Here we have assessed the role of a type III translocator protein, BipB, in the cell biology and virulence of Burkholderia pseudomallei. Genetic inactivation of bipB reduced multinucleated giant cell formation, cell-to-cell spreading of bacteria, and induction of apoptosis of J774A.1 macrophages. The bipB mutant was also significantly attenuated following intranasal challenge of BALB/c mice, whereas virulence was fully restored by complementation with a functional bipB gene.
Burkholderia pseudomallei, the etiological agent of melioidosis in humans and animals, is a gram-negative bacterium. Melioidosis is endemic in southeast Asia and tropical Australia and has been reported sporadically elsewhere (6). Currently, there is no vaccine against melioidosis. Uniquely among intracellular bacterial pathogens, B. pseudomallei induces host cell fusion leading to multinucleated giant cell (MNGC) formation in tissue culture models of infection (14). This novel phenotype may be relevant to pathogenesis, since granuloma formation and generation of MNGC are also found in tissues of humans with melioidosis (23). In addition to inducing MNGC formation, B. pseudomallei is able to spread from cell to cell and induce apoptotic death in infected host cells (14). The molecular mechanisms of these pathogenic characteristics have not been elucidated.
Analysis of the B. pseudomallei genome and several other studies have demonstrated the presence of a type III secretion system (TTSS) (for reviews, see references 3, 12, 17, 20, and 22). A knockout mutant of B. pseudomallei lacking a functional bipD gene, a homologue of Salmonella enterica serovar Typhimurium sipD, on the TTSS3/bsa cluster of TTSS exhibited reduced replication in murine macrophage-like cells (20), was significantly attenuated in BALB/c mice and gave partial protection against subsequent challenge with wild-type B. pseudomallei (19). These data correlated with the recent report that the TTSS3/bsa cluster is required for the pathogenicity of B. pseudomallei (21). In addition to BipD, B. pseudomallei BipB and BipC (46 and 30% amino acid identity to Salmonella SipB and SipC, respectively) have been identified in the TTSS3/bsa cluster (3). Here, we report on the role of BipB in the pathogenesis of infection with B. pseudomallei. With Salmonella organisms, purified SipB integrates into artificial membranes and induces liposome fusion (10), and it is required for inducing apoptosis in murine macrophages (11). By analogy with SipB, therefore, we investigated the role of BipB for MNGC formation, cell-to-cell spreading, and induction of apoptosis in infected host cells. We also examined the virulence of a B. pseudomallei bipB mutant in a murine model of melioidosis.
Construction of a B. pseudomallei bipB mutant.
Analysis of the B. pseudomallei genome (http://www.sanger.ac.uk/Projects/B_pseudomallei), by use of the sipB sequence from S. enterica serovar Typhimurium as the query in a TBLASTX search, identified a coding sequence of 1,860 bp encoding the predicted BipB protein of 620 amino acids. In order to determine the function of BipB in B. pseudomallei, a chromosomal bipB mutant of B. pseudomallei was constructed. In brief, a 250-bp internal fragment of the bipB gene was amplified from B. pseudomallei K96243 genomic DNA by use of primers BipB-45 (5′-AACCAGGCCACGCAGCAG-3′) and BipB-46 (5′-CGTCTTCTGCATCTCCTC-3′). The amplified fragment was cloned into a suicide vector, pKNOCK-Tc (1), kindly provided by M. F. Alexeyev. This constructed plasmid was introduced from Escherichia coli S17-1λpir (7) into B. pseudomallei K96243 by conjugation. Transconjugants were selected by plating on pseudomonas agar supplemented with SR103 (Oxoid, United Kingdom) containing tetracycline. The isolated mutant, designated B. pseudomallei BS46 (bipB::pSSB-1), was verified by PCR and Southern blot hybridization to ensure insertion of the bipB suicide plasmid at the correct location (data not shown). For complementation analysis, the amplified bipB gene was cloned into pBBR1MCS (15) and introduced into B. pseudomallei BS46. To confirm that B. pseudomallei BS46pBipB contained the bipB gene, the DNA plasmid was extracted and sequenced (data not shown).
To determine whether bipB was cotranscribed with the downstream genes bipC-bprA-bipD, reverse transcription-PCR (RT-PCR) was undertaken. Extraction of total RNA, by use of the modified hot acid phenol method, was carried out as described previously (2). In brief, mid-exponential-phase cultures were harvested and extracted with hot acid phenol. Total RNA was precipitated and resuspended with RNase-free distilled water. For RT-PCR analysis, bipB-bipC-bprA-bipD was reversed transcribed into cDNA (Invitrogen) and then amplified with different primers, namely, BipB-73 (5′-CTGCTCGGCGATCTGCTCAA-3′), BipC-72 (5′-ACCGCCTTGTCGCCCTG-3′), BipC-80 (5′-GAGCAGAAAGAGGACGAGA-3′), and BipD-77 (5′-CGCAGATCGTCGTCGGTCA-3′) (Fig. 1A).
FIG. 1.
The B. pseudomallei bipB operon. (A) Physical map of bipB-bipC-bprA-bipD gene organization together with locations of primer pairs BipB-73-BipC-72 and BipC-80-BipD-77 for RT-PCR analysis of B. pseudomallei bipB operon. (B) Ethidium bromide-stained gel showing the amplified DNA of RT-PCR products from primer pairs BipB-73-BipC-72 (lane 1) and BipC-80-BipD-77 (lane 2). Lane 3 is an RNA sample subjected to PCR to ensure no DNA contamination in the RNA preparation. Lane M shows lambda DNA markers.
As depicted in Fig. 1B, B. pseudomallei bipB-bipC-bprA-bipD was transcribed in a single transcriptional unit. It is likely that B. pseudomallei BS46 is a polar bipB mutant. To investigate whether this mutation does not have effect on expression of other secreted proteins, Western blot analysis using anti-BopE (kindly provided by M. P. Stevens, United Kingdom) to detect BopE in whole-cell and secreted protein fractions of B. pseudomallei BS46 and wild-type strains was undertaken. BopE, homologous to the Salmonella SopE, was an effector protein secreted by the B. pseudomallei TTSS (18). BopE was detected in both whole-cell and secreted protein fractions of B. pseudomallei BS46 (data not shown). This suggests that the TTSS of B. pseudomallei BS46 is still functional to express and secrete other proteins such as BopE.
The polar bipB mutant is defective in MNGC formation.
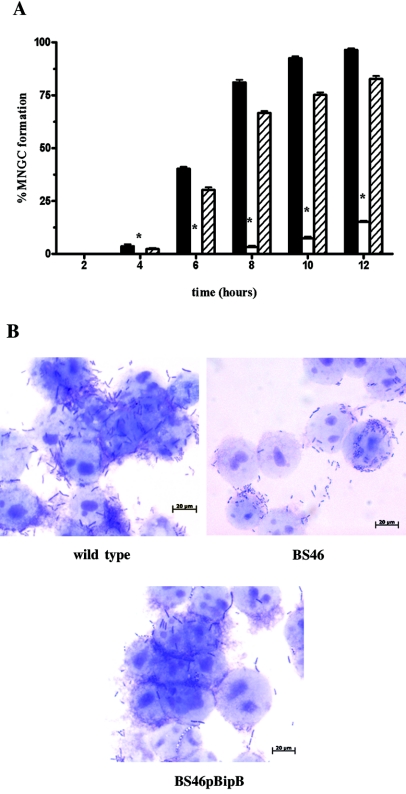
To investigate the potential role of BipB in MNGC formation, B. pseudomallei K96243 (wild type), BS46 (bipB::pSSB-1), and BS46pBipB (BS46 harboring pBipB) were used to infect J774A.1 murine macrophage-like cells as described previously (14). At different times after initiation of the challenge, the infected cells were fixed, Giemsa stained, and evaluated for MNGC formation. Figure 2 shows that BipB protein plays a role in B. pseudomallei-induced MNGC formation. At 12 h postinfection (Fig. 2A), wild-type bacteria induced extensive MNGC formation (96.46%), while BS46 did not (15.12%). The formation of MNGC was restored in a complementation assay using strain BS46pBipB (82.7%). Figure 2B shows that MNGC loaded with numerous bacilli could be readily observed at 6 h after infection with wild-type bacteria but that this was abolished in the bipB mutant BS46. However, this defective phenotype was transcomplemented by reintroduction of the plasmid-born bipB gene. However, when the observation period was extended to 24 h, formation of MNGC in BS46-infected macrophage did occur but was still significantly less than the wild-type strain. Thus, BipB is necessary for optimal MNGC induction, but BipB-independent fusion can also occur, albeit at a reduced efficiency.
FIG. 2.
MNGC formation of B. pseudomallei. (A) The percentages of MNGC formation of J774A.1 cells infected with B. pseudomallei K96243 (wild type; solid bars), BS46 (bipB::pSSB-1; open bars), and BS46pBipB (BS46 harboring pBipB; striped bars) were determined every 2 h. Asterisks indicate significant differences (P < 0.05, t test) between the wild type and BS46 at 4 h (P = 0.0142) and 6 to 12 h (P < 0.0001) and between BS46 and BS46pBipB at 4 h (P = 0.0155) and 6 to 12 h (P < 0.0001). Percentage of MNGC formation was determined by the following equation: MNGC formation = (number of nuclei within multinucleated giant cells/total number of nuclei counted) × 100. Error bars represent standard errors of the means for experiments performed in triplicate. (B) Giemsa staining of MNGC formation of J774A.1 cells infected with wild type, BS46, or BS46pBipB. Bars, 20 μm.
The mechanism for the MNGC formation is still unknown, and to our knowledge, this altered phenotype has not been observed in other intracellular bacteria that possess the TTSS. Based on the Salmonella SipB-induced fusion events in vitro (10) and those that would be transient in vivo (9), we hypothesize that BipB may have membrane fusion activity as well. It may act in concert with other proteins to induce fusion of host cell membranes. A combination of biochemistry, cell biology, and proteomics will be required to unveil the detailed pathways of MNGC formation.
The polar bipB mutant is defective in cell-to-cell spread and invasion into epithelial cells.
The observation of MNGC led us to look closely at cell-to-cell spread of infected host cells by using a plaque assay previously described (14). HeLa cells were infected with B. pseudomallei and overlaid with an agarose medium containing kanamycin (250 μg/ml). To enhance visualization, plaques were overlaid with agarose containing an additional 0.01% neutral red and observed 4 h later. Figure 3A demonstrates that plaque-forming efficiencies for B. pseudomallei wild type (2.66) and BS46pBipB (0.68) were significantly higher than that for BS46 (0.2). It is possible that only partial complementation in BS46pBipB could have resulted from a polar effect that disrupted downstream bipC and bipD genes also participating in cell-to-cell spreading. This hypothesis is supported by a previous report, from Stevens et al. (20), that a bipD mutant exhibited an inability to escape from endocytic vacuoles, a requirement for cell-to-cell spread. If so, it would indicate that BipB works cooperatively with BipC and BipD in a manner similar to that of SipABCD in Salmonella (4).
FIG. 3.
Plaque formations, invasion, and apoptosis induction. (A) Plaque formations of HeLa cells by B. pseudomallei K96243 (wild type; solid bars), BS46 (bipB::pSSB-1; open bars), and BS46pBipB (BS46 harboring pBipB; striped bars). Asterisks indicate significant differences (P < 0.05, t test) between wild type and BS46 (P = 0.0001) and between BS46 and BS46pBipB (P = 0.0031). Plaque-forming efficiency was determined by the following equation: plaque-forming efficiency = number of plaques/bacterial CFU added per well. Error bars represent standard errors of the means for experiments performed in triplicate. (B) Invasion of A549 cells by B. pseudomallei K96243 (wild type; solid bars), BS46 (bipB::pSSB-1; open bars), and BS46pBipB (BS46 harboring pBipB; striped bars) strains. Asterisks indicate significant differences (P < 0.05, t test) between wild type and BS46 (P = 0.0050) and between BS46 and BS46pBipB (P = 0.0173). Percent invasion was determined by the following equation: invasion = (number of intracellular bacteria postinfection/number of CFU added) × 100. Error bars represent standard errors of the means for experiments performed in triplicate. (C) Effect of bipB mutation on induction of apoptosis. J774A.1 cells were infected with B. pseudomallei K96243 (wild type; solid bars), BS46 (bipB::pSSB-1; open bars), BS46pBipB (BS46 harboring pBipB; striped bars), and uninfected cells (checkered bars). The percentages of J774A.1 cells stained fluorescein isothiocyanate positive and propidium iodide negative by flow cytometry were analyzed. Asterisks indicate significant differences (P < 0.05, t test) between wild type and BS46 at 2 h (P = 0.0123), 3 h (P = 0.0004), and 4 to 6 h (P < 0.0001) and between BS46 and BS46pBipB at 2 h (P = 0.1064), 3 h (P = 0.0006), and 4 to 6 h (P < 0.0001). Error bars represent standard errors of the means for experiments performed in triplicate.
The strategies that intracellular bacteria, i.e., Listeria sp. and Shigella sp., use to spread from cell to cell via interepithelial protrusion are quite similar (8). The process depends on the efficiency of bacterial invasion into the epithelial cytosol, protrusion formation, and the lysis of the double-membrane-bound protrusion vacuole to release bacteria into the adjacent cell. To investigate whether defective cell-to-cell spread (as detected by plaque assay) was due to an invasion defect, invasion efficiency was determined by using human respiratory epithelial cell line A549 challenged with B. pseudomallei as described earlier. This cell line was chosen because it is more susceptible to invasion than HeLa cells. Intracellular bacteria were counted after lysing of infected cells. Invasion efficiency of BS46 was severely restricted (0.09%) when compared to that of the wild type (0.39%), but invasion efficiency was restored to nearly normal levels in BS46pBipB (0.28%) (Fig. 3B). These data correlated with those for the bipD mutant that exhibited impaired entry into nonphagocytic host cells (18). In this scenario, we believe that several effector proteins, such as BopE, that contribute to invasion (18) would not be delivered into the host cell cytoplasm, even though it was expressed. This proposed mechanism is based on the study of Salmonella in which inactivation of sip genes resulted in impaired invasion efficiency due to the lack of translocation of effector proteins, such as SopE, into host cells (4, 13, 24). In addition to invasion, BipB may play a role in other steps involved in cell-to-cell spreading. Further experiments are required to investigate this possibility.
The polar bipB mutant is defective in induction of apoptosis.
B. pseudomallei can induce apoptotic death in infected macrophages (14). To determine the role of BipB in this process, J774A.1 cells were infected with B. pseudomallei strains. At different time intervals, the supernatant and cells were collected to quantify the apoptosis level by using an annexin V-fluorescein isothiocyanate detection kit (BD Biosciences, CA). At 6 h postinfection (Fig. 3C), cells infected with wild-type B. pseudomallei yielded significantly higher numbers of positive cells (10.20%) than those infected with BS46 (3.21%). Infection with BS46pBipB restored cytotoxicity (6.77%). These data indicated that BipB was required for efficient induction of apoptosis in host cells, although a low level of apoptosis may occur via a BipB-independent mechanism, since the level of apoptosis in uninfected cells is 2.3%. This is the first report identifying a B. pseudomallei virulence factor that mediates apoptosis. Interestingly, this finding joins a growing list of bacteria, including Pseudomonas aeruginosa, Yersinia sp., Salmonella sp., and Shigella flexneri, that kill host cells via apoptotic death through a type III secretion-mediated mechanism. In Salmonella and Shigella, SipB and IpaB have been shown to induce macrophage apoptotic death by activating caspase-1 (11, 25). Here, we also expect that apoptosis induced by B. pseudomallei will involve BipB interaction with the caspase pathway (14).
Effect of bipB mutation on virulence of B. pseudomallei in vivo.
The finding that BipB is important in induction of MNGC, plaque formation, bacterial invasion, and killing of phagocytic cells in vitro led to the hypothesis that a mutant unable to produce this protein could be less virulent than the wild-type strain in vivo. We therefore assayed the virulence of the bipB mutant in a pulmonary model of melioidosis in BALB/c mice as previously described (19). B. pseudomallei strains were administered via the intranasal route. Viable counts were performed to confirm the inoculation dose, and the mice were monitored twice daily for signs of infection. There was a significant difference in percentage survival (the P value was <0.05, as determined by a log rank test) for mice infected with wild-type B. pseudomallei versus mice infected with BS46 (Fig. 4). All mice given the wild-type strain died within 5 to 11 days, whereas five of six mice infected with the bipB mutant survived until day 42 (termination of experiment). To confirm that attenuation resulted from the inactivation of bipB, we also challenged mice with strain BS46pBipB, and all died by day 4 postchallenge (Fig. 4), which was not significantly different from the wild-type strain. These observations indicated that a functional bipB gene was required for full virulence of B. pseudomallei in mice. This result is supported by previous reports (19, 21) that TTSS3/Bsa plays an important role for maximal virulence in all of its animal hosts.
FIG. 4.

Survival of BALB/c mice (six mice per group) inoculated intranasally with 103 CFU of B. pseudomallei K96243 (▪) or BS46 (▴) or BS46pBipB (⧫). Mice were observed daily, and percent survival was plotted against time.
Delivery of virulence-associated effector proteins into eukaryotic cells requires a set of translocator proteins. The translocons are components of oligomeric protein channels that insert themselves into the eukaryotic cell membrane to form a pore which effector proteins can pass through to gain access to the cytosolic host targets (5, 16). We have shown here that BipB translocator plays a critical role in the intracellular lifestyle of B. pseudomallei (i.e., MNGC formation, invasion of nonphagocytic cells, and induction of apoptotic death). We hypothesize that the bipB mutant is unable to deliver the effector proteins into the host cell cytoplasm and was thus impaired in invasion efficiency and ability to induce apoptosis. However, it is also possible that BipB acts as an effector protein to induce apoptotic death. Deletion of BipB clearly also reduces the efficiency of MNGC formation; however, the relationship between BipB protein and the fusion process is still under investigation. In vivo, BipB was required for full virulence of B. pseudomallei in mice, thus further confirming the importance of BipB for virulence in murine models of melioidosis.
Acknowledgments
This work was supported by the Thailand Research Fund (TRF) grant PHD/0093/2546 through the Royal Golden Jubilee Ph.D. program to S. Suparak and S. Korbsrisate, grant RSA4580034 from the TRF to S. Korbsrisate, and a Senior Research Scholar grant (RTA4580010) to S. Mongkolsuk.
We thank S. Lerdwana for flow cytometric analysis, P. Vattanaviboon for his suggestion, and T. W. Flegel for critical reading of the manuscript. We also acknowledge the staff from the Medical Molecular Biology Unit, Siriraj Hospital, Thailand, for assistance in cell culture techniques.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 2.Ambulos, N. P., Jr., E. J. Duvall, and P. S. Lovett. 1987. Method for blot-hybridization analysis of mRNA molecules from Bacillus subtilis. Gene 51:281-286. [DOI] [PubMed] [Google Scholar]
- 3.Attree, O., and I. Attree. 2001. A second type III secretion system in Burkholderia pseudomallei: who is the real culprit? Microbiology 147:3197-3199. [DOI] [PubMed] [Google Scholar]
- 4.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 5.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dance, D. A. 2000. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop. 74:159-168. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., and P. Cossart. 1998. Intracellular pathogens and the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:137-166. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. B., and S. Falkow. 1990. Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J. Infect. Dis. 162:1096-1106. [DOI] [PubMed] [Google Scholar]
- 10.Hayward, R. D., E. J. McGhie, and V. Koronakis. 2000. Membrane fusion activity of purified SipB, a Salmonella surface protein essential for mammalian cell invasion. Mol. Microbiol. 37:727-739. [DOI] [PubMed] [Google Scholar]
- 11.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kespichayawattana, W., S. Rattanachetkul, T. Wanun, P. Utaisincharoen, and S. Sirisinha. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68:5377-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 17.Rainbow, L., C. A. Hart, and C. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374-384. [DOI] [PubMed] [Google Scholar]
- 18.Stevens, M. P., A. Friebel, L. A. Taylor, M. W. Wood, P. J. Brown, W. D. Hardt, and E. E. Galyov. 2003. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J. Bacteriol. 185:4992-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens, M. P., A. Haque, T. Atkins, J. Hill, M. W. Wood, A. Easton, M. Nelson, C. Underwood-Fowler, R. W. Titball, G. J. Bancroft, and E. E. Galyov. 2004. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150:2669-2676. [DOI] [PubMed] [Google Scholar]
- 20.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 21.Warawa, J., and D. E. Woods. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101-108. [DOI] [PubMed] [Google Scholar]
- 22.Winstanley, C., B. A. Hales, and C. A. Hart. 1999. Evidence for the presence in Burkholderia pseudomallei of a type III secretion system-associated gene cluster. J. Med. Microbiol. 48:649-656. [DOI] [PubMed] [Google Scholar]
- 23.Wong, K. T., S. D. Puthucheary, and J. Vadivelu. 1995. The histopathology of human melioidosis. Histopathology 26:51-55. [DOI] [PubMed] [Google Scholar]
- 24.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 25.Zychlinsky, A., B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti. 1994. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol. 11:619-627. [DOI] [PubMed] [Google Scholar]