Abstract
The obligate intracellular pathogen Chlamydia trachomatis expresses a type III secretion system (T3SS) which has the potential to contribute significantly to pathogenesis. Based on a demonstrated role of type III secretion (T3S)-specific chaperones in the secretion of antihost proteins by gram-negative pathogens, we initiated a study of selected putative Chlamydia T3S chaperones in an effort to gain mechanistic insight into the Chlamydia T3SS and to potentially identify Chlamydia-specific secreted products. C. trachomatis Scc2 and Scc3 are homologous to SycD of Yersinia spp. Functional studies of the heterologous Yersinia T3SS indicated that although neither Scc2 nor Scc3 was able to fully complement a sycD null mutant, both have SycD-like characteristics. Both were able to associate with the translocator protein YopD, and Scc3 expression restored limited secretion of YopD in in vitro studies of T3S. CopB (CT578) and CopB2 (CT861) are encoded adjacent to scc2 and scc3, respectively, and have structural similarities with the YopB family of T3S translocators. Either Scc2 or Scc3 coprecipitates with CopB from C. trachomatis extracts. Expression of CopB or CopB2 in Yersinia resulted in their type III-dependent secretion, and localization studies with C. trachomatis-infected cells indicated that both were secreted by Chlamydia.
Chlamydia trachomatis is a medically significant gram-negative bacterium commonly associated with human ocular (serovars A to C) and sexually transmitted (serovars D to K) diseases or, more rarely, with systemic disease (lymphogranuloma venereum; LGV1, -2, and -3) (38). Chlamydia spp. are obligate intracellular parasites that invade host cells as metabolically inert particles termed elementary bodies (EBs), which subsequently develop into vegetative yet noninfectious forms termed reticulate bodies (RBs). After multiple rounds of division, an undefined signal triggers a subset of RBs to differentiate asynchronously back to EBs which, when liberated from the host cell, initiate subsequent rounds of infection (31). This developmental cycle occurs entirely within a parasitophorous vacuole termed an inclusion. Although sequestered within the membrane-bound inclusion, chlamydial development and pathogenesis require intimate association with and active manipulation of host cell processes. This ability to subvert host cell processes from within a privileged niche is clearly a hallmark of chlamydial pathogenesis (reviewed in reference 11), yet the full extent to which Chlamydia spp. modulate host cell activities and potential mechanisms by which these alterations are affected are poorly understood.
One mechanism to induce host cell alterations is through the use of a type III secretion system (T3SS). Type III secretion (T3S) is a recognized virulence determinant among multiple gram-negative pathogens of both plants and animals, where contributions to pathogenesis are manifested primarily by the deployment of antihost proteins termed effectors (24). This complex machinery enables secretion of effectors from bacteria, followed by translocation through a host membrane barrier to the host cell cytoplasm, where the effectors target specific cellular processes relevant to the respective pathogen. Chlamydiae express a functional T3SS (12, 23) that is available to deploy effector proteins throughout the developmental cycle (10). Chlamydia spp. exploit a host niche unique among type III-expressing pathogens. Although components of the Chlamydia T3SS basal apparatus are identifiable in sequenced genomes (25, 36, 42) due to homology to components in other T3SSs, gene products with obvious similarity to known effector proteins are not evident. Identification of Chlamydia effectors is further complicated by the fact that the primary sequences of T3S substrates lack obvious consensus secretion signals. Significantly, chaperone-mediated secretion of effectors is a common theme in characterized T3SSs, and chlamydial genomes contain genes encoding proteins with both structural and sequence similarities to characterized T3S chaperones.
T3S-specific chaperones are a diverse group of proteins that share limited sequence similarity but function by associating with cytoplasmic pools of T3S substrates to promote their secretion or prevent premature association with one another (9). These secretion substrates include antihost effectors as well as proteins involved in translocating those secreted effectors into host cells. Unlike general chaperones, T3S chaperones are specific, associating with one or sometimes two secretion substrates, resulting in each secreted effector having a dedicated chaperone. T3SSs are exquisitely controlled such that secretion activity of the apparatus is intimately coupled to gene regulation. In addition to facilitating secretion, a subset of described T3S chaperones is also required for the proper regulation of T3S expression through involvement in complex feedback cascades (16). These chaperones are proposed to be fundamentally different from those exerting activity solely as facilitators of secretion (33).
Scc1, Scc2, and Scc3 (specific chlamydia chaperones) have been implicated as T3S-associated chaperones, based on sequence and structural similarities to known chaperones in other T3SSs (10, 12, 23). Scc1 is homologous to the monospecific effector chaperone SycE of Yersinia spp., whereas both Scc2 and Scc3 are homologous to a family of bifunctional chaperones exemplified by SycD of Yersinia spp., SicA of Salmonella spp., and IpgC of Shigella spp. (34). In these systems, the chaperone interacts with and serves as a secretion pilot for a conserved set of translocator proteins termed YopB and YopD for Yersinia, SipB and SipD for Salmonella, and IpaB and IpaC for Shigella. Once secreted, respective translocator pairs may associate, are essential for effector protein translocation, and have been implicated as components of a host membrane-localized pore (24). SycD and its Salmonella or Shigella homologs remain in the bacterial cytoplasm and contribute to repression (SycD) or activation (SicA and IpgC) of T3S gene expression (reviewed in reference 34).
Given the importance of the SycD class of T3S-specific chaperones and the lack of identified Chlamydia T3S substrates, we examined the possibility of exploiting the potential chaperone activities of Scc2 and Scc3 to identify novel secreted Chlamydia proteins. We investigated whether Scc2 and Scc3 have SycD-like activities in the heterologous Yersinia T3SS and report here results that are consistent with Scc2 and Scc3 having active roles in T3S. Both Scc2 and Scc3 interacted with the translocator protein YopD in Yersinia, and further analysis of Scc2 and Scc3 led to the identification of CopB and CopB2 as components of the Chlamydia T3SS. Both a CopB-containing protein and a truncated CopB2 were type III secreted when expressed in Yersinia, and based on localization in infected cells, native CopB and CopB2 were translocated by Chlamydia. These results extend the characterization of the Chlamydia T3SS and demonstrate the utility of chaperones as tools in the discovery of T3S-specific substrates.
MATERIALS AND METHODS
Cell culture and organisms.
Bacteria and recombinant plasmid constructs are listed in Table 1. Escherichia coli TOP10 F′ was cultivated at 37°C either in Luria-Bertani broth or on Luria-Bertani agar plates. Yersinia pseudotuberculosis serovar III and Yersinia enterocolitica serovar O:9 were cultivated at 26°C in heart infusion broth (HIB; Difco) or on tryptose blood agar plates (Difco) for routine growth and as described previously (19) in HIB supplemented either with 0.4% glucose (wt/vol), 5 mM EGTA, and 20 mM MgCl2 (without Ca2+ [−Ca2+]) or with 0.4% glucose (wt/vol) and 5 mM CaCl2 (with Ca2+ [+Ca2+]) for physiological studies. E. coli and yersiniae were routinely cultured in the presence of appropriate antibiotics at 150 μg ml−1 for carbenicillin (Invitrogen), 100 μg ml−1 for ampicillin (Sigma), 30 μg ml−1 for kanamycin (U.S. Biochemical), or 35 μg ml−1 for nalidixic acid (Sigma). Where appropriate, bacteria were cultivated in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) to achieve induction of trans genes. C. trachomatis serovar L2 was cultivated in HeLa 229 epithelial cells maintained at 37°C in the presence of 5% CO2-95% humidified air in RPMI 1640 (Gibco BRL) supplemented with 10% (vol/vol) fetal bovine serum and 10 μg ml−1 gentamicin (Gibco BRL) as described previously (7). Infection of HeLa cells was done at 37°C in Hanks' balanced salt solution (HBSS; Gibco BRL) with RenoCal-76 (Bracco Diagnostics) density gradient-purified chlamydiae, as described previously (7, 18).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Property(ies) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 | ||
| TOP10 F′ | F′ [lacIq Tn10(Tetr)]mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Y. enterocolitica | ||
| W22703 | O:9 serotype, pYVe227 | 8a |
| CT133 | pYVe227 sycD [SycDΔ7-49]a | 2 |
| VTL2 | pYVe227 yopD [YopDΔ9-306] | 27a |
| Y. pseudotuberculosis | ||
| YPIII pIB102 | pIB yadA::Tn5 | 6a |
| YPIII pIB68 | pIB yscS [YscSΔ19-59] yadA::Tn5 | 5a |
| Plasmids and constructs | ||
| pTrcHisB | Apr; expression vector for creation of N-terminal His6-containing fusions | Invitrogen |
| pFlag-CTC | Apr; expression vector for creation of C-terminal Flag-tagged fusions | Sigma |
| pEF/myc/cyto | Neor; eukaryotic expression vector used as source of neomycin phosphotransferase (npt) | Invitrogen |
| pHT-LcrH | lcrH PCR amplified from Y. pseudotuberculosis YPIII pIB102 (nt 4-504) in pTrcHisB fused in frame with the His6-containing leader sequence | This study |
| pSycD-FT | PCR-amplified Y. enterocolitica sycD (nt 4-504) in pFlag-CTC fused in frame with C-terminal Flag epitope tag | This study |
| pScc1-FT | PCR-amplified scc1 (CT088; nt 4-438)b in pFlag-CTC fused in frame with C-terminal Flag epitope tag | This study |
| pScc2-FT | PCR-amplified scc2 (CT576; nt 4-696) in pFlag-CTC fused in frame with C-terminal Flag epitope tag | This study |
| pScc3-FT | PCR-amplified scc3 (CT862; nt 4-594) in Flag-CTC fused in frame with C-terminal Flag epitope tag | This study |
| pCopB2NT | PCR-amplified copB2 N terminus (CT861; nt 4-951) in pFlag-CTC in frame with C-terminal Flag epitope tag | This study |
| pCopBnpt | pFlag-CTC expressing a chimeric product consisting of CopB (aa 1-120)c, Npt, and a C-terminal Flag epitope tag | This study |
Numbers in brackets correspond to residues deleted from the gene product.
The C. trachomatis genome designation and the respective nucleotides (nt) amplified are provided.
Numbers refer to residues of the C. trachomatis product present in the respective chimeric protein. aa, amino acids.
DNA methods.
Specific genes were amplified from genomic (C. trachomatis serovar L2) or virulence plasmid (Yersinia) DNA in a GeneAmp model 2400 thermocycler (Perkin-Elmer Cetus) by using Pfu DNA polymerase (Promega) and custom oligonucleotide primers synthesized by DNagency. Cloning techniques were performed essentially as described previously (29) Coding sequences for C. trachomatis L2 scc1, scc2, and scc3 or a portion of copB2 were amplified using sense primers (KFSCC-4A, KFSCC2-4A, KF862-1A, and KF861-6A, respectively) and antisense primers (KFSCC-4B, KFSCC2-4B, KF862-1B, and KF861-6B, respectively). Y. enterocolitica sycD was amplified with KFLCRH-2A and KFLCRH-1B. A chimeric gene construct containing a 5′ portion of chlamydial copB (amplified with KF578-1A and KF578npt-1B) fused to neomycin phosphotransferase (npt amplified from pEF/myc/cyto with KFnpt578-1A and KFnpt-6B) was generated by overlap extension PCR. All products contained engineered 5′ XhoI and 3′ KpnI sites. XhoI- and KpnI-treated products were ligated into pFlag-CTC (Sigma), resulting in the addition of a 3′ Flag epitope tag to respective genes. Construction also resulted in the replacement of native initiating methionine codons with a vector-encoded Met-Leu-Leu coding sequence. N-terminally His6-tagged Y. pseudotuberculosis SycD was generated by amplification with sense (KFLCRH-1A) and antisense (KFLCRH-1B) primers also containing engineered XhoI and KpnI sites and ligation of XhoI- and KpnI-treated PCR products into XhoI- and KpnI-digested pTrcHisB. Precise regions amplified from respective genes are provided in Table 1, and specific primer sequences are listed in Table 2. Chemically competent E. coli strain TOP10 F′ was transformed according to the manufacturer's instructions (Invitrogen), and Y. pseudotuberculosis or Y. enterocolitica was transformed by electroporation as described by Perry et al. (35).
TABLE 2.
Cloning primer sets
| Gene | Designation | Sequence (5′ → 3′)a |
|---|---|---|
| sycD | KFLCRH-1A | AAACCGCTCGAGACAACAAGAGACGACAGACACTCAAGAATACC |
| KFLCRH-2A | AAACCGCTCGAGCAACAAGAGACGACAGACACTCAAGAATACC | |
| KFLCRH-1B | ATCTGGTACCTCATGGGTTATCAACGCACTCATGTTCCATCTCC | |
| scc1 | KFSCC-4A | AAACCGCTCGAGCAAAATCAATTTGAACAACTCC |
| KFSCC-4B | ATCTGGTACCCAGGTGATACATACCTAGAGCATGTAAATCTGG | |
| scc2 | KFSCC2-4A | AAACCGCTCGAGAGCACTCCATCTTCTAATAATTC |
| KFSCC2-4B | ATCTGGTACCCTTTGCTTTTTTCTTGTTAGAAGCAGC | |
| scc3 | KF862-1A | AAACCGCTCGAGACCACCAAGCAAGATCCAATGTCTTG |
| KF862-1B | ATCTGGTACCGGCGTACGATTTGATCGCCAAAATTTCTTCTTTTAAAGAGC | |
| copB | KF578-1A | AAACCGCTCGAGGTCCCTTTCATCTTCTTCGTCTTCCG |
| KF578npt-1B | GCGTGCATTCCATCTTGTTCAATGCTAGTATCTTCTATGCATTGTTGCG | |
| copB2 | KF861-6A | AAACCGCTCGAGTCATCCTGGTTTGCTCAAGCGACAGACGTCGC |
| KF861-6B | CGGAGGTACCGCTCGCTAGAGAATCCCAAAATCGCG | |
| npt | KFnpt-6B | AAAGGTACCGAAGAACTCGTCAAGAAGGCGATAGAAGGCG |
| KFnpt578-1A | CGCAACAATGCATAGAAGATACTAGCGATTGAACAAGATGGATTGCACGC |
Engineered restriction sites are indicated in bold.
Protein purification.
Flag-tagged C. trachomatis L2 Scc1, Scc2, and Scc3 used in coprecipitation experiments were expressed in E. coli TOP10 F′ cultivated in the presence of 0.1 mM IPTG. Bacterial lysates were prepared as described previously (13), and Flag-tagged proteins were purified in one step by passage over α-FlagM2 resin (Sigma) columns, followed by elution of bound protein with 100 mM glycine, pH 3.0. Eluates were neutralized by addition of Tris (Sigma), pH 8.0, to 50 mM and then dialyzed against phosphate-buffered saline (PBS) (135 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). Single-step purification of His6-tagged SycD used in antibody production was accomplished by using Talon (CLONETECH, Palo Alto, CA) metal affinity resin essentially as described previously (12), followed by dialysis into PBS. All isolated recombinant proteins were assayed for purity by Coomassie staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-resolved proteins (data not shown). Identities of respective proteins were confirmed by mass spectroscopy of gel-extracted, trypsin-treated proteins as described previously (46), using a Voyager-DE STR matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (PerSeptive Biosystems) and the Protein Prospector MS-Fit (University of California, San Francisco) database.
Yersinia physiological studies.
Physiological studies examining potential roles of chlamydial type III chaperones were performed with Y. enterocolitica. Strains were cultivated in HIB (without Ca) at 26°C for ca. 10 generations, diluted into fresh −Ca2+ and +Ca2+ HIB to an optical density at 620 nm (OD620) (A620) of 0.05, and cultivated for an additional 2 h at 26°C. Cultures were then shifted to 37°C, and IPTG was added to appropriate levels. At 4 h after temperature shift to 37°C, total-culture (TC) samples were harvested or cultures were separated into cell-free supernatants (CS) and whole-cell (WC) pellets by centrifugation. Cell pellets were treated for coprecipitation studies (see below), or total proteins were precipitated by addition of trichloroacetate (TCA; Fisher) to 10% (vol/vol) to respective culture fractions. TCA-precipitated proteins were solubilized in electrophoresis sample buffer (2.3% [wt/vol] SDS, 5% [vol/vol] β-mercaptoethanol, 25% [vol/vol] glycerol, and 60 mM Tris [pH 6.8]). Type III secretion of chlamydial proteins was tested as described previously (12), by heterologous expression in Y. pseudotuberculosis cultivated in +Ca2+ or −Ca2+ HIB, and processed exactly as described above.
Chlamydia coprecipitation assays.
A mixture of C. trachomatis L2 RBs and EBs was harvested by release from 24-h cultures with a Kontes homogenization and purification in RenoCal-76 density gradients as described previously (7). Purified chlamydiae were suspended in ice-cold lysis solution (5 mM dithiothreitol, 200 mM NaCl) containing protease inhibitors, incubated on ice for 2 h, and homogenized through a 25-gauge needle. Debris were precipitated by centrifugation for 15 min at 16,000 × g, and cleared lysates were combined with 5 μg of Flag-tagged Scc1, Scc2, or Scc3 and dialyzed against PBS overnight at 4°C. Flag-tagged proteins were purified over α-FlagM2 resin as described above, and resulting material was analyzed by immunoblotting.
Antibody production and immunodetection.
SycD- and Scc3-specific polyclonal antibodies were raised by immunizing female New Zealand White rabbits with pure HT-SycD or Scc3-FT, respectively, as described previously (39). Rabbit polyclonal antibodies specific for C. trachomatis L2 CADD (NH2-DEENGYPNHIDLWKQC-COOH), CopB (NH2-TRQNRDDLSMESDVA-COOH), and CopB2 (NH2-HSQQPSHKIQRRKERC-COOH) peptides conjugated to keyhole limpet hemocyanin were generated commercially by Sigma Genosys. Specificities of antibody preparations were confirmed by immunoblot analysis of SDS-PAGE-resolved whole-cell lysates of E. coli TOP10 F′ expressing the respective target protein or a vector-only control (data not shown).
Proteins were analyzed by immunoblotting with resolution in SDS-PAGE (27) gels (12% [vol/vol] polyacrylamide) and subsequent transfer to Immobilon-P (Millipore Corp.) in carbonate buffer (10 mM NaHCO3, 3 mM Na2CO3, 10% methanol, pH 9.9). Specific proteins from Yersinia cultures were detected by probing with α-Flag M2 (Sigma), α-β-lactamase (5′→3′; Boulder, CO), α-HT-SycD, or α-YopD and α-YopE (kind gifts from G. Plano). Material from C. trachomatis coprecipitation assays was probed with α-HT-CopB (10), α-CopB2, or α-Hsp60 (47). Unless otherwise noted, proteins were visualized by using alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG; Sigma), followed by development with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP; Gibco-BRL). Indirect immunofluorescence of methanol-fixed material was employed as described previously (12) to test protein localization in C. trachomatis L2-infected HeLa monolayers cultivated on 12-mm coverslips. Scc2, Scc3, CopB, and CopB2 were detected with α-HT-Scc2 (10), α-Scc3-FT, α-CopB (α-peptide), and α-CopB2, respectively, and visualized by probing with Texas Red-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch). Fluorescent and Nomarski images were acquired using a 60× apochromat objective on an FXA photomicroscope (Nikon) and digitized via a Dage-MTI CCD 72 camera with a DSP2000 image processor. CopB distribution was further examined via immunoelectron microscopy. HeLa cells were cultured on Thermonox coverslips (Nunc) and infected with C. trachomatis L2 for ca. 18 h. Cultures were fixed with periodate-lysine-paraformaldehyde and processed essentially as described previously (39). CopB was detected using α-CopB (α-peptide), peroxidase-conjugated F(ab′)2 donkey α-rabbit IgG (Jackson ImmunoResearch Laboratories), and Immunopure metal enhancement DAB substrate (Pierce Chemical). An RMC MT-7000 ultramicrotome was used to cut ca.-70-nm sections that were counterstained with 1% (wt/vol) uranyl acetate and Reynolds lead acetate. Samples were observed at 80 kV with a Philips CM-10 transmission electron microscope. Micrographs were processed using Adobe Photoshop 5.0 (Adobe Systems).
Alpha-toxin permeabilization.
A biochemical assay to examine proteins exposed to the host cytoplasmic compartment was developed by using selective permeabilization of the plasma membranes with staphylococcal alpha-toxin. HeLa cell monolayers were cultivated in 6-well cluster dishes and mock treated or infected with C. trachomatis L2 for 24 h. Monolayers were washed twice with 37°C HBSS and then treated with 1.0 ml HBSS or HBSS containing 50 μg alpha-toxin (Calbiochem) for 2 h at 37°C. Culture supernatants were removed, passed through a 0.22-μm filter, and stored on ice for lactose dehydrogenase (LDH) activity assays. Monolayers were gently washed with 37°C HBSS and then treated for 30 min at ambient temperature with 1.0 ml HBSS with or without 750 μg NHS-S-S-biotin [sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate; Pierce Chemical]. Culture supernatants were removed and replaced with 2.0 ml RPMI plus 5% bovine serum albumin (wt/vol) for 5 min to quench unreacted biotinylating reagent. Monolayers were gently washed two additional times with HBSS, and cells were lysed by treatment with 650 μl/well of ice-cold H2O containing 0.1% Triton X-100 (vol/vol) and 150 mM NaCl. Lysates were collected and centrifuged at 16,000 × g for 15 min, and 150 μl was stored on ice for ATP assays, while the remaining 500 μl was combined with an equal volume of 100 mM Tris, pH 7.4, for precipitation of biotinylated proteins. NHS-S-S-biotin-labeled proteins were specifically purified by incubating lysates with NeutrAvidin-agarose beads at 4°C overnight, followed by extensive washing with PBS and solubilization in electrophoresis sample buffer. Isolated material was resolved by SDS-PAGE, and detection of specific proteins was accomplished by using α-MOMP (3), α-EF-Tu (48), α-Hsp60, α-CADD, or α-CopB2, followed by peroxidase-conjugated goat anti-rabbit IgG and chemiluminescent development with Super Signal reagent (Pierce Chemical). ATP and LDH concentrations were determined according to the manufacturer's instructions by use of an ATP bioluminescence assay kit (CLS II; Roche) and an LDH diagnostic kit (Sigma), respectively.
RESULTS
Scc2 and Scc3 similarity to SycD.
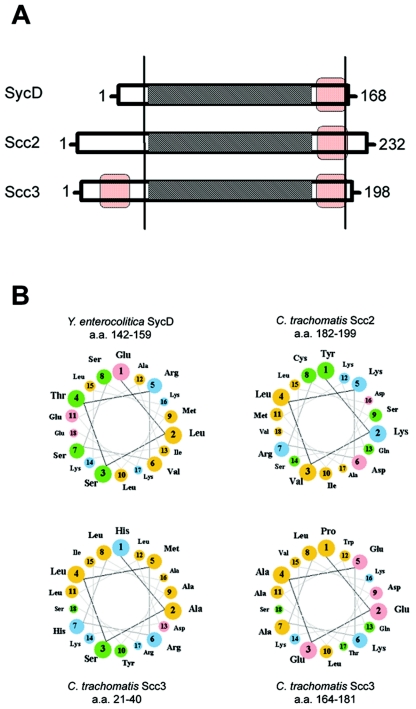
Chlamydia Scc2 and Scc3 have been described as T3S-associated chaperones, based on homology to SycD of Yersinia spp. (10, 42). SycD is multifunctional in Yersinia, acting both as a direct secretion chaperone and as a component in T3S regulatory cascades. Scc2 and Scc3 are paralogously related, sharing 45% overall sequence similarity and displaying 51% and 48%, respectively, overall sequence similarity to SycD. Closer examination reveals that sequence similarities are confined to internal domains representing residues 23 to 153 of SycD, 66 to 195 of Scc2, and 68 to 198 of Scc3 (Fig. 1A). Tetratricopeptide repeats (TPR) are implicated in protein-protein interactions, and all three proteins possess these repeats within the ca.-130-residue region of similarity. Use of PSIPRED (30) revealed that Scc2 and Scc3 possess predicted C-terminally localized helices that, like the helix found in SycD, have an imperfect amphipathic character (Fig. 1B). Despite these striking similarities, Scc2 and Scc3 possess qualities that clearly distinguish them from SycD and each other. The predicted pIs of Scc2 and Scc3 are 9.58 and 6.71, respectively, whereas the predicted pI of SycD is an acidic 4.70. At 232 and 198 residues, respectively, Scc2 and Scc3 are significantly larger than the 168-residue SycD. Most of this additional length corresponds to a unique N-terminal domain of ca. 60 residues, yet Scc2 and Scc3 do not exhibit significant sequence similarity within this domain. Interestingly, the N-terminal region (residues 21 to 40) of Scc3, but not of Scc2, contains a canonical amphipathic α helix. Taken together, the sequence data suggest that Scc2 and Scc3 may share basic functions with Yersinia SycD but have the capacity to mediate unique and possibly novel effects.
FIG. 1.
Comparison of C. trachomatis Scc2 and Scc3 with Yersinia SycD. (A) Schematic representations of proteins, indicating numbers of total residues. Vertical bars bracket domains in which sequence similarities are found. Respective TPR are shown as shaded areas, and amphipathic helices are represented by red boxes. (B) Amphipathic characters of predicted α helices are shown in helical wheel projections (http://cti.itc.virginia.edu/∼cmg/Demo/wheel/wheelApp.html), where nonpolar residues are shown in orange, polar, uncharged residues in green, acidic residues in pink, and basic residues in blue. Residue numbers of corresponding domains are provided for respective proteins. a.a., amino acids.
Activities of chlamydial gene products in sycD null Yersinia.
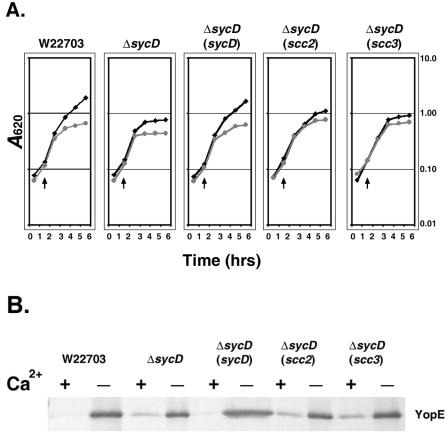
A direct test of Scc2 and Scc3 function is not possible in the absence of a tractable genetic system for Chlamydia spp. However, evidence indicates that some conserved components of T3SSs can be functionally exchanged (1, 17, 20, 37), and we have successfully employed Yersinia as a heterologous T3SS to identify Chlamydia T3S substrates (10, 12). Yersinia SycD is essential for negative regulation of T3S gene expression in the absence of inductive signals (−Ca2+ in vitro and cell contact in vivo), and as a consequence, strains lacking sycD display constitutive growth restriction (indicative of T3S activation) and deregulated secretion of Yops (5). We therefore assayed potential function by testing the ability of ectopically expressed Scc2 or Scc3 to compensate for loss of SycD activity in a Y. enterocolitica ΔsycD strain with in vitro growth assays commonly used to study regulation of T3S expression and activity (Fig. 2). Y. enterocolitica ΔsycD or Y. enterocolitica ΔsycD expressing pSycD-FT, pScc2-FT, or pScc3-FT was cultivated in Ca2+-supplemented or -deficient HIB, and bacterial growth was evaluated by measuring culture optical density (Fig. 2A). As expected, Yersinia ΔsycD displayed growth restriction in both the presence and the absence of Ca2+, whereas the wild-type strain maintained logarithmic growth in the presence of Ca2+. Ectopic expression of SycD in the ΔsycD strain resulted in full complementation of the growth phenotype. Although Yersinia ΔsycD expressing Scc2 or Scc3 displays growth restriction at a slightly higher A620 than does ΔsycD, Ca2+-dependent growth was not restored. Yersinia YopE is an endogenous, Ca2+-regulated T3S substrate. We examined levels of YopE secretion by immunoblotting of SDS-PAGE-resolved cell-free culture supernatants (Fig. 2B) to test directly whether Scc2 or Scc3 compensates for SycD in Ca2+ regulation. Consistent with growth data, expression of SycD-FT restored Ca2+-regulated secretion of YopE in the ΔsycD strain, since YopE was only detected in the −Ca2+ culture supernatant. In contrast, the presence of neither Scc2-FT nor Scc3-FT prevented YopE secretion in the presence of Ca2+. Based on these data, we conclude that neither Scc2 nor Scc3 was able to restore full Ca2+ regulation in Yersinia ΔsycD, indicating that the chlamydial proteins cannot substitute for the role of SycD in Ca2+ regulation.
FIG. 2.
Complementation analysis of Y. enterocolitica sycD. The Y. enterocolitica wild type (W22703) and sycD (ΔsycD) expressing vector alone or SycD-FT, Scc2-FT, or Scc3-FT were cultivated with 0.1 mM IPTG and in the presence (⧫ [black lines] or +) or absence (• [grey lines] or −) of 2.5 mM Ca2+. Bacterial growth experiments were repeated four times, and a representative data set in which growth was plotted hourly by measuring optical density at A620 is shown (A). Cultures were harvested 4 h after temperature shift (indicated by arrows) to 37°C, and material corresponding to 0.05 OD620/ml of cell-free culture supernatants was resolved in 12% (wt/vol) polyacrylamide gels (B). The immunoblot was analyzed using α-YopE, and proteins were visualized by probing with alkaline phosphatase-conjugated secondary antibodies, followed by development with NBT-BCIP.
SycD is a specific secretion chaperone for YopB and YopD. Strains lacking sycD contain low steady-state levels of Yops B and D due to decreased intracellular stability and are unable to secrete these Yops (32). Yersinia SycD-FT or C. trachomatis Scc2-FT or Scc3-FT was expressed in Y. enterocolitica ΔsycD cultivated in conditions repressive (+Ca2+) or inductive (−Ca2+) for T3S expression. TC levels of Flag-tagged proteins and Yops E and D, and CS levels of YopD, were assayed by immunoblotting as a sensitive test for the ability of expressed gene products to complement the sycD null phenotype (Fig. 3). Similarly, Flag-tagged C. trachomatis Scc1, which is not homologous to SycD, was expressed as a negative control. Comparable levels of all Flag-tagged proteins were detected by Flag-specific immunoblotting with respective culture lysates. As expected, YopD was detected in culture supernatants and YopE and YopD levels were elevated in −Ca2+ cultures containing wild-type Y. enterocolitica, whereas no significant levels of YopD secretion or increases in YopE or YopD levels were detected in the sycD null strain. Wild-type, Ca2+-regulated secretion of YopD and expression patterns of Yops E and D and were restored in the presence of SycD-FT. Compared to the sycD parent, YopE levels remained unaltered in the presence of Scc1-FT, Scc2-FT, and Scc3-FT. Significantly, an overall increase in YopD was detected in cultures expressing Scc3-FT, and Ca2+-regulated secretion of YopD was restored, though much lower levels of secreted YopD were detected in cultures expressing Scc3-FT than in the wild type. Immunoblots were subsequently probed with β-lactamase-specific antibodies to test for nonspecific leakage of bacterium-associated proteins into culture supernatants. Although significant signal was detected in bacterium-containing lysates, none was detected in culture supernatants. These data suggest that, like SycD, the presence of Scc3 results in stabilization of intracellular YopD, yet Scc3 fails to elicit the regulatory effects manifested by SycD in this system.
FIG. 3.
Complementation of YopD secretion. Y. enterocolitica sycD expressing SycD-FT, Scc1-FT, Scc2-FT, and Scc3-FT or Y. enterocolitica WT and sycD (ΔsycD) were cultivated in the presence (+) or absence (−) of 2.5 mM Ca2+. Cultures were incubated at 26°C for 2 h and then shifted to 37°C. IPTG was added to 0.1 mM at the time of temperature shift to achieve induction of trans genes. After 4 h of growth at 37°C, 1.0-ml volumes were harvested and directly precipitated with TCA for TC samples or fractionated into samples representing CS prior to concentration. Material corresponding to 0.05 OD620/ml of original cultures for CS and 0.02 OD620/ml for TC was resolved in 12% (wt/vol) polyacrylamide gels. Flag-tagged SycD, Scc1, Scc2, and Scc3 were detected by immunoblotting with α-Flag M2, and YopD and YopE were detected with α-YopD and α-YopE, respectively. Immunoblots were probed with β-lactamase-specific antibodies as a control for cell lysis. Proteins were visualized by probing with alkaline phosphatase-conjugated secondary antibodies and development with NBT-BCIP.
Protein interactions with YopD.
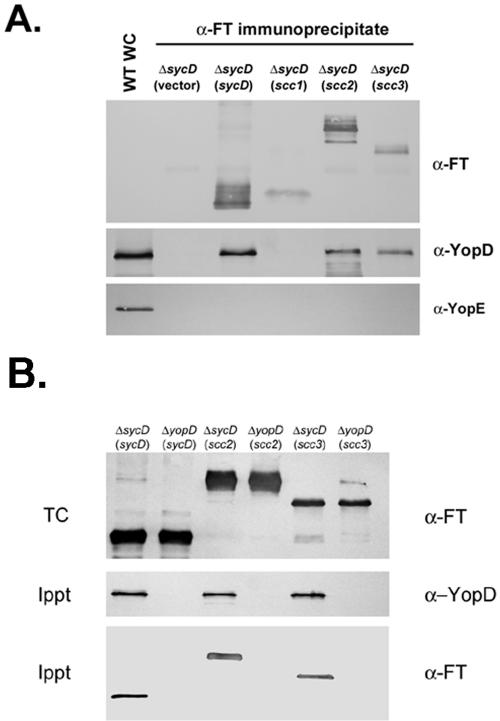
Intracellular stability and type III-mediated secretion of YopD depend on direct interaction with SycD. We examined the abilities of our Flag-tagged proteins to interact with YopD to test whether the Scc3-mediated increase in detectable YopD may occur by the same mechanism. Ca2+-containing cultures of Y. enterocolitica wild type or Y. enterocolitica ΔsycD expressing SycD-FT, Scc1-FT, Scc2-FT, or Scc3-FT were harvested, and proteins were immunoprecipitated from cleared lysates of disrupted cells by using Flag-tag-specific antibodies (Fig. 4A). All Flag-tagged proteins were detected by immunoblotting of specifically precipitated material. One lane of the immunoblot was loaded with a whole lysate from the Y. enterocolitica wild type as a control for antibody reactivity, and a YopD-specific signal was detected in this lane and in lanes containing SycD-FT, Scc2-FT, and Scc3-FT, but not Scc1-FT. The presence of YopD was specific since YopE was detected only in the whole-lysate control and not in immunoprecipitated material. A reciprocal experiment was performed, in which YopD was specifically immunoprecipitated to confirm the copurification of Scc2-FT and Scc3-FT (Fig. 4B). Flag-tagged proteins were additionally expressed in Y. enterocolitica ΔyopD as an antibody specificity control. Recovery of YopD was detected by immunoblotting of precipitated material from the yopD-containing strain. Scc2-FT, Scc3-FT, and the positive control SycD-FT were readily detectable in preimmunopreciptate lysates and in material containing YopD. Precipitation was YopD dependent, since none of these proteins were detected in samples derived from Y. enterocolitica ΔyopD. Similar results were obtained from immunoprecipitations from −Ca2+ cultures (data not shown). Taken together, these data provide strong evidence that Chlamydia Scc2 and Scc3 interact with the T3S substrate YopD.
FIG. 4.
Scc2 and Scc3 coprecipitate with YopD. Cleared lysates of Y. enterocolitica sycD (ΔsycD) or yopD (ΔyopD) expressing vector or Flag-tagged proteins were generated from +Ca cultures (cultivated in the presence of 0.1 mM IPTG) harvested 4 h after temperature shift. (A) Flag-tagged proteins were immunoprecipitated from lysates with α-FlagM2 resin, and purified material was examined by immunoblotting with Flag-tag-specific antibodies (α-FT), α-YopD, or α-YopE. A lane containing a WC lysate from WT Y. enterocolitica was included as a positive control for α-YopD or α-YopE reactivity. (B) YopD was specifically immunoprecipitated by addition of α-YopD protein A-conjugated beads to lysates. Proteins in precipitate material (Ippt) were detected by immunoblotting with α-YopD or α-FT. Parallel TC lysates were also generated from cultures and probed by immunoblotting with α-FT. Proteins were visualized by probing with alkaline phosphatase-conjugated secondary antibodies, followed by development with NBT-BCIP.
Coprecipitations from C. trachomatis lysates.
Although only Scc3 partially compensated for the lack of SycD, the surprising observation that both Scc2 and Scc3 interacted with YopD raised the possibility that these proteins may possess cognate partners in Chlamydia. The SycD binding partners YopB and YopD are encoded in the same operon and immediately downstream of sycD. We therefore considered the possibility that gene products encoded adjacent to scc2 or scc3 could be T3S substrates. Scc2 is encoded upstream of three putative open reading frames, designated CT577, CT578, and CT579, in the C. trachomatis genome database, while scc3 is positioned upstream of two open reading frames, designated CT861 and CT860. In both cases, the next predicted 5′-localized gene is oriented in the opposite direction, probably not cotranscribed, and predicted functions do not indicate association with the T3S mechanism. CT578 and CT861 encode proteins with predicted masses of 50.2 and 56.4 kDa, respectively, and have structural features, including two predicted N-terminal and one C-terminal coiled-coil domains separated by large hydrophobic domains, that make them reminiscent of the YopB family of proteins.
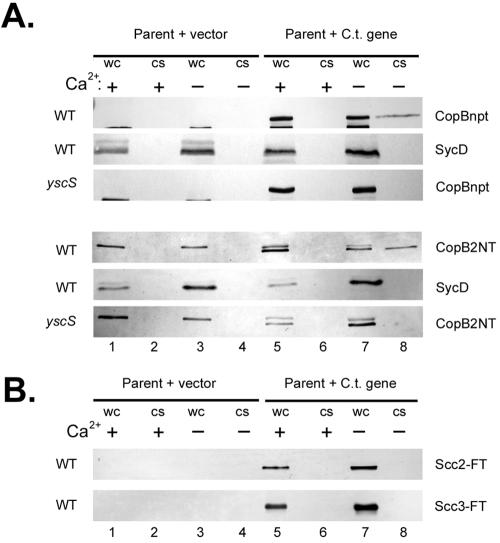
Given their conserved positioning near a sycD-like gene and structural similarities to the YopB family of proteins, we tentatively designated CT578 and CT861 CopB and CopB2, respectively, and then tested whether either associated with Scc2 or Scc3 (Fig. 5). To test for specific interactions, Scc2-FT and Scc3-FT were purified from E. coli and equivalent amounts were separately mixed with cleared C. trachomatis L2 culture lysates. Treatments with additions of PBS (mock) or Scc1-FT served as negative controls. After overnight incubation, Flag-tagged proteins were purified over α-FTM2 affinity columns and eluted material was resolved in SDS-PAGE gels for immunoblot analysis. Lysates from whole uninfected or C. trachomatis-infected HeLa cultures were resolved as antibody specificity controls. Scc1-FT, Scc2-FT, and Scc3-FT were detected in α-FT immunoblots, indicating successful isolation from C. trachomatis extracts. We were unable to detect copurification of CopB2 in any treatment, and this was not due to protein instability, since CopB2 was detectable in column flowthrough fractions (data not shown). CopB, however, was present in material containing either Scc2 or Scc3 but not in material containing Scc1 or in mock samples. This copurification was specific since C. trachomatis Hsp60 was not detectable in corresponding material. (A crossreactive band corresponding to antibody heavy chain is faintly visible in all precipitated material probed with Hsp60- and CopB2-specific antibodies.) Although these data do not differentiate between direct and indirect interactions, they do indicate that either Scc2 or Scc3 can associate with CopB.
FIG. 5.
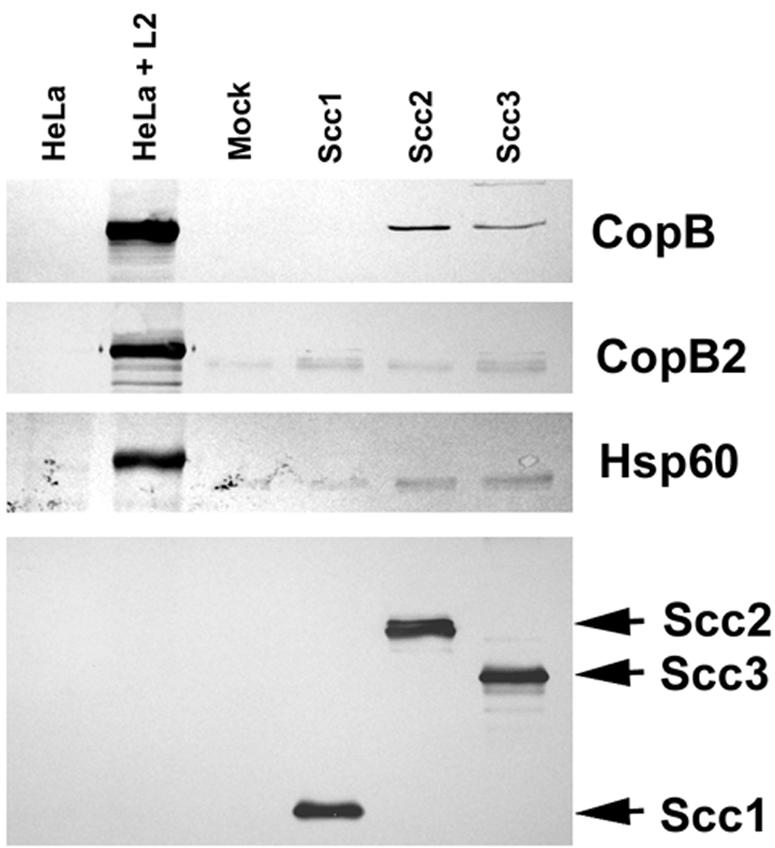
Scc2 and Scc3 coprecipitate CopB from C. trachomatis-containing lysates. Scc1-FT, Scc2-FT, Scc3-FT, and PBS as a mock-treated control (Mock) were incubated with cleared lysates from purified C. trachomatis RB and EB developmental forms. Flag-tagged proteins were purified by addition of α-FlagM2 resin, and material was analyzed by immunoblotting with CopB-, CopB2-, Hsp60-, or FT-specific antibodies. Lanes containing whole-culture lysates from mock-infected (HeLa) or C. trachomatis-infected (HeLa + L2) HeLa cell cultures were included as a control for antibody specificity. Proteins were visualized by probing with alkaline phosphatase-conjugated secondary antibodies, followed by development with NBT-BCIP.
Secretion of CopB and CopB2 by the Yersinia T3SS.
We extended the analysis of CopB and CopB2 to test whether they may be T3S substrates in the heterologous Yersinia T3SS. We were unable to express full-length CopB or CopB2, possibly due to the large predicted hydrophobic domains in each protein. Instead, a chimeric gene encoding the first 120 residues of CopB fused to npt or a truncated gene encoding residues 2 to 317 of CopB2 was constructed in the Flag-tag expression vector. The resulting CopBnpt and CopB2NT along with Scc2-FT and Scc3-FT were expressed separately in T3S competent (wild-type [WT]) or null (yscS) Y. pseudotuberculosis under conditions inductive (−Ca2+) or repressive (+Ca2+) for T3S, and protein localization was analyzed by immunoblotting of lysates from fractionated cultures (Fig. 6). CopBnpt and CopB2NT were detectable in the bacterial cell pellets (WC) of WT and yscS yersiniae, regardless of Ca2+ levels. Significantly, both were secreted into the CS fraction from −Ca2+ cultures of WT Yersinia. Export was specific since the cytoplasmically localized SycD was not detected in any supernatant fraction. Secretion was Ca2+ regulated and T3S dependent since no CopBnpt- or CopB2NT-specific signal was detected in CS fractions from +Ca2+ WT and yscS or −Ca2+ yscS cultures. Scc2 and Scc3 were not T3S substrates for Yersinia since, even though detectable in WC samples, neither was detected in CS fractions (Fig. 5B). Hence, CopB and CopB2 represent two newly recognized T3S substrates.
FIG. 6.
CopB and CopB2 are T3S substrates. Y. pseudotuberculosis YPIII pIB102 (WT) expressing pCopBnpt, pCopB2NT, pScc2-FT, pScc3-FT, or YPIII pIB68 (yscS) expressing pCopBnpt or pCopB2NT were cultivated in HIB in the presence (+) or absence (−) of 2.5 mM Ca2+ (lanes 5 to 8). Strains expressing vector only as a negative control were included (lanes 1 to 4). After an initial 2-h incubation at 26°C, IPTG was added to 0.1 mM and cultures were shifted to 37°C. Cultures were harvested after 4 h of growth at 37°C and fractionated into samples representing CS and WC. Material corresponding to 0.10 OD620/ml of original cultures for CS and 0.02 OD620/ml for WC was resolved in 12% (wt/vol) polyacrylamide gels and analyzed by immunoblotting with α-Flag M2 (A and B) or α-SycD (A). For immunoblotting, proteins were visualized by probing with alkaline phosphatase-conjugated secondary antibodies, followed by development with NBT-BCIP. C.t., C. trachomatis.
Immunolocalization of Scc2, Scc3, CopB, and CopB2 in C. trachomatis-infected cultures.
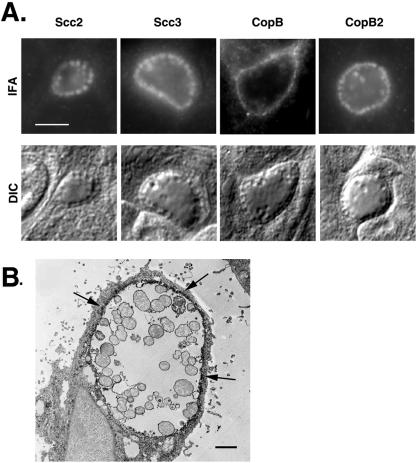
Distribution of Scc2, Scc3, CopB, and CopB2 in C. trachomatis-infected cultures was examined to test whether localizations were consistent with proposed roles as cytoplasmic chaperones and secreted effectors, respectively. Fixed samples of C. trachomatis-infected HeLa cultures were probed with specific polyclonal antibodies, and Scc2, Scc3, CopB, and CopB2 were visualized by indirect immunofluorescence microscopy (Fig. 7A). Staining patterns for Scc2, Scc3, and CopB2 are consistent with labeling of chlamydiae which are clearly visible in corresponding DIC images. CopB staining, however, displayed a rim-like pattern indicative of inclusion membrane-localized proteins. Possible CopB-specific staining of some bacteria was detected, but this was faint and not visible on all chlamydiae within a given inclusion. Immunoelectron microscopy was performed to more precisely examine the localization of CopB (Fig. 7B). A CopB-specific signal was evident on or in close proximity to the inclusion membrane and did not appear to be confined to particular zones. Significant staining of chlamydiae was not apparent. These data are consistent with Scc2 and Scc3 functioning within the chlamydial cytosol and CopB exerting an activity as a secreted protein. The CopB2 staining pattern was less revealing, given that only bacterium-associated signal was detected, even when CopB2 localization was examined at later time points (data not shown).
FIG. 7.
Immunolocalization of Scc2, Scc3, CopB, and CopB2 in C. trachomatis-infected HeLa monolayers. Proteins were detected 20 h postinfection via (A) indirect immunofluorescence or (B) immunoelectron microscopy. (A) Immunofluorescent (IFA) and Nomarski (DIC) images of individual inclusions are shown. Proteins were visualized by probing with Texas Red-conjugated secondary antibodies. Bar = 5 μm. (B) CopB was specifically stained by the immunoperoxidase method and examined by transmission electron microscopy. Arrows indicate the CopB-specific signal localized to the inclusion membrane. Bar = 1 μm.
Alpha-toxin permeabilization assay of protein localization.
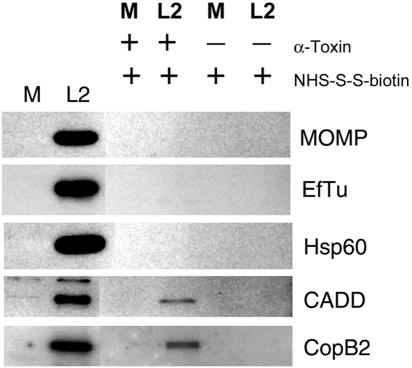
Heterologous expression in Yersinia indicated that CopB2 can be secreted, yet we were unable to detect CopB2 localization outside the chlamydial inclusion by indirect immunofluorescence. It is possible that signal strength or accessibility of the peptide epitope could be a confounding factor contributing to a lack of detection. To address this issue, we developed a sensitive biochemical assay to test specifically for accessibility of Chlamydia proteins to the host cytoplasm. Plasma membranes of C. trachomatis- or mock-infected HeLa cells were permeabilized by addition of staphylococcal alpha-toxin or mock treated with HBSS as a control, and accessible proteins were specifically labeled by addition of the cross-linking agent NHS-S-S-biotin. The release of intracellular ATP and retention of LDH were measured to ensure successful membrane permeabilization in the absence of general lysis (Table 3). Indeed, monolayers receiving alpha-toxin retained significantly decreased amounts of ATP compared to mock-treated controls. This was not due to cell lysis, since visual inspection revealed intact monolayers (data not shown) and since LDH activity in culture supernatants was not significantly elevated compared to that of mock-treated controls. Biotinylated proteins were purified, resolved via SDS-PAGE, and examined for protein content by immunoblotting (Fig. 8). Whole-culture lysates from parallel, untreated wells of mock- and C. trachomatis-infected(L2) HeLa cells were resolved as antibody specificity controls and as an indicator of total levels of respective proteins in unfractionated cultures. All proteins were detected in significant levels in whole-culture lysates. Chlamydial MOMP, EfTu, and Hsp60 were not detected in purified material from any treatment. One possibility for the lack of MOMP signal could be due to its insolubility during our cell lysis procedure. Although MOMP is a membrane-associated protein, a proportion of it is soluble in Triton X-100 (7) and we were able to detect MOMP in the soluble material (data not shown). C. trachomatis CADD has been shown to gain access to the host cytoplasm, where it interacts with host tumor necrosis factor family receptors (41). CADD-specific antibodies were therefore employed as a positive control for an extrainclusion-localized chlamydial protein and detected a protein only in purified material derived from cultures treated with alpha-toxin and NHS-biotin. CopB2-specific signal was also detected in this sample. CADD and CopB2 were not detected in the absence of NHS-S-S-biotin (data not shown) or alpha-toxin treatment. These data indicate that, like CADD, at least a portion of CopB2 is localized outside the chlamydial inclusion and is accessible to exogenous NHS-S-S-biotin. We propose that the pool of CopB2 detected by this assay is intracellularly (host) localized, since alpha-toxin permeabilization was required, but is not intrainclusion localized, since the prevalent Chlamydia-associated proteins MOMP, EfTu, and Hsp60 were not accessible to biotin labeling. Hence, we believe that CopB2 is secreted by Chlamydia and is exposed to the host cell cytoplasm.
TABLE 3.
ATP retention and LDH release
| Treatment | Alpha-toxin (μg/ml) | Biotin (μg/ml) | [ATP] (relative units [± SD])a | % Controlb | LDH activity (U/ml [± SD])c | % Controlb |
|---|---|---|---|---|---|---|
| Mock | 0.00 | 750.0 | 82.2 (± 1.22) | 100 | 615.5 (± 133) | 5.92 |
| Mock | 50.0 | 750.0 | 15.1 (± 1.08) | 18.4 | 1912 (± 6.36) | 18.4 |
| Mock | 50.0 | 0.000 | 11.4 (± 2.42) | 13.9 | 1867 (± 43.3) | 17.9 |
| L2 | 0.00 | 750.0 | 117 (± 7.52) | 100 | 1147 (± 135) | 11.0 |
| L2 | 50.0 | 750.0 | 15.2 (± 2.01) | 12.9 | 1904 (± 20.5) | 18.3 |
| L2 | 50.0 | 0.000 | 13.3 (± 1.22) | 11.4 | 1901 (± 13.4) | 18.3 |
Values correspond to quantitation of ATP-dependent bioluminescence in sample dilutions falling within the linear range of a standard curve and reflect levels of ATP retained by respective cultures.
Percent control represents activity in a fractionated culture sample compared to activity in an unfractionated, parallel culture.
Values represent LDH activity released by HeLa monolayers into the culture medium.
FIG. 8.
Accessibility of CopB2 to NHS-S-S-biotin in cells permeabilized with staphylococcal alpha-toxin. C. trachomatis-infected (L2) or mock-infected (M) HeLa cultures were treated with HBSS with (+) or without (−) alpha-toxin (α-Toxin), and susceptible proteins were labeled by addition of NHS-S-S-biotin. Material from whole-culture lysates (first lane M and first lane L2) or containing purified, biotinylated proteins from disrupted cultures was resolved in 12% (wt/vol) polyacrylamide gels and analyzed by immunoblotting using α-CopB2 or antibodies specific for bacterium-associated proteins MOMP, EfTu, and Hsp60 or extrainclusion-localized Chlamydia CADD. Proteins were visualized by probing with horseradish peroxidase-conjugated secondary antibodies, followed by chemiluminescent development with Super Signal substrate. All images are taken from equivalent exposures.
DISCUSSION
Although a basic understanding of the Chlamydia T3SS is beginning to emerge, the full complement of components contributing to this mechanism, including those with effector functions, remains to be identified. In the absence of a tractable genetic system, expression of putative T3S-associated proteins in heterologous T3SSs is one mechanism to identify chlamydial T3SS components. Some Chlamydia T3S components are sufficiently divergent that previous attempts to complement null mutations in heterologous T3SSs with corresponding Chlamydia genes were not successful (reference 23; K. A. Fields, unpublished data). However, the similarities of Chlamydia Scc2 and Scc3 with each other and with Yersinia SycD are especially intriguing. As a group, T3S chaperones are typically small (ca. 15 to 20 kDa), have an acidic pI, and contain a predicted C-terminally localized amphipathic alpha helix, yet they lack significant primary sequence similarity (reviewed in reference 9). Existence of multiple SycD homologs in a T3SS is unusual, and among all characterized T3SSs, only Chlamydia spp. and Bordetella spp. (26) exhibit this quality. A recently proposed classification (34) of T3S chaperones places SycD and its homologs in a distinct category due to their ability to serve as specific chaperones for translocator proteins and their direct involvement in regulation of T3SS gene expression. Both Scc2 and Scc3, like SycD and its homologs, possess a TPR domain. TPR domains represent structural motifs found in a variety of eukaryotic and prokaryotic proteins, where they are involved in protein-protein interactions (6). Homology modeling suggests that these domains in T3S chaperones enable a fundamentally unique interaction with binding partners and may contribute to the functional demarcation between “regulator” and “effector” chaperones (33).
By focusing on Chlamydia SycD homologs, we were afforded the opportunity to examine multiple functions in evaluating the capacity of Scc2 or Scc3 to complement sycD null Y. enterocolitica. SycD binds the translocator proteins YopB and YopD and is required for their intracellular stability and eventual secretion. SycD also contributes to repression of T3S gene expression directly (15) or in conjunction with YopD (2, 45). During in vitro growth, ΔsycD yersiniae display growth restriction and secretion of a subset of Yops regardless of Ca2+ levels (5). Expression of neither Scc2 nor Scc3 restored a Ca2+-dependent growth phenotype or Ca2+-regulated secretion of the effector protein YopE (Fig. 2). We chose to test effects on YopD to more closely examine potential Scc2- or Scc3-specific effects, since SycD interacts with discreet YopD domains (14) but associates with YopB more promiscuously (32) and possibly with lower affinity (14). Indeed, our coprecipitation analysis (Fig. 4) indicated that both Scc2 and Scc3 interact with YopD. The interaction between Scc3 and YopD seems to be functional, since secretion of YopD was detected in the presence of Scc3 (Fig. 3). Interestingly, secretion of YopD was observed only in cultures lacking Ca2+, raising the possibility of Scc3-dependent, Ca2+-regulated secretion. This possibility, however, is difficult to ascertain given the phenotype of ΔsycD yersiniae. Although Yops are expressed constitutively in ΔsycD strains and secretion of some proteins (YopH, LcrV, and YopK) occurs completely independently of Ca2+, secretion of other T3S substrates (YopM, YopN, and YpKA) remains regulated comparable to secretion in the wild type (15). We saw no effect of Scc3 on YopE secretion (Fig. 2B), and analysis of culture supernatants by Coomassie staining of proteins indicated that Scc3 did not alter the deregulated secretions of YopH, LcrV, and YopK (data not shown). Hence, the secretion effect was specific to YopD. We believe that, given the lack of a global effect on the regulation of Yop secretion, the presence of YopD in −Ca2+ culture supernatants most likely reflects only a secretion chaperone activity of Scc3. According to our current data, neither Scc2 nor Scc3 is able to complement the regulatory defects of ΔsycD yersiniae. Like SycD, Scc2 and Scc3 possess the ability to interact with T3S substrates and could therefore serve as chaperones.
We also observed a Scc3-dependent increase in levels of cell-associated YopD irrespective of Ca2+. Although we did not rule out an effect of Scc3-FT on yopD expression, it seems likely that, given the known role of SycD and our coprecipitation data, the observed effect is manifested through an interaction of Scc3 with YopD. We cannot definitively explain why trans expression of Scc3 resulted in higher levels of detectable YopD than in the presence of SycD. Immunoblots with Flag-specific antibodies show possibly higher absolute amounts of Scc3 than of SycD in this experiment. However, this effect on YopD levels was reproducible, and given that SycD is multifunctional and binds several T3S-associated proteins, it is possible that there is less SycD (than Scc3) available to stabilize YopD. If Scc3 is only capable of binding YopD, this could result in greater amounts of YopD. Scc2 clearly did not have this effect. YopD contains at least two SycD binding sites (14), and it is possible that that the inability of an Scc2 interaction to stabilize YopD reflects an association with a different domain than that associated with Scc3. Levels of coprecipitated protein did not differ significantly (Fig. 5), suggesting that lack of YopD stability and secretion is not due to a fundamentally “weaker” interaction with Scc2.
Scc2 and Scc3 are significantly divergent from each other and SycD such that it is probable that they contribute uniquely to the Chlamydia T3SS. It was therefore surprising that our data indicated that either Scc2 or Scc3 could interact with CopB in Chlamydia. However, scc2 is a late-cycle gene, whereas scc3 is first expressed during mid-cycle development of the L2 serovar (10). Moreover, microarray analyses indicate differences in the expression kinetics of scc2 and scc3 in C. trachomatis serovar D (4). It is possible that interactions of Scc2 or Scc3 with CopB occur at different times in the developmental cycle and have distinct effects. The N-terminal domains unique to Scc2 and Scc3 do not share significant sequence similarity and could potentially confer the capacity for additional interactions specific to the Chlamydia T3SS. The amphipathic alpha helix predicted in the N terminus of Scc3 supports this hypothesis. The inability of Scc2-FT and Scc3-FT to be secreted by the Yersinia T3SS and their immunolocalization in Chlamydia (Fig. 7) indicate that, like SycD, they function in the bacterial cytosol. However, the deduced Scc3 sequence contains a predicted membrane-interactive hydrophobic domain (residues 104 to 124), and biochemical fractionation indicates that a portion of Scc2 is associated with Chlamydia membranes (10), again raising the possibility of unique character. It should be noted, however, that the corresponding SycD homolog in enteropathogenic E. coli, CesD, is membrane associated (44).
We expressed truncated versions of CopB and CopB2 in Yersinia to avoid toxicity and solubility issues typical of this family of membrane-interactive proteins. Moreover, both full-length CopB and CopB2 contain a high proportion of atypical codons further complicating their characterization. Given the stability of our chimeric CopB-containing protein, we were unable to address potential contributions of Scc2 or Scc3 to CopB turnover. Slepenkin et al. (40) have reported that Scc3 has the capacity to interact with CopN, raising the possibility that a complex cascade of chaperone interactions contributes to a functional T3SS in chlamydiae. We are currently developing a system capable of addressing the functions of these chaperones as they pertain to identified substrates.
We were able to employ the affinities of Scc2 and Scc3 for their effectors in conjunction with expression of putative T3S substrates in the heterologous Yersinia T3SS to identify two new chlamydial type III effector proteins. CopB has low sequence similarity to the pore-forming translocator YopB family of proteins. The rim-like, inclusion membrane-localized distribution of CopB confirms that it is secreted by Chlamydia and is consistent with CopB functioning as an extended component of the T3S apparatus. We were surprised that neither Scc2 nor Scc3 was able to coprecipitate CopB2. Our data, however, indicate that CopB2 gains access to the host cell cytoplasm during a chlamydial infection and that, although this protein lacks similarity to YopB-like proteins, it does possess similar structural characteristics. Interestingly, the Salmonella SipB (21) and Shigella IpaB (8, 22, 43) translocator proteins additionally interact with and activate caspase-1 in eukaryotic cytoplasms. Since Chlamydia infection results in caspase-1 activation (28), it is tempting to speculate that CopB2 may have a role in this process.
Identification of novel T3S substrates is of particular importance and has the capability to significantly advance our understanding of chlamydial pathogenesis. The C. trachomatis genome contains at least seven predicted T3S chaperones (42), and our success in the identification of CopB and CopB2 through investigation of Scc2 and Scc3 demonstrates the utility of studying these proteins. Moreover, the biochemical approach using staphylococcal alpha-toxin permeabilization provides a sensitive means to determine exposure of Chlamydia proteins to the host cytoplasm. Future experiments will exploit these advances to identify additional host-interactive Chlamydia proteins.
Acknowledgments
We thank O. Schneewind (University of Michigan) and H. Wolf-Watz (University of Umea) for kindly providing Y. enterocolitica and Y. pseudotuberculosis strains, respectively. We also thank G. Plano (University of Miami) for the kind gift of α-YopD and α-YopE and G. Munson and Katerina Wolf for critical reading of the manuscript.
REFERENCES
- 1.Anderson, D. M., D. E. Fouts, A. Collmer, and O. Schneewind. 1999. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc. Natl. Acad. Sci. USA 96:12839-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. M., K. S. Ramamurthi, C. Tam, and O. Schneewind. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind yopQ RNA. J. Bacteriol. 184:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman, T., S. Håkansson, Å. Forsberg, L. Norlander, A. Macellaro, A. Bäckman, I. Bölin, and H. Wolf-Watz. 1991. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J. Bacteriol. 173:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bergman, T., K. Erickson, E. Galyov, C. Persson, and H. Wolf-Watz. 1994. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J. Bacteriol. 176:2619-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blatch, G., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 6a.Bolin, I., and H. Wolf-Watz. 1984. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 43:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., M. Smith, K. Thirumalai, and A. Zychlinksy. 1996. A bacterial invasin induces macrophage apoptosis by directly binding ICE. EMBO J. 15:3853-3860. [PMC free article] [PubMed] [Google Scholar]
- 8a.Cornelis, G. R., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by the recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87:285-291. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, M. F., and G. R. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151-158. [DOI] [PubMed] [Google Scholar]
- 10.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671-683. [DOI] [PubMed] [Google Scholar]
- 11.Fields, K. A., and T. Hackstadt. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 14:14. [DOI] [PubMed] [Google Scholar]
- 12.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048-1060. [DOI] [PubMed] [Google Scholar]
- 13.Fields, K. A., A. W. Williams, and S. C. Straley. 1997. Failure to detect binding of LcrH to the V antigen of Yersinia pestis. Infect. Immun. 65:3954-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, M. S., M. Aili, M. Wiklund, and H. Wolf-Watz. 2000. A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38:85-102. [DOI] [PubMed] [Google Scholar]
- 15.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 16.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 17.Ginocchio, C., and J. Galan. 1995. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect. Immun. 63:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 19.Hakansson, S., T. Bergman, J. Vanooteghem, G. Cornelis, and H. Wolf-Watz. 1993. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect. Immun. 61:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ham, J. H., D. W. Bauer, D. E. Fouts, and A. Collmer. 1998. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc. Natl. Acad. Sci. USA 95:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hersh, D., D. Monack, M. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilbi, H., J. Moss, D. Hersch, Y. Chen, A. Banerjee, R. Flavell, J. Yuan, P. Sansonetti, and A. Zychlinsky. 1998. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J. Biol. Chem. 273:32895-32900. [DOI] [PubMed] [Google Scholar]
- 23.Hsia, R. C., Y. Pannekoek, E. Ingerowski, and P. M. Bavoil. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25:351-359. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 26.Kerr, J. R., G. P. Rigg, R. C. Matthews, and J. P. Burnie. 1999. The Bpel locus encodes type III secretion machinery in Bordetella pertussis. Microb. Pathog. 27:349-367. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27a.Lee, V. T., and O. Schneewind. 1999. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31:1619-1629. [DOI] [PubMed] [Google Scholar]
- 28.Lu, H., C. Shen, and R. C. Brunham. 2000. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J. Immunol. 165:1463-1469. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.McGuffin, L., K. Bryson, and D. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 31.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neyt, C., and G. R. Cornelis. 1999. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 31:143-156. [DOI] [PubMed] [Google Scholar]
- 33.Pallen, M. J., M. S. Francis, and K. Futterer. 2003. Tetratricopeptide-like repeats in type III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53-60. [DOI] [PubMed] [Google Scholar]
- 34.Parsot, C., C. Hamiaux, and A. Page. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7-14. [DOI] [PubMed] [Google Scholar]
- 35.Perry, R. D., M. Pendrak, and P. Schuetz. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosqvist, R., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1995. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 14:4187-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schachter, J. 1988. Overview of human diseases, p. 153-165. In A. L. Barron (ed.), Microbiology of Chlamydia. CRC Press, Inc., Boca Raton, Fla.
- 39.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753-765. [DOI] [PubMed] [Google Scholar]
- 40.Slepenkin, A., L. M. de la Maza, and E. M. Peterson. 2005. Interaction between components of the type III secretion system of Chlamydiaceae. J. Bacteriol. 187:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenner-Liewen, F., H. Liewen, J. Zapata, K. Pawlowski, A. Godzik, and J. Reed. 2002. CADD, a Chlamydia protein that interacts with death receptors. J. Biol. Chem. 277:9633-9636. [DOI] [PubMed] [Google Scholar]
- 42.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 43.Thirumalai, K., K.-S. Kim, and A. Zychlinsky. 1997. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect. Immun. 65:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wainwright, L. A., and J. B. Kaper. 1998. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol. Microbiol. 27:1247-1260. [DOI] [PubMed] [Google Scholar]
- 45.Williams, A. W., and S. C. Straley. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180:350-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf, K., E. Fischer, D. Mead, G. Zhong, R. Peeling, B. Whitmire, and H. D. Caldwell. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69:3082-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, Y., K. Lyng, Y. X. Zhang, D. D. Rockey, and R. P. Morrison. 1992. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect. Immun. 60:2288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y.-X., Y. Shi, M. Zhou, and G. A. Petsko. 1994. Cloning, sequencing, and expression in Escherichia coli of the gene encoding a 45-kilodalton protein, elongation factor Tu, from Chlamydia trachomatis serovar F. J. Bacteriol. 176:1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]