transformation and expanded as described previously (18, 52).
Western immunoblotting.
Samples were prepared for sodium dodecyl sulfate polyacrylamide gel electrophoresis as described previously (31) and Western immunoblotting (54) as follows. After blocking in 5% skim milk in Tris-buffered saline (TBS; 0.2 M Tris-HCl, 0.85% NaCl, pH 8.2), membranes were incubated with primary antibody for 2 h at room temperature, washed five times for 5 min each in TBS-0.05% Tween 20, probed with secondary AP-conjugated antibody (Bio-Rad, Hercules, CA, or Promega, Madison, WI) for 1 h at room temperature, and washed again five times for 5 min each in TBS-0.05% Tween 20. Monoclonal P30-specific antibody (44) was used at a dilution of 1:750, rabbit anti-P65 serum (40) was used at 1:3,000, anti-P1 serum (28) was used at 1:1,000, and anti-GFP (Clontech; Palo Alto, CA) was used at 1:1,000.
Generation of a P30-EYFP fusion.
The gene for wild-type P30 was amplified by PCR using 5′ primer BamHIp21pos (5′-GTAGCTGGATCCACTTGGTCT-3′) and 3′ primer NcoIp30neg (5′-AAGCACCATGGAGCGTTTTGGTGGA-3′), generating BamHI and NcoI sites (underlined) near the 5′ and 3′ ends of the product, respectively, and replacing the stop codon with ATG of the NcoI recognition sequence. The resulting product was digested with BamHI and NcoI (Promega) and ligated into the corresponding sites of pEYFP (Clontech). The resulting plasmid was digested with BamHI and EcoRI to liberate p30-eyfp, which was cloned into the corresponding sites in the Tn4001 derivative in plasmid pMT85 (E. Pirkl and R. Herrmann, unpublished data), and then transformed into E. coli DH5α. The resulting plasmid was sequenced and electroporated into M. pneumoniae as described previously (18). Genomic DNA from mycoplasma transformants was digested with HindIII (Promega), religated, and transformed into E. coli. Plasmid DNA from kanamycin-resistant transformants was sequenced to determine the transposon junction by use of pMT85-specific primer pMT85seq (5′-CCGCGCGTTGGCCGATTCATTAATGCACGC-3′). An intergenic insertion was identified for two of eight transformants sequenced, one of which (an insertion at nucleotide 372735, between MPN312 and MPN313) is described here.
EYFP fluorescence microscopy.
Mycoplasma cultures were inoculated directly into 600 μl of SP-4 medium (55) in four-well borosilicate-glass chambers (Nalge/Nunc, Naperville, Ill.) and incubated overnight at 37°C. Phase and fluorescence images captured on a Leica DM IRB inverted microscope (Leica Microsystems, Wetlzlar, Germany) at an exposure of 0.4 sec by use of phase-contrast optics (100× oil-immersion objective, 1.4 numerical aperture) through a Chroma EYFP filter set (Chroma, Rockingham, VT) were recorded and digitized with a Hamamatsu Orca ER charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) using the computer program Openlab v3.0-4.0 (Improvision, Lexington, MA). Images were merged using Openlab or Adobe Photoshop v6.0-7.0 (Adobe Systems, Inc., San Jose, CA).
Analysis of satellite growth.
Motility stocks of each strain (see below) were diluted serially to a dilution of 10−2 to 10−5 in SP-4 medium-3% (wt/vol) gelatin and inoculated into adjacent wells of four-well borosilicate-glass chambers. At 12 h intervals postinoculation the appearance of satellite motility around colonies was recorded digitally with phase-contrast optics (40× objective) as described above.
Microcinematography and quantitation of cell gliding.
In order to quantitate gliding velocities for individual cells we modified a microcinematographic approach described over 30 years ago for M. pneumoniae (6) to take advantage of new technologies. Mycoplasmas grown in SP-4 medium in tissue-culture flasks for 72 h, until approximately the mid-logarithmic growth phase, were harvested by centrifugation at 17,000 × g for 25 min at 4°C, suspended in 2.5 ml fresh SP-4 medium supplemented with gelatin (3% wt/vol) by passage through a 25-gauge needle seven times, divided into motility stocks, and stored at −80°C. Motility stocks were thawed, mixed with 500 μl of fresh SP-4-3% gelatin-0.05 M HEPES (pH 7.2), passed through a 25-gauge needle five times to disperse cell aggregates, and inoculated into four-well borosilicate glass chambers, which were placed onto the observation stage of a Leica DM IRB microscope enclosed within an incubation chamber (Solent Scientific Limited, Portsmouth, United Kingdom) preheated to 37°C. After 10 min at 37°C, images were recorded and digitized at automated frame intervals as described above. The number of cells per field at the start of image capture was 30 to 60, with gliding velocities measured for a minimum of 200 cells per strain from no less than five separate studies and various motility stocks and media batches for control purposes. Gliding was quantitated using Openlab measurements module v3.51-4.0 (Improvision). Briefly, digitally recorded images of each frame were compiled into an image stack, with motility quantitated by recording the x and y coordinates of the leading edge of each motile cell in the observation field for each frame. An intrinsic time stamp was assigned per frame during capture, and the gliding velocity of each cell was calculated by dividing the distance covered (change in x and y coordinates) by the absolute time between frames. A cell resting period was assigned when there was no net cell movement greater than 1 pixel (0.0645 μm) between sequentially captured frames. Corrected gliding velocities were calculated as the total distance traveled by a cell divided by the total time of the field interval minus the amount of time spent in resting periods.
Analysis of HA and attachment to glass.
HA is a convenient indicator for M. pneumoniae cytadherence (50). Qualitative assessment of HA for mycoplasma colonies was performed as described previously (29) using sheep or chicken blood. HA was measured quantitatively with [3H]thymidine-labeled mycoplasmas (13), cultured in SP-4 rather than Hayflick medium. For quantitation of attachment to glass, frozen stocks of radiolabeled mycoplasma cultures were thawed and centrifuged for 5 min at 123 × g to remove cellular aggregates, and 150-μl aliquots of the resulting suspension were added to 500 μl of HEPES-buffered SP-4 medium-3.9% gelatin, pH 7.2 (final gelatin concentration, 3.0%). Suspensions were inoculated in triplicate onto sterile glass coverslips in 24-well plates and incubated for 30 min at 37°C. Coverslips were then washed three times in phosphate-buffered saline and analyzed by liquid scintillation spectrometry. Experiments were repeated four times per strain per condition tested.
Statistical analyses.
Quantitative motility, HA, and glass-binding data were assessed for statistical significance. Differences between strains were considered significant when data from comparisons made using a two-tailed Student's t test had a P value of <0.05.
RESULTS
Evaluation of gliding motility by satellite growth.
Several bacterial species manifest gliding motility as satellite growth or colony spreading over time. Although there are exceptions, satellite growth usually correlates with a gliding-competent phenotype (34, 51). The known gliding mycoplasma species are unique among gliding prokaryotes in that their motility requires a solid-liquid interface (1, 6, 26), but we were unable to observe satellite growth around wild-type microcolonies at an agar/broth interface with agar concentrations ranging from 0.5 to 12.5% or with embedding in soft agar as described for M. mobile (36). However, satellite growth was evident on polystyrene overlaid with agar at concentrations of <0.5% or on polystyrene or glass when 3% gelatin was included in broth medium to increase viscosity. Using the latter approach we compared satellite growth levels for wild-type and P30 mutant M. pneumoniae.
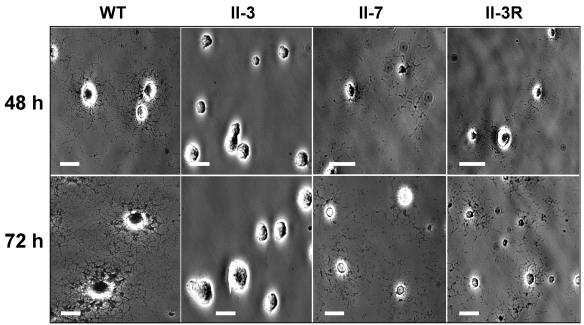
Wild-type satellite growth was most clearly observed after 48 to 72 h with stocks diluted sufficiently to allow unimpeded spreading (Fig. 2). HA mutant II-3 exhibited no satellite growth for up to 120 h postinoculation but rather formed microcolonies having a smooth leading edge (Fig. 2). In contrast, HA mutant II-7 and revertant II-3R exhibited limited satellite growth by 48 h, albeit much less dense than the wild-type growth (Fig. 2). Satellite growth increased by 72 h but remained considerably less abundant than for wild-type M. pneumoniae cultured for the same time period. By 96 to 120 h, satellite growth for both II-7 and II-3R approximated that seen at 72 h for the wild type (data not shown). The extended time scale required for wild-type satellite growth levels for these two strains suggested lower gliding velocities and/or gliding frequencies than for the wild type.
FIG. 2.
Analysis of wild-type and mutant M. pneumoniae satellite growth. Colony morphology of wild-type, mutant II-3, mutant II-7, and revertant II-3R cultured on glass in SP-4 medium-3% gelatin. Images were captured at 48 h and 72 h postinoculation as indicated. Bar, 30 μm.
Quantitation of cellular gliding.
Wild-type stocks prepared without centrifugation exhibited a mean gliding velocity of 0.29 ± 0.0069 μm/sec with a mean corrected velocity of 0.36 ± 0.0082 μm/sec, consistent with data published previously (42). However, wild-type stocks prepared with a centrifugation step exhibited significantly lower mean gliding and corrected gliding velocities of 0.18 ± 0.0079 μm/sec and 0.26 ± 0.0068 μm/sec, respectively (Table 1). Jaffe et al. likewise noted that centrifugation during mycoplasma stock preparation affected gliding by M. mobile (22). However, as stock preparation for M. pneumoniae HA mutants required centrifugation, motility stock cultures for all strains in subsequent studies here were prepared in that manner.
TABLE 1.
Mean gliding and corrected gliding velocities and percent time resting for wild-type M. pneumoniae, cytadherence mutant and revertant strains, and transformants
| Mycoplasma strain | Mean gliding velocity (μm/sec) | Mean corrected gliding velocity (μm/sec) | % Time resting |
|---|---|---|---|
| Wild-type M129 | 0.18 ± 0.0079 | 0.26 ± 0.0068 | 34.6 |
| Mutant II-3 | 0 | 0 | NA |
| Transformant II-3 + P30-WT | 0.21 ± 0.028 | 0.29 ± 0.019 | 31.9 |
| Transformant II-3 + P30-EYFP | 0.10 ± 0.025 | 0.19 ± 0.020 | 50.3 |
| Revertant II-3R | 0.0058 ± 0.00091 | 0.012 ± 0.0023 | 47.8 |
| Transformant II-3 + P30-II-3R | 0.011 ± 0.0049 | 0.018 ± 0.0051 | 48.3 |
| Mutant II-7 | 0.0035 ± 0.0011 | 0.0059 ± 0.00093 | 48.8 |
| Transformant II-3 + P30-II-7 | 0.0056 ± 0.0028 | 0.011 ± 0.0081 | 54.7 |
Wild-type cellular gliding was recorded at 1 frame per sec over 30-sec intervals, but no motility was observed for mutant II-7 or revertant II-3R by these parameters (motility being defined as the translocation of at least 1 μm per field interval). However, cellular motility for II-7 and II-3R was evident with image capture at 1 frame per min and field intervals of 20 min, with velocities approximately 20- to 50-fold slower than wild-type velocities (Table 1). Wild-type controls exhibited no reduction in motility after 20 min and remained steady over 3 h (data not shown). Mutant II-3 exhibited no motility even when observed over 3-h field intervals and was deemed nonmotile under these conditions.
Wild-type M. pneumoniae cells commonly exhibit resting periods of various lengths and frequencies during their motility tracks (6, 42). Because reduced satellite growth could result from increased resting frequencies rather than or in addition to reduced gliding velocities, we also examined the percentage of time that individual cells spent in intermittent resting periods during their motility tracks (percent time resting). The slower-gliding II-3R and II-7 strains also exhibited significantly higher percent time resting values than did wild-type M. pneumoniae, reflecting longer and/or more frequent resting periods (Table 1).
Analysis of recombinant P30 derivatives.
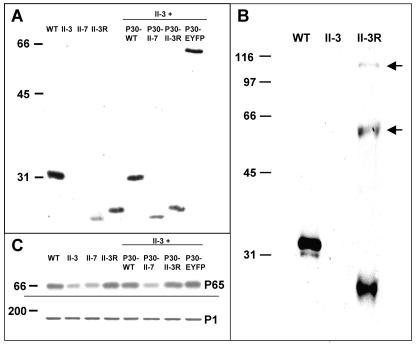
To confirm that loss or alteration in P30 was directly responsible for the observed motility defects, mutant II-3 transformants with recombinant P30 alleles introduced via transposon delivery were generated and analyzed. Multiple transformants were examined for each construct to control for the site of recombinant transposon insertion, and representative steady-state levels for each are shown by Western immunoblotting in Fig. 3. As expected, no P30 was detected in mutant II-3 while the truncated P30 in II-7 migrated faster than wild-type P30 (Fig. 3A) (10, 44). Unexpectedly, the truncated P30 in II-7 was consistently less abundant than its wild-type counterpart. Similar results were observed using an antibody specific for the N terminus of P30, indicating that epitope loss was not responsible for the reduced band intensity (data not shown). The 17-residue change in P30-II-3R increased its electrophoretic mobility to approximately 28,000, compared to 32,000 for wild-type P30 (Fig. 3A), while minor species were also observed at approximately 56,000 and 114,000 (Fig. 3B), corresponding to dimeric and tetrameric forms. Recombinant P30 levels in mutant II-3 transformants with the wild-type (II-3 + P30-WT), the II-7 (II-3 + P30-II-7), or the II-3R (II-3 + P30-II-3R) allele were generally comparable to that in the corresponding strains from which each was derived. Thus, P30 was produced at wild-type levels in II-3 + P30-WT and near-wild-type levels in II-3R and II-3 + P30-II-3R, and the truncated P30 was observed at similarly reduced levels in II-7 and II-3 + P30-II-7 (Fig. 3A).
FIG. 3.
Western immunoblot analysis of P30 and P65 in wild-type, mutant, revertant, and transformant M. pneumoniae. (A and B) Western immunoblot analysis of the indicated strains probed with P30-specific antibodies. II-3 + P30, mutant II-3 transformed with the indicated recombinant P30 allele by transposon delivery. (B) Samples processed to allow resolution of the multimeric forms of P30 from II-3R (arrows). (C) Western immunoblot analysis of parallel samples from panel A by use of P65-specific antibodies (top panel) or P1-specific antibodies as an internal control (bottom panel).
The II-3 transformants producing recombinant P30 were evaluated for satellite growth and cellular gliding phenotypes. Satellite growth for wild-type M. pneumoniae and II-3 + P30-WT was indistinguishable over the same time frame; likewise, II-3 + P30-II-7 and II-3 + P30-II-3R exhibited limited levels of satellite growth comparable to their II-7 and II-3R counterparts (data not shown). In addition, gliding velocities and resting frequencies for transformants with P30-WT, P30-II-7, and P30-II-3R were comparable to those in wild-type, II-7, and II-3R M. pneumoniae, respectively (Table 1). Thus, the defects in gliding correlated specifically with loss or alteration in P30.
Analysis of P30-EYFP.
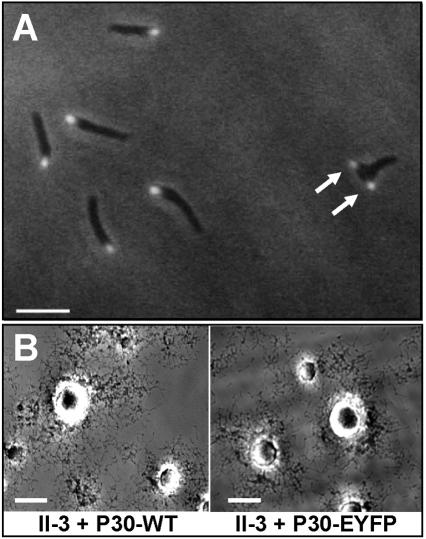
The P30-EYFP fusion of the predicted size (approximately 60,000) was produced in mutant II-3 transformants at levels near that of wild-type P30 and reacted with antibodies to both P30 (Fig. 3) and GFP (data not shown). Like P30 (reference 3 and data not shown), the P30-EYFP fusion was observed by fluorescence microscopy to localize predominantly at a single cell pole (Fig. 4A), which by time-lapse microcinematography was shown to be the leading end of motile cells (data not shown). Many nonmotile cells exhibited two P30 foci (Fig. 4A, arrows), probably reflecting attachment organelle duplication, which is thought to precede cell division (47). Satellite growth for II-3 transformants with P30-EYFP was indistinguishable from wild-type growth (Fig. 4B). Although nearly double the size of wild-type P30, the P30-EYFP fusion only reduced gliding velocity slightly (to 65 to 73% of wild-type velocity) compared to the reduction to 2 to 5% of the wild-type level with the P30 defects in II-7 and II-3R (Table 1).
FIG. 4.
Analysis of P30-EYFP in mutant II-3. (A) Fluorescence/phase-contrast microscopy established that the P30-EYFP fusion localizes to the terminal organelle, and cells with a second fluorescent focus were apparent (arrows). Bar, 2 μm. (B) Satellite growth in mutant II-3 + recombinant P30-EYFP was indistinguishable from growth in mutant II-3 + recombinant wild-type P30. Bar, 30 μm.
P65 stability.
P65 is found at reduced steady-state levels in mutants II-3 and II-7 (25). The reversion in II-3 to yield II-3R restored P65 to wild-type levels (Fig. 3C), as did the recombinant wild-type, II-3R, and P30-EYFP alleles but not the II-7 allele when introduced into mutant II-3. Thus, II-3R was gliding defective despite the presence of P65 at wild-type levels.
Analysis of HA and glass binding.
HA provides a convenient indicator for adherence to respiratory epithelium (50). As expected (29), when examined qualitatively the HA mutants II-3 and II-7 failed to adsorb sheep erythrocytes, while revertant II-3R (44) was clearly HA positive, although not at the high erythrocyte density of the wild type (Fig. 5A). A similar pattern was observed with chicken erythrocytes (data not shown). The quantitative HA assay reinforced in part the qualitative observations but also revealed a difference between II-3 and II-7 (Fig. 5B). While mutant II-3 bound to erythrocytes minimally (5% of wild-type levels), HA by mutant II-7 was significantly higher than that of mutant II-3, at approximately 35% of the wild-type level. Revertant II-3R HA was substantially higher than the II-7 level but still only approximately 60% of the wild-type level. The contribution of P30 to the HA phenotype of each was reinforced by the quantitative HA data obtained using transformants producing recombinant P30 (Fig. 5B). Somewhat surprisingly, however, the recombinant P30-EYFP fusion restored HA in mutant II-3 to a level significantly higher than in the wild type or II-3 + P30-WT.
FIG. 5.
Assessment of HA with wild-type M. pneumoniae and P30 mutant, revertant, and transformant strains. (A) Qualitative HA by the indicated strains. Bar, 60 μm. (B) Quantitative HA by the indicated strains normalized to wild-type binding. Error bars, standard errors of the means; WT, wild type.
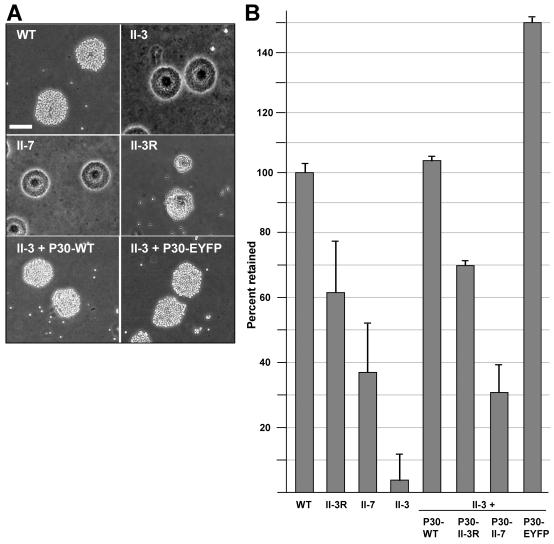
By definition, attachment to a surface is a prerequisite for gliding motility. Given the variability in HA and gliding motility with the loss or alteration in P30, we examined the glass binding of each under conditions identical to those used to assess cellular gliding velocities and frequencies. Unlike the HA data, all strains, including the nonmotile mutant II-3, exhibited levels of glass attachment that were statistically comparable to the wild-type level (Table 2). Thus, under the conditions used here to measure M. pneumoniae cell motility, the gliding deficiencies were not simply a function of glass-binding ability.
TABLE 2.
Wild-type and mutant glass binding under conditions used for quantitation of gliding velocities and frequenciesa
| Strain | % Retained ± SEM |
|---|---|
| Wild type | 100 ± 10 |
| HA mutant II-3 | 84 ± 10 |
| HA mutant II-7 | 93 ± 7 |
| HA revertant II-3R | 87 ± 8 |
| II-3 + P30-WT | 90 ± 7 |
| II-3 + P30-II-7 | 93 ± 20 |
| II-3 + P30R | 97 ± 6 |
| II-3 + P30-EYFP | 102 ± 7 |
All strains exhibited levels of binding that were indistinguishable from wild-type levels (P > 0.05). Attachment data shown normalized to wild type.
DISCUSSION
Mycoplasmas are considered to be among the smallest and simplest known cells capable of self-replication, and yet M. pneumoniae and related species exhibit distinct cellular polarity characterized by the presence of a complex terminal organelle that is the leading end of gliding cells and initiates mycoplasma binding to host cell receptors. However, the mechanism by which M. pneumoniae glides is unknown, and its genome contains no homologs of known motility proteins of any type in walled bacteria or other mycoplasmas. Furthermore, very little is known about specific terminal organelle components with respect to their function in adherence and gliding (46).
In the current study we examined M. pneumoniae satellite growth around microcolonies as a possible indicator of gliding motility. Attempts to promote satellite growth using soft agar as described for gliding walled bacteria (34, 51), Mycoplasma mobile (36), and Spiroplasma citri (21) were unsuccessful, but satellite growth was apparent when M. pneumoniae was cultured on plastic or glass in growth medium containing gelatin. At the concentrations examined gelatin is visibly less viscous than soft agar and hence probably requires less force for gliding to be achieved. The inability to observe M. pneumoniae satellite growth in soft agar at concentrations of ≥0.5% may therefore reflect a lower force-generating capacity than for other gliding species. Regardless, our finding that HA mutant II-7 but not HA mutant II-3 exhibited satellite growth, albeit much more slowly than with the wild type, was unexpected, as was the finding that satellite growth by the HA-positive revertant II-3R was not like wild-type growth. A requirement for P30 in cytadherence was established previously (3, 29); the current studies indicated an additional role for P30 in gliding motility.
We modified existing microscopy protocols (6) to take advantage of digital technology and measure mycoplasma gliding on the cellular level. Gliding velocities for wild-type strain M129 were comparable to those reported previously (42) for the wild-type FH strain (data not shown) and 20- to 50-fold faster than mutant II-7 and revertant II-3R velocities, while mutant II-3 was nonmotile. Thus, the cell velocities and gliding frequencies for wild-type M. pneumoniae, mutants II-3 and II-7, and revertant II-3R correlated directly with their satellite growth phenotypes, demonstrating that satellite growth is a good indicator for identifying potential gliding-defective mutants in this species.
Transformation of mutant II-3, having no detectable P30, with recombinant wild-type, mutant, or revertant alleles for P30 by transposon delivery, yielded motility phenotypes that were comparable to those of the strains from which each recombinant allele originated (Table 1). Therefore, it was possible to correlate the specific defects in P30 with the altered motility phenotypes, underscoring a role for P30 in M. pneumoniae gliding motility. Our results from complementation analysis also confirmed that the stability of cytadherence-associated protein P65 is affected specifically by the loss or alteration of P30. However, as gliding remained deficient despite restoration of P65 to wild-type levels in II-3R and mutant II-3 producing recombinant P30-II-3R, the motility defects in P30 mutants cannot be attributed to an effect on P65 levels.
For mutant II-7 and revertant II-3R the reduced gliding velocities correlated specifically with a 48-residue deletion and a 17-residue substitution, respectively, in the putative cell surface domain of P30 (Fig. 1). The latter was associated with the formation of multimers that were stable to detergent denaturation (Fig. 3B). Wild-type P30 has a single Cys residue located in the predicted transmembrane domain, while the substitution in P30R introduced two additional Cys residues (Fig. 1B). P30 is predicted to assume a coiled-coil conformation from residues 90 to 150 (data not shown), which includes the region encompassed by the substitution in P30R. Additional Cys residues in this region might promote multimerization in the oxidizing environment of the cell exterior. However, the P30R multimers were resistant to reducing agents dithiothreitol and β-mercaptoethanol, with and without alkylation with iodoacetamide (data not shown), suggesting that non-disulfide-covalent or strong hydrophobic interactions may contribute to multimerization. Somewhat surprisingly, while the relatively minor substitution in II-3R reduced gliding velocity markedly, fusion of EYFP to P30, which approximately doubled its size, impaired P30 function very little and actually increased HA by 50%. The reason for enhanced HA with this fusion is not clear, but the fact that gliding was not similarly enhanced reinforces the impression of dual but distinct functions of P30 in cytadherence and cell gliding.
The observation that HA mutants II-3 and II-7 differed with respect to gliding motility prompted a closer examination of their HA phenotypes. As expected, the two were indistinguishable by qualitative screening, whereby mycoplasma colonies adsorb erythrocytes (Fig. 5A). However, as determined by the more sensitive measure of attachment of radiolabeled mycoplasmas to erythrocytes in suspension, mutant II-7 adherence was six- to sevenfold higher than that of mutant II-3. Furthermore, closer inspection of revertant II-3R revealed intermediate levels of HA by both qualitative and quantitative techniques (Fig. 5). The relative levels of HA actually paralleled the gliding velocities, with wild-type ≫ revertant II-3R > mutant II-7 ≫ mutant II-3, raising the possibility that the gliding defect might actually be an extension of the HA deficiency. However, this conclusion was not supported by the observation that under the conditions used to assess cell gliding, all strains attached to glass at comparable levels.
The role of P30 in gliding motility is not clear. The major adhesin protein P1 localizes to the terminal organelle in wild-type M. pneumoniae and in P30 mutants but is nonfunctional in the latter, raising the possibility that P30 is required to stabilize the adhesin in the appropriate conformation in the mycoplasma membrane (44). Antibodies to P1 cause gliding mycoplasmas to detach from the glass surface (46), but detachment was not pronounced in our studies with P30 mutants. Taken together, these observations suggest that P30 may contribute to the linkage between surface-binding moieties and the force-generating mechanism of M. pneumoniae.
Finally, individual cells of mutants II-3 and II-7 (44) and revertant II-3R (data not shown) exhibit a branched morphology rather than the filamentous spindle-like morphology of wild-type M. pneumoniae after attaching to plastic or glass. Whereas the nonmotile mutant II-3 retains this branched morphology, mutant II-7 (44) and revertant II-3R (data not shown), for which the gliding machinery is indeed active though less efficient, eventually assume a filamentous shape over time. Thus, based on the current study, the filamentous, extended morphology of wild-type cells is likely a function of cell gliding, in which case the filamentous appearance of M. pneumoniae cells on ciliated respiratory epithelium (8, 16) suggests that gliding indeed occurs on the surface of host cells as well.
In summary, the current report provides the first genetic evidence linking a specific protein to gliding motility by M. pneumoniae. Furthermore, specific domains of P30 thought to be oriented on the mycoplasma cell surface were implicated in gliding function. Surprisingly, fusion of EYFP to the C terminus of P30 had little impact on gliding and actually enhanced HA. Additional studies, including structural analysis of the surface domain of P30, will be required to establish in more detail how P30 functions in both gliding and cytadherence.
Acknowledgments
We thank R. Herrman for providing P65-specific antisera as well as transposon pMT85 prior to publication, J. Baseman for providing P30 antibody, and E. Sheppard and R. Krause for their technical contributions to data analysis.
This work was supported by Public Health Service grant AI49194 from the National Institute of Allergy and Infectious Diseases to D.C.K.
REFERENCES
- 1.Andrewes, C. H. 1946. A motile organism of the pleuropneumoniae group. J. Pathol. Bacteriol. 58:578-580. [DOI] [PubMed] [Google Scholar]
- 2.Balish, M. F., and D. C. Krause. 2002. Cytadherence and the cytoskeleton, p. 491-518. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 3.Baseman, J. B., J. Morrison-Plummer, D. Drouillard, B. Puleo-Scheppke, V. V. Tryon, and S. C. Holt. 1987. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr. J. Med. Sci. 23:474-479. [PubMed] [Google Scholar]
- 4.Biberfield, G., and P. Biberfield. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biscardi, S., M. Lorrot, E. Marc, F. Moulin, B. Boutonnat-Faucher, C. Heilbronner, J. L. Iniguez, M. Chaussain, E. Nicand, J. Raymond, and D. Gendrel. 2004. Mycoplasma pneumoniae and asthma in children. Clin. Infect. Dis. 38:1341-1346. [DOI] [PubMed] [Google Scholar]
- 6.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. 32:321-326. [DOI] [PubMed] [Google Scholar]
- 7.Clyde, W. A., Jr., and F. W. Denny. 1967. Mycoplasma infections in childhood. Pediatrics 40:669-684. [PubMed] [Google Scholar]
- 8.Collier, A. M., and W. A. Clyde, Jr. 1974. Appearance of Mycoplasma pneumoniae in lungs of experimentally infected hamsters and sputum from patients with natural disease. Am. Rev. Respir. Dis. 110:765-773. [DOI] [PubMed] [Google Scholar]
- 9.Collier, A. M., W. A. Clyde, Jr., and F. W. Denny. 1971. Mycoplasma pneumoniae in hamster tracheal organ culture: immunofluorescent and electron microscopic studies. Proc. Soc. Exp. Biol. Med. 136:569-573. [DOI] [PubMed] [Google Scholar]
- 10.Dallo, S. F., A. L. Lazzell, A. Chavoya, S. P. Reddy, and J. B. Baseman. 1996. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect. Immun. 64:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny, F. W., W. A. Clyde, Jr., and W. P. Glezen. 1971. Mycoplasma pneumoniae disease: clinical spectrum, pathophysiology, epidemiology, and control. J. Infect. Dis. 123:74-92. [DOI] [PubMed] [Google Scholar]
- 13.Fisseha, M., H. W. Gohlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foy, H. M. 1999. Mycoplasma pneumoniae pneumonia: current perspectives. Clin. Infect. Dis. 28:237. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 16.Gabridge, M. G., M. J. Bright, and H. R. Richards. 1982. Scanning electron microscopy of Mycoplasma pneumoniae on the membrane of individual ciliated tracheal cells. In Vitro 18:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Gobel, U., V. Speth, and W. Bredt. 1981. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J. Cell Biol. 91:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 19.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, P. C., A. M. Collier, and J. B. Baseman. 1976. Interaction of virulent Mycoplasma pneumoniae with hamster tracheal organ cultures. Infect. Immun. 14:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, C., F. Nouzieres, S. Duret, J. M. Bove, and J. Renaudin. 1997. Isolation, characterization, and complementation of a motility mutant of Spiroplasma citri. J. Bacteriol. 179:4802-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, S. L., and R. J. Martin. 16. June 2005, posting date. Chlamydophila pneumoniae and Mycoplasma pneumoniae: a role in asthma pathogenesis? Am. J. Respir. Crit. Care Med. [Online.]. [DOI] [PubMed]
- 25.Jordan, J. L., K. M. Berry, M. F. Balish, and D. C. Krause. 2001. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J. Bacteriol. 183:7387-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhoff, H., and R. Rosengarten. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130(Pt. 9):2439-2445. [DOI] [PubMed] [Google Scholar]
- 27.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 28.Krause, D. C., and J. B. Baseman. 1983. Inhibition of Mycoplasma pneumoniae hemadsorption and adherence to respiratory epithelium by antibodies to a membrane protein. Infect. Immun. 39:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause, D. C., and D. Taylor-Robinson. 1992. Mycoplasmas which infect humans, p. 417-444. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman, (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Layh-Schmitt, G., R. Himmelreich, and U. Leibfried. 1997. The adhesin related 30-kDa protein of Mycoplasma pneumoniae exhibits size and antigen variability. FEMS Microbiol. Lett. 152:101-108. [DOI] [PubMed] [Google Scholar]
- 33.Lipman, R. P., W. A. Clyde, Jr., and F. W. Denny. 1969. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 100:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 35.Meng, K. E., and R. M. Pfister. 1980. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J. Bacteriol. 144:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 37.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 38.Park, S. J., Y. C. Lee, Y. K. Rhee, and H. B. Lee. 2005. Seroprevalence of Mycoplasma pneumoniae and Chlamydia pneumoniae in stable asthma and chronic obstructive pulmonary disease. J. Korean Med. Sci. 20:225-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell, D. A., P. C. Hu, M. Wilson, A. M. Collier, and J. B. Baseman. 1976. Attachment of Mycoplasma pneumoniae to respiratory epithelium. Infect. Immun. 13:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proft, T., and R. Herrmann. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol. Microbiol. 13:337-348. [DOI] [PubMed] [Google Scholar]
- 41.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radestock, U., and W. Bredt. 1977. Motility of Mycoplasma pneumoniae. J. Bacteriol. 129:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-Dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of a 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobeslavsky, O., B. Prescott, and R. Chanock. 1968. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J. Bacteriol. 96:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talkington, D. F., K. B. Waites, S. B. Schwartz, and R. E. Besser. 2001. Emerging from obscurity: understanding pulmonary and extrapulmonary syndromes, pathogenesis, and epidemiology of human Mycoplasma pneumoniae infections, p. 57-84. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infections 5. American Society for Microbiology, Washington, D.C.
- 54.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tully, J. G., R. F. Whitcomb, H. F. Clark, and D. L. Williamson. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892-894. [DOI] [PubMed] [Google Scholar]
- 56.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldo, R. H., III, P. L. Popham, C. E. Romero-Arroyo, E. A. Mothershed, K. K. Lee, and D. C. Krause. 1999. Transcriptional analysis of the hmw gene cluster of Mycoplasma pneumoniae. J. Bacteriol. 181:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]