Abstract
The universal stress protein (UspA) superfamily encompasses a conserved group of proteins that are found in bacteria, archaea, and eukaryotes. Escherichia coli harbors six usp genes—uspA, -C, -D, -E, -F, and -G—the expression of which is triggered by a large variety of environmental insults. The uspA gene is important for survival during cellular growth arrest, but the exact physiological role of the Usp proteins is not known. In this work we have performed phenotypic characterization of mutants with deletions of the six different usp genes. We report on hitherto unknown functions of these genes linked to motility, adhesion, and oxidative stress resistance, and we show that usp functions are both overlapping and distinct. Both UspA and UspD are required in the defense against superoxide-generating agents, and UspD appears also important in controlling intracellular levels of iron. In contrast, UspC is not involved in stress resistance or iron metabolism but is essential, like UspE, for cellular motility. Electron microscopy demonstrates that uspC and uspE mutants are devoid of flagella. In addition, the function of the uspC and uspE genes is linked to cell adhesion, measured as FimH-mediated agglutination of yeast cells. While the UspC and UspE proteins promote motility at the expense of adhesion, the UspF and UspG proteins exhibit the exact opposite effects. We suggest that the Usp proteins have evolved different physiological functions that reprogram the cell towards defense and escape during cellular stress.
The levels of the universal stress protein UspA of Escherichia coli become elevated in response to a large variety of stress conditions, including starvation for carbon, nitrogen, phosphate, sulfate, and amino acids and exposure to heat, oxidants, metals, uncouplers of the electron transport chain, polymyxin, cycloserine, ethanol, and antibiotics (9, 13, 17; T. Nyström, unpublished data). The protein has given its name to the orthologous group of proteins called the UspA superfamily of proteins (the COG0589 cluster in GenBank and the PF00582 protein family of alignments at the Sanger Centre). This superfamily encompasses an ancient group of proteins that are found in all the major branches of the evolutionary tree (1). Usp-containing organisms are usually equipped with several usp genes despite the fact that they exhibit extensive similarity. They encode either a small Usp protein (around 14 to 15 kDa) harboring one Usp domain, a larger version (around 30 kDa) consisting of two Usp domains in tandem, or large proteins in which the Usp domain is present together with other functional domains (13). E. coli has five small Usp proteins and one tandem-type Usp protein (UspE). Based on structure analysis and their amino acid sequence, the Usp proteins have been divided into four different classes. In E. coli, UspA, UspC (yecG), and UspD (yiiT) belong to class I, UspF (ynaF) and UspG (ybdQ) belong to class II, and the two Usp domains of UspE (ydaA) separate into classes III and IV (13, 23, 26; L. Brive, personal communication).
The Usp paradigm, UspA, is an autophosphorylating serine and threonine phosphoprotein (6). Phosphorylation occurs in response to stasis and is, in part, dependent on the tyrosine phosphoprotein TypA (7). Mutants devoid of UspA die prematurely during stasis (17), whereas overproduction blocks the cells in a growth-arrested state (18). In addition, deletion of class I or class III/IV usp genes results in sensitivity to DNA-damaging agents (9). In contrast, cells lacking the class II UspG (called UP12 in reference 2) are sensitive to uncouplers (e.g., carbonyl cyanide m-chlorophenylhydrazone) but not DNA-damaging agents (2), indicating that the functions of the class I and II Usps may be distinct. To approach this notion we created mutants of different classes, i.e., the ΔuspA ΔuspC ΔuspD triple mutant (class I), the ΔuspF ΔuspG double mutant (class II), and the ΔuspE mutant (class III/IV). Phenotypic analysis of these mutants revealed hitherto unknown Usp functions, including those related to iron scavenging, oxidative stress defense, cell adhesion, and motility. Moreover, the analysis of class I to IV and individual usp mutants demonstrates that functions may overlap between Usp classes and also that the function of Usps within a class is distinct.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All strains used in this study are listed in Table 1. Strains were cultivated at 37°C in Luria-Bertani (LB) medium. When required, the medium was supplemented with antibiotics at the following concentrations: ampicillin (50 μg ml−1), kanamycin (50 μg ml−1), spectinomycin (100 μg ml−1), and chloramphenicol (30 μg ml−1). The strains LN81, LN32, and LN82 were obtained by P1 transduction of the deletion mutation from the strains AD546, NGN183, and AD547, respectively.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| MG1655 | Wild type | Lab collection |
| KM354 | recJ | 15 |
| AD546 | MC4100 ΔuspC::aadA (Spcr) | 9 |
| NGN183 | MC4100 ΔuspD::cat (Camr) | 9 |
| AD547 | MC4100 ΔuspE::aadA (Spcr) | 9 |
| LN23 | KM354 ΔuspA::aphA-3 (Kanr) | This study |
| LN24 | KM354 ΔuspF::aphA-3 (Kanr) | This study |
| LN25 | KM354 ΔuspG::cat (Camr) | This study |
| LN29 | MG1655 ΔuspA::aphA-3 (Kanr) | This study |
| LN81 | MG1655 ΔuspC::aadA (Spcr) | This study |
| LN32 | MG1655 ΔuspD::cat (Camr) | This study |
| LN82 | MG1655 ΔuspE::aadA (Spcr) | This study |
| LN30 | MG1655 ΔuspF::aphA-3 (Kanr) | This study |
| LN31 | MG1655 ΔuspG::cat (Camr) | This study |
| LN86 | MG1655 ΔuspA::aphA-3 (Kanr) ΔuspC::aadA (Spcr) ΔuspD::cat (Camr) | This study |
| LN87 | MG1655 ΔuspF::aphA-3 (Kanr) ΔuspG::cat (Camr) | This study |
| Plasmids | ||
| pUC18 | Cloning vector; Ampr | |
| pUC18K | Carries the aphA-3 gene conferring kanamycin resistance | 14 |
| pTP223 | Expresses the λ recombination function (exo, bet, gam) under control of Plac | 19 |
| pNGN27 | pBluescript carrying the cat gene conferring chloramphenicol resistance | 9 |
| p5A | pUC18 carrying the 5′-flanking region of uspA | This study |
| p5-3A | pUC18 carrying the 5′- and 3′-flanking regions of uspA | This study |
| pΔA | pUC18 ΔuspA::aphA-3 (Kanr) | This study |
| p5F | pUC18 carrying the 5′-flanking region of uspF | This study |
| p5-3F | pUC18 carrying the 5′- and 3′-flanking regions of uspF | This study |
| pΔF | pUC18 ΔuspF::aphA-3 (Kanr) | This study |
| p5G | pUC18 carrying the 5′-flanking region of uspG | This study |
| p5-3G | pUC18 carrying the 5′- and 3′-flanking region of uspG | This study |
| pΔG | pUC18 ΔuspG::cat (Camr) | This study |
Construction of the uspA, uspF, and uspG deletion mutants.
The chromosomal deletion mutations were created stepwise. The 5′- and 3′-flanking regions of uspA, -F, and -G were PCR amplified using the 5F/5R and 3F/3R primers, respectively (Table 2). First, the 5′ regions were cloned into the pUC18 plasmid, generating the p5 plasmid series (Table 1). In a second step, the 3′-flanking regions were cloned into the p5 plasmids, generating the p5-3 plasmid series (Table 1). Third, the aphA-3 gene (conferring kanamycin resistance [Kanr]) and the cat gene (conferring chloramphenicol resistance [Camr]) were cut out from plasmids pUC18K and pNGN27, respectively, and ligated into the SmaI site of the p5-3 plasmids, thus creating pΔA (Kanr), pΔF (Kanr), and pΔG (Camr). The plasmids were purified, and the cloned deletion constructs were excised and introduced into the E. coli strain KM354 by linear transformation. This strain contains the plasmid pTP223 to promote gene replacement by recombination (16). The chromosomal deletion mutations were then moved from the KM354 to the MG1655 background by transduction using the phage P1.
TABLE 2.
Primers used in this study
| Namea | 5′-3′ sequenceb |
|---|---|
| uspA5F | gcatcGAATTCEctaccgctcccgatacgc |
| uspA5R | tccCCCGGGSagtgttactccttccataaag |
| uspA3F | tccCCCGGGStcttccctctacgacgtgttcc |
| uspA3R | acgcGTCGACSlctcttgcgagaatccaagc |
| uspF5F | gcatcGAATTCEatgaccccgtttggcgacag |
| uspF5R | tccCCCGGGSaaaacctcctgttttagtatcc |
| uspF3F | tccCCCGGGScactaacgcccgcacattgc |
| uspF3R | acgcGTCGACSlaccgcagtcacgtcgtatgc |
| uspG5F | cagcGAGCTCScccactcggtcgcctttttgc |
| uspG5R | tccCCCGGGSaaccctttctccctgtt |
| uspG3F | tccCCCGGGSttgccagaataagtatcccgcc |
| uspG3R | aaCTGCAGPgacggcagcaaagtcgttgg |
F, forward primer; R, reverse primer.
Restriction sites used in the different steps of the cloning procedures are indicated in capital letters. The superscripts in the primer sequences indicate the restriction enzyme recognizing the site. E, EcoR1; S, SmaI; Sl, SalI; Sc, SacI; P, PstI.
Growth assays.
Overnight culture of E. coli wild-type and usp mutant strains were used to inoculate fresh LB medium. During the exponential growth phase (optical density at 600 nm [OD600], 0.5) the precultures were diluted 10 times and split into two cultures of equal volume. To one, phenazine methosulfate (PMS; 15 μM) or streptonigrin (1 μg ml−1) was added. Growth was monitored by measuring OD600.
t-BOOH sensitivity.
Aliquots of overnight cultures grown in LB medium were incubated with different concentrations of t-BOOH (tert-butyl hydroperoxide) for 1 h at 37°C without shaking. The viability of the cells after exposure was determined as the number of CFU ml−1 on LB plates.
Autoaggregation assay.
Overnight cultures of the E. coli strains were adjusted to approximately the same OD600 (4.0), and 4 ml of each was placed in a sterile 15-ml tube at room temperature. At the beginning of each experiment, all cultures were vigorously shaken. Cell settlings were visually observed. Samples were also taken for observation using standard phase-contrast microscopy.
Ag43 immunodetection.
Cells grown overnight in LB were tested for the presence of the protein Ag43. For each culture, the equivalent of 0.2 OD600 units was analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis, followed by immunodetection using a 1:3,000 dilution of polyclonal rabbit antiserum raised against the α domain of Ag43 and a secondary goat anti-rabbit antibody conjugated to alkaline phosphatase.
Yeast cell agglutination assay.
Saccharomyces cerevisiae and E. coli cells from overnight culture were pelleted, washed, and resuspended in phosphate-buffered saline to OD600 of 5 and 3, respectively. Equal volumes of E. coli and yeast cell suspensions were mixed on a glass slide. Aggregation was monitored visually, and the titer was recorded as the last dilution of bacteria giving a positive agglutination reaction. To control that the agglutination observed was indeed due to the adhesion of bacterial fimbriae to mannose residues of the yeast cell surface, we also performed the experiments in the presence of d-mannose (25 mg ml−1). In this condition, fimbria-dependent agglutination is abolished. All assays were performed at least three times and gave similar results.
Motility assay.
Bacterial cells were picked from colonies grown on LB agar plates and inoculated onto low-agar-concentration (0.3%) plates. The plates were incubated at room temperature for 48 h, and motility was assessed qualitatively by examining the circular halo formed by the growing motile bacterial cells.
Scanning electron microscopy (SEM).
Strains were grown overnight in LB medium at 37°C without shaking. From these cultures, 30 μl was taken, and cells were allowed to sediment and adhere to Au sputtered Thermanox plates for 15 min. The plates were then transferred to 2.5% glutaraldehyde for 15 min, rinsed with Na-cacodylate buffer, and postfixed in 1% OsO4 (15 min). Specimens were then dehydrated in graded series of ethanol and dried with hexamethyldisilazane for 2 × 5 min. All specimens were then sputter coated with Palladium and examined under a Zeiss DSM 982 Gemini scanning electron microscope.
RESULTS
The class I proteins UspA and UspD and class II proteins UspF and UspG confer resistance against oxidative stress.
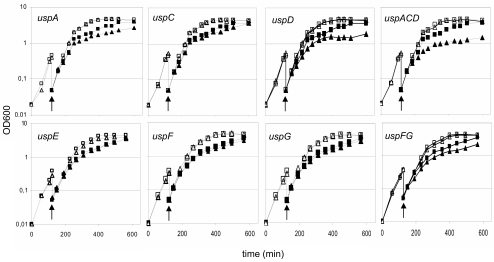
Production of UspA has been shown to drastically increase when exponentially growing E. coli cells are challenged with hydrogen peroxide (H2O2) (9, 17). Moreover, the viability of growing uspA mutant cells was found to be markedly more reduced than that of the wild-type strain after H2O2 exposure (17). Thus, we assayed the sensitivity of the different usp mutants toward the superoxide-generating agent, PMS, added during exponential growth of cells. Cells lacking all class I (uspACD) genes were drastically more sensitive than the wild-type strain to such oxidative stress (Fig. 1). When analyzing the single-mutant strains of this class, it appeared that the sensitivity of the triple mutant was due to the additive effect of the two deletions of uspA and uspD; both single mutants were more sensitive than the wild-type strain but less so than the triple mutant (Fig. 1). In contrast, the lack of uspC did not affect the sensitivity of cells towards PMS. The class II genes, uspF and uspG, seemed to play a minor role in oxidative stress resistance. Cells lacking both these genes displayed a slightly increased sensitivity toward PMS. The sensitivity of this double mutant appears to be due to the simultaneous absence of uspF and uspG since the single mutants exhibited no significant sensitivity to PMS. The class III/IV mutant, uspE, was indistinguishable from the wild-type strain with respect to PMS sensitivity.
FIG. 1.
Effect of oxidative stress on growth of usp mutants. Fresh LB medium was inoculated with overnight culture of the different strains. In exponential phase, the precultures were diluted and split in two cultures, one of which was treated with 15 μM PMS. Open symbols, growth without PMS; filled symbols, growth in the presence of PMS; squares, wild-type strain MG1655; triangles, usp mutant strain. The mutant tested is shown on each panel, and arrows indicate the time of PMS addition. All experiments were performed at least three times to assess reproducibility. The figure presents the results of one typical experiment.
The class I protein UspD and class II proteins UspF and UspG confer resistance against streptonigrin.
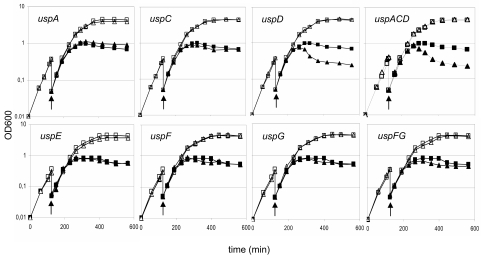
In many cases oxidative stress and iron homoeostasis are intimately linked. The iron exacerbates the effects of the reactive oxygen species through the Fenton reaction producing hydroxyl radicals in the presence of oxygen (11). Overproduction of the UspA protein has been demonstrated to reduce the synthesis of iron siderophore uptake proteins (18). Thus, it is possible that the lack of Usp proteins could cause a decontrolled influx of iron, leading to an excess in the cell. We tested this hypothesis by analyzing the effects of streptonigrin on cell growth. Streptonigrin is an antibiotic whose toxicity is dependent on intracellular iron concentrations; increased sensitivity to this drug is a sign of increased availability of intracellular free iron (25). Based on PMS sensitivity data, we expected the class I uspACD and class II uspFG mutants to be more sensitive to streptonigrin. This was indeed the case, and the class III/IV uspE mutant, again, showed no sensitivity (Fig. 2). We also found a good correlation between the degree of streptonigrin and PMS sensitivity: in both cases the lack of UspFG is less dramatic than the lack of class I Usps. When testing the single deletion mutants we observed, again, an additive effect of the lack of UspF and UspG (Fig. 2). As expected, the lack of the class I protein UspC did not affect sensitivity towards streptonigrin. However, somewhat surprisingly, the lack of only uspD and not uspA resulted in an increased streptonigrin sensitivity (Fig. 2). Thus, the sensitivity of uspD and uspFG but not uspA mutants to oxidative agents is associated with streptonigrin sensitivity, i.e., increased intracellular iron availability.
FIG. 2.
Effect of streptonigrin on growth of usp mutants. Fresh LB medium was inoculated with overnight culture of the different strains. In exponential phase, the precultures were diluted and split in two cultures, one of which was treated with streptonigrin (1 μg ml−1). Open symbols, growth without streptonigrin; filled symbols, growth in the presence of streptonigrin; squares, wild-type strain MG1655; triangles, usp mutant strain. The mutant tested is shown on each panel, and arrows indicate the time of streptonigrin addition. All experiments were performed at least three times to assess reproducibility. The figure presents the results of one typical experiment.
Deficiencies in the class I protein UspC, the class II protein UspG, and the class III/IV protein UspE affect cell-to-cell aggregation.
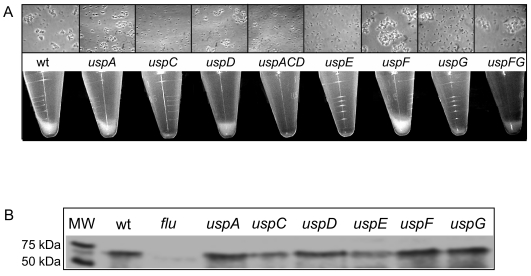
In the course of characterizing the E. coli usp mutants, we made the serendipitous observation that uspE mutant cells failed to sediment in liquid medium after letting the culture stand still for hours. A similar phenotype has been described previously for Azospirillum brasilense carrying a mutation in a usp-like gene (8). Microscopic observations of overnight cultures of the different E. coli usp mutants revealed differences in the strains' capacities to form cell aggregates (Fig. 3A, upper panel). In parallel, the cell settlings of the usp mutants were assayed by observing the formation of a cell pellet in the overnight cultures (Fig. 3A, lower panel). The uspE mutant was completely unable to aggregate, and only single cells were observed in all the uspE cultures assayed. Also, even after prolonged incubation (several hours) no cell pellet was visible in the uspE culture. The uspC, uspACD, and uspG mutants were also impaired in their ability to form aggregates, but a few could be observed, albeit smaller than those formed by the wild-type cells. After 1 h, the cells of these mutants had not settled, but they eventually did so after prolonged static incubation (Fig. 3A and data not shown). Ectopic expression of the uspC gene complemented the aggregation defect of the uspC mutant (data not shown). No other usp gene significantly affected cell flocculation (Fig. 3A).
FIG. 3.
Effect of usp deletions on cellular autoaggregation. (A) Overnight cultures were vigorously shaken and subsequently incubated statically at room temperature. Upper panel: an aliquot of each culture was taken at the beginning of the assay for microscopic observation. Images were captured with a magnification of ×1,000. Lower panel: cell settling state of each strain after 1 h. (B) Ag43 production. For each strain, the equivalent of 0.2 OD600 unit of an overnight culture was analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and immunodetection using a polyclonal rabbit antiserum raised against the α domain of Ag43.
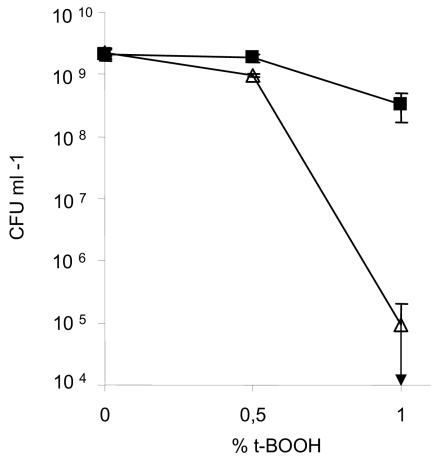
In E. coli, cell-to-cell aggregation (as described above) can be mediated by the outer membrane protein Ag43 encoded by the flu gene. Using antibody raised against the α domain of Ag43, we could detect this protein in the wild type and all the single usp mutants (Fig. 3B). However, we noticed that Ag43 levels were lower in the mutants lacking uspE or uspC (Fig. 3B). Thus, the failure of the uspE and uspC mutants to aggregate can, in part, be explained by a lower production of Ag43. An E. coli flu mutant is devoid of the capacity to aggregate and has also been shown to be more sensitive to H2O2 killing (5, 21). Therefore, we decided to test the capacity of the uspE mutant, which is totally unable to aggregate in stationary phase, to cope with lethal doses of oxidative stress, and we determined the rate of die-off during such conditions. Overnight cultures were challenged with t-BOOH, and the number of CFU was determined after exposure to the oxidant (Fig. 4). After exposure to 0.5% t-BOOH the viability of the mutant strain was significantly reduced compared to that of the wild-type strain. An even more drastic difference was obtained when the cells were exposed to 1% t-BOOH: only 0.003% of the uspE cells survived compared to 15% of the wild-type cells (Fig. 4). When we assayed the sensitivity of the uspE mutant toward PMS during growth, we did not observe any difference compared to the wild-type strain (Fig. 1). We wondered then if this was due to a specific sensitivity of the uspE mutant to peroxide agents (t-BOOH) rather than superoxide-generating agents (PMS). Therefore, we challenged the uspE mutant with t-BOOH during growth, the same way we did with PMS, but could not see any significant sensitivity (data not shown). Thus, we detected an oxidative stress sensitivity of the uspE mutant only when growth-arrested cells were exposed to lethal doses of t-BOOH.
FIG. 4.
Survival of stationary-phase cells after challenge with the oxidative agent t-BOOH. Cells from overnight culture were incubated for 1 h at 37°C with different concentrations of t-BOOH. The survival is expressed as the number of CFU per ml on the LB plate after t-BOOH exposure. Symbols: ▪, wild-type strain MG1655; ▵, LN82 (uspE). Each point represents the average value calculated from the results of at least three experiments, and error bars indicate the calculated standard deviation.
Fimbria-mediated adhesion is increased by the class II UspF and UspG proteins and decreased by the class I UspC and class III/IV UspE proteins.
The Ag43-dependent aggregation of E. coli has been shown to be antagonized by the presence of type 1 fimbriae on the bacterial cell surface (10). Thus, we tested the fimbriation of the usp mutants by their ability to agglutinate yeast cells, a fimbria-dependent process (the FimH subunit binds to mannose residues on the yeast cell surface [12]). To confirm that the formation of yeast aggregates was indeed due to the presence of bacterial fimbriae, we verified that the agglutination observed was abolished in the presence of d-mannose (see Materials and Methods) (data not shown). We found that the E. coli cells' efficiency of yeast cell agglutination was markedly enhanced when either uspC or uspE was inactivated (Table 3), consistent with the fact that cell-cell aggregation was markedly reduced in these mutants (Fig. 3A). On the other hand, although the double mutant uspFG exhibits the same ability as the wild-type strain to agglutinate yeast, cells lacking one or the other of these two class II Usps were greatly impaired (Table 3). Thus, the single deletion of uspF or uspG displayed a negative dominant effect on fimbria-dependent adhesion.
TABLE 3.
Yeast agglutination capacity of usp mutants
| Titer | Wild type | Strain containing mutation:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| uspA | uspC | uspD | uspACD | uspE | uspF | uspG | uspFG | ||
| Agglutinationa | 1/4 | 1/4 | 1/10 | 1/5 | 1/10 | 1/10 | 1/2 | 1 | 1/4 |
The agglutination titer refers to the highest dilution of bacteria giving a positive yeast agglutination reaction. A value of 1 corresponds to a nondiluted bacterial suspension.
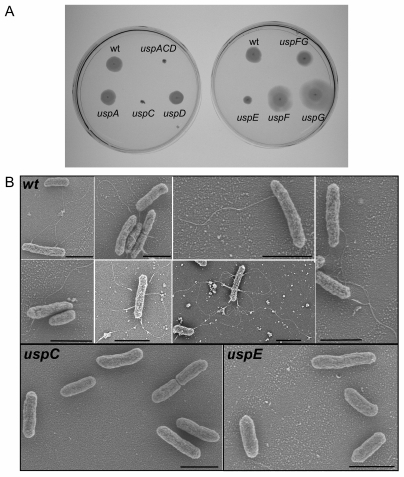
Inactivation of the class I uspC or the class III/IV uspE inhibits motility.
Adhesion and motility represent two dynamic aspects of the bacterial life cycle. These two fundamental properties have been found to exhibit reciprocal regulation to avoid simultaneous expression of counterproductive functions/organelles, i.e., adhesion/fimbriae and motility/flagella (4, 24). Such coordinated regulation may occur through two-component systems (22), direct cross talk between fimbrial gene products and bacterial motility genes (4), and small signaling molecules, such as acetyl phosphate (22, 24). We wondered whether the increased ability of uspC and uspE mutants to perform fimbria-dependent adhesion was associated with a decreased motility and whether the opposite was true for the uspF and uspG mutants. This was indeed the case. As seen in Fig. 5A, uspE mutant cells are severely restricted in their ability to swim and the uspC mutant (as well as the triple uspACD mutant) is completely nonmotile, whereas the uspF and uspG mutants displayed an enhanced swimming capacity. Ectopic expression of uspC caused an enhanced motility exceeding that of the wild-type cells (data not shown). The uspA and uspD mutants displayed no apparent defects in swimming ability (Fig. 5A).
FIG. 5.
(A) Effect of usp deletions on cell motility. Low-agar-concentration (0.3%) plates were inoculated with each strain as indicated. The halo of growth was observed after 48 h incubation at room temperature. (B) Detection of flagella by scanning electron microscopy. The observed strain is indicated in each panel. The black bar on each panel represents 2 μm.
The nonmotile phenotype of uspC and uspE mutants could be a consequence of a failure in making flagella or an inability to properly use these organelles. SEM revealed the first possibility to be true (Fig. 5B). Quantitative analysis of SEM data demonstrated that, while 4% of the wild-type cells had visible flagella, under the conditions used, no uspC cell (in 10,000) and only one uspE cell (in 10,000) displayed flagella. Although the uspF and uspG mutants displayed an enhanced motility we did not observe an increased number of flagella on the cells by SEM (data not shown).
DISCUSSION
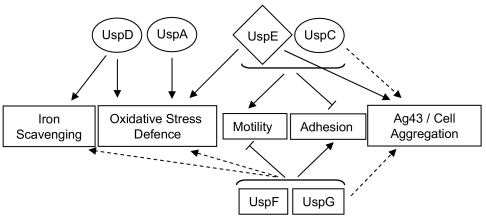
The large number of usp genes in many bacteria and the coordinated induction of usp genes upon cellular stress (9) has previously appeared somewhat redundant in view of the sequence similarity of Usp proteins, especially within a class (13). However, the phenotypes of the different usp mutants reported here provide new insights into the role of universal stress proteins in the life cycle of bacteria. It is now clear that the function of Usps in E. coli is multifaceted (Fig. 6). Class I UspA and UspD appear involved in regulating the cells' capacity to withstand oxidative agents and, in the case of UspD, intracellular iron availability. The functions of other Usps, i.e., class I UspC and class II UspF and UspG, are instead mainly related to adhesion and motility. In addition, the role of the unique Usp protein of class III/IV, UspE, overlaps the functions of all the other classes (Fig. 6).
FIG. 6.
Role of the 6 E. coli Usps in oxidative stress defense, iron metabolism, and cell surface properties. The name of each Usp is surrounded by a shape symbolizing the class it belongs to: a circle for class I, a square for class II, and a diamond for class III and IV. An arrow represents a positive effect of the Usp protein in a specific function, whereas a T shape signifies a negative effect. Major and minor effects of the Usps in the different functions are represented by solid and dashed lines, respectively. The brackets indicate that both of the included proteins are involved in the indicated process. For example, UspC and UspE both affect motility positively and adhesion negatively.
With respect to oxidative stress resistance, UspA has previously been shown to be important for H2O2 resistance during growth (17). Here, we tested the resistance of all usp mutants of E. coli to oxidative agent exposure during growth. The results suggest a major role for the class I proteins, UspA and UspD, in the PMS (superoxide) resistance of growing cells (Fig. 1). The contribution of UspD to PMS resistance may be associated with a role in iron scavenging, i.e., the effect of reactive oxygen species in the uspD mutant could be exacerbated by an excess of free intracellular iron, as indicated by the elevated sensitivity towards streptonigrin (Fig. 2). The PMS sensitivity of the uspA mutant, in contrast, was not associated with elevated sensitivity to streptonigrin. Thus, the functions of UspA and UspD in PMS resistance are probably not identical or entirely overlapping. This notion is supported also by the fact that the uspA and uspD mutants acted in an additive manner (Fig. 1). A minor role in oxidative stress resistance can be attributed to the class II proteins UspF and UspG (Fig. 1). Like for uspD, the minor sensitivity of the double mutant uspFG was associated with signs of elevated levels of free iron in the cell (streptonigrin sensitivity) (Fig. 2). In addition, UspE appears to play a crucial role in oxidative stress resistance, but this role was restricted to defense against high concentrations of oxidative agent (Fig. 4). The exact function of the tandem UspE protein is not known, but one interesting and unique phenotype of the uspE mutant is its total inability to form cell-cell interactions and cell aggregates in stationary phase (Fig. 3A). The outer membrane protein Ag43 has been identified as responsible for cell aggregation in E. coli, and such aggregation had been shown to confer protection against H2O2 killing (5, 21). This has been mechanistically explained by the fact that cells in direct contact with the oxidative agent will be more affected than cells located inside an aggregate. Thus, the elevated sensitivity of the uspE mutant to oxidative agents might be related to its failure to form aggregates under these conditions. The aggregation defect of the uspE mutant could be linked to a lower production of the Ag43 protein (Fig. 3B) and an enhanced capacity of adhesion promoted by the FimH protein (Table 3); indeed, the Ag43-mediated aggregation of E. coli cells has previously been shown to be blocked by type 1 fimbriation (10).
Despite the fact that the UspC protein clearly belongs to the class I subfamily of Usp proteins based on sequence analysis, it is functionally distinct from its family members, UspA and UspD. In fact, the function of UspC appears closer to that of the class III/IV protein UspE (Fig. 6). For example, both uspC and uspE mutants are nonmotile due to the lack of flagella (Fig. 5); the residual motility of the uspE mutant could be due to the few cells possessing flagella (1 out of 10,000) or may be accomplished by a sliding phenomenon, which is linked to lipopeptide, lipopolysaccharide, and glycolipid production (3). In addition, both the uspC and uspE mutants display an enhanced capacity for adhesion, linked to the presence of fimbriae (Table 3). Intriguingly, the class II proteins, UspF and UspG, appear to have the exact opposite effects, i.e., both deletion mutations enhanced motility (Fig. 5A) and resulted in poor fimbriation (yeast agglutination data are shown in Table 3). Using DNA microarray analysis, the expression of uspG was reported as being induced 21 times in a fimH mutant (20). In the same study, the authors observed only a twofold induction of uspG expression when using a mutant deleted of the entire fim operon. FimH is the protein responsible for the initiation of the fimbria formation. In a fimH mutant, the major fimbrial subunit protein FimA accumulates in the periplasm, but this will, of course, not occur when the entire fim operon is deleted. Thus, it is possible that the gene encoding UspG (and perhaps also the gene encoding UspF) responds to the accumulation of surface-destined proteins in the periplasm and that this class of Usps is involved in the export of such proteins.
In light of the data presented in this paper, the coordinated induction of Usps can now be partly explained: elevated levels of Usp proteins during a shift from feast to famine conditions are expected to enhance the cell's capacity to withstand different stresses and modulate activities related to motility and adhesion. Thus, the Usps are reprogramming the cell towards defense and escape. Notably, the functions provided by the Usp proteins, oxidative stress defense, iron homeostasis, and motility/adhesion, are known to be essential in bacterial pathogenesis, and it would therefore not be surprising if usp mutants of different pathogens displayed a reduced virulence. However, we suggest that creating cells lacking the whole complement of their Usp repertoire may be the only means of revealing the true importance of Usp proteins in the bacterial life cycle and virulence.
Acknowledgments
This work was sponsored by grants from the Swedish Natural Research Council, the Inga-Britt and Arne Lundberg Research Foundation, and an award from the Göran Gustafsson Foundation for Scientific Research in Molecular Biology.
We thank all members, past and present, of the Thomas Nyström group for fruitful discussions, Laurent Loiseau for providing plasmids, Peter Owen for antibodies against Ag43, and Karen Otto for help with the yeast agglutination assays.
REFERENCES
- 1.Aravind, L., V. Anantharaman, and E. V. Koonin. 2002. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins 48:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Bochkavera, E. S., A. S. Girshovich, and E. Bibi. 2002. Identification and characterization of the Escherichia coli stress protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 269:3032-3040. [DOI] [PubMed] [Google Scholar]
- 3.Brown, I. I., and C. C. Hase. 2001. Flagellum-independent surface migration of Vibrio cholerae and Escherichia coli. J. Bacteriol. 183:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freestone, P., T. Nystrom, M. Trinei, and V. Norris. 1997. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274:318-324. [DOI] [PubMed] [Google Scholar]
- 7.Freestone, P., M. Trinei, S. C. Clarke, T. Nyström, and V. Norris. 1998. Tyrosine phosphorylation in Escherichia coli. J. Mol. Biol. 279:1045-1051. [DOI] [PubMed] [Google Scholar]
- 8.Galindo-Blaha, C. A., and I. S. Schrank. 2003. An Azospirillum brasilense Tn5 mutant with modified stress response and impaired in flocculation. Antonie Leeuwenhoek 83:35-43. [DOI] [PubMed] [Google Scholar]
- 9.Gustavsson, N., A. Diez, and T. Nyström. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107-117. [DOI] [PubMed] [Google Scholar]
- 10.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 12.Korhonen, T. K., H. Leffler, and C. Svanborg-Eden. 1981. Binding specificity of piliated strains of Escherichia coli and Salmonella typhimurium to epithelial cells, Saccharomyces cerevisiae cells, and erythrocytes. Infect. Immun. 32:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kvint, K., L. Nachin, A. Diez, and T. Nyström. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 14.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy, K. C. 1991. λ Gam protein inhibits the helicase and χ-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 173:5808-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyström, T., and F. C. Neidhart. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537-544. [DOI] [PubMed] [Google Scholar]
- 18.Nyström, T., and F. C. Neidhart. 1996. Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J. Bacteriol. 178:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poteete, A. R., and A. C. Fenton. 1984. λ Red-dependent growth and recombination of phage P22. Virology 134:161-167. [DOI] [PubMed] [Google Scholar]
- 20.Schembri, M. A., D. W. Ussery, C. Workman, H. Hasman, and P. Klemm. 2002. DNA microarray analysis of fim mutations in Escherichia coli. Mol. Genet. Genomics 267:721-729. [DOI] [PubMed] [Google Scholar]
- 21.Schembri, M. A., L. Hjerrild, M. Gjermansen, and P. Klemm. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sousa, M. C., and D. B. McKay. 2001. Structure of the universal stress protein of Haemophilus influenzae. Structure 9:1135-1141. [DOI] [PubMed] [Google Scholar]
- 24.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 25.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarembinski, T. I., L. W. Hung, H. J. Mueller-Dieckmann, K. K. Kim, H. Yokota, R. Kim, and S. H. Kim. 1998. Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc. Natl. Acad. Sci. USA 95:15189-15193. [DOI] [PMC free article] [PubMed] [Google Scholar]