Abstract
We report here studies of expression and functional analysis of a Bacillus subtilis gene, ywcE, which codes for a product with features of a holin. Primer extension analysis of ywcE transcription revealed that a single transcript accumulated from the onset of sporulation onwards, produced from a σA-type promoter bearing the TG dinucleotide motif of “extended” −10 promoters. No primer extension product was detected in vivo during growth. However, specific runoff products were produced in vitro from the ywcE promoter by purified σA-containing RNA polymerase (EσA), and the in vivo and in vitro transcription start sites were identical. These results suggested that utilization of the ywcE promoter by EσA during growth was subjected to repression. Studies with a lacZ fusion revealed that the transition-state regulator AbrB repressed the transcription of ywcE during growth. This repression was reversed at the onset of sporulation in a Spo0A-dependent manner, but Spo0A did not appear to contribute otherwise to ywcE transcription. We found ywcE to be required for proper spore morphogenesis. Spores of the ywcE mutant showed a reduced outer coat which lacked the characteristic striated pattern, and the outer coat failed to attach to the underlying inner coat. The mutant spores also accumulated reduced levels of dipicolinic acid. ywcE was also found to be important for spore germination.
During the initial stages of sporulation in Bacillus subtilis, the rod-shaped cell is asymmetrically divided into a smaller prespore and a larger mother cell. Migration of the septal membranes towards the proximal cell pole, a process called engulfment, and their eventual fusion at the cell pole release the prespore as a free protoplast within the mother cell cytoplasm (15). The engulfed prespore (or forespore) is separated from the mother cell cytoplasm by two membranes of opposing polarities, which derive from the invagination of the cell envelope earlier during polar division (15). The arrangement of the spore membranes dictates the pattern of synthesis of the various spore structures, whose assembly may start soon after activation of the mother cell line of gene expression but becomes more evident following forespore engulfment (13, 15, 16, 28). Synthesis of an inner layer of peptidoglycan, known as the primordial cell wall, occurs across the prespore inner membrane and requires prespore-specific gene expression (16). In contrast, synthesis of the spore cortex peptidoglycan occurs across the prespore outer membrane and requires the expression of several mother cell-specific genes (16). Another protective layer that forms around the developing spore is a protein coat that confers protection against peptidoglycan-breaking enzymes and noxious chemicals and is also important for spore germination (13, 28). Formation of both the cortex and coat layers requires the synthesis of SpoIVA (49, 55), which localizes at or near the forespore outer membrane (14). It is not known how SpoIVA influences cortex synthesis. However, SpoIVA is required for proper localization and maintenance around the developing spore of several morphogenetic proteins that act to guide assembly of the spore coat (14, 41).
SpoVID localizes to the surface of the developing spore in a SpoIVA-dependent manner and is required for attachment of the nascent coat to the spore surface (41). In spoVID mutants, the coat forms swirls throughout the mother cell cytoplasm, and the resulting spores, which have an exposed cortex layer, are highly susceptible to lysozyme (5). SpoVID acts in part through another coat morphogenetic protein called SafA (40, 56). SafA has a cell wall-binding LysM domain (4) encompassing its first 50 amino-terminal residues and has been shown by immunogold labeling to localize at the interface between the cortex and coat regions (40). The C-terminal half of SafA shares similarity to known inner coat proteins, and it has been proposed that SafA helps to link the cortex, to which it would bind via its LysM domain, to the nascent coat via interactions with other coat proteins involving its C-terminal portion (41). SpoVID directly interacts with SafA and is required for the targeting of SafA to the surface of the developing spore (40). One intriguing aspect of the function of SafA is how its LysM domain is put in contact with the spore cortex peptidoglycan. SafA is made in the mother cell, whereas the cortex accumulates in the compartment defined by the inner and outer spore membranes (15, 16). Several proteins with secretory signal sequences are made in either the prespore or the mother cell and are presumably translocated across the prespore membranes into the cortical region. The cortex-lytic enzyme SleB, which also has a potential peptidoglycan-binding domain, for example, is synthesized in the forespore with a signal peptide that is cleaved, and mature SleB is found associated with spore membranes, cortexes, and coats (38). SpoIVH, which is required for cortex synthesis, is produced in both the prespore and the mother cell with a signal peptide which can be replaced by that of SleB (31). SafA does not have a signal sequence, and it has been suggested that other mechanisms could facilitate its transfer from the mother cell cytoplasm into the cortex region of the developing spore, including the involvement of holins (40). Holins are involved in the precise temporal control of cell lysis upon bacteriophage infection by forming a pore that permits access of phage-encoded lysins to the cell wall compartment (59). There are two main classes of holins. Class I includes proteins with three membrane-spanning domains, a dual start motif, and a highly charged C-terminal region. The prototype member of this class is the λ S protein (18, 59) (see Fig. 1C). Class II holins, such as the S protein from the lambdoid bacteriophage 21, are smaller (60 to 85 residues), with only two predicted transmembrane domains (3, 59).
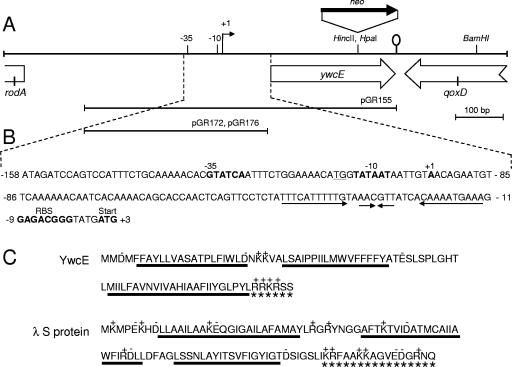
FIG. 1.
ywcE locus and YwcE protein. Panel A shows the ywcE region of the B. subtilis chromosome. The ywcE gene is divergently oriented relative to rodA (only the 5′ end of this gene is represented) and convergent with qoxD (only its 3′ end is shown). A stem-loop structure between ywcE and qoxD represents a transcriptional terminator. Restriction enzyme sites used to insert a neomycin resistance cassette within the ywcE gene or used for in vitro transcription experiments are represented. The relative positions of the ywcE −10 and −35 promoter elements as well as that of the transcriptional start site are shown. The lines below the partial physical and genetic map represent inserts in the indicated plasmids. Panel B shows the sequence of the ywcE promoter. The −35 and −10 elements, the transcriptional start site (+1), the ribosome binding site, and the initiation codon are shown in bold. The arrows represent two regions of dyad symmetry located just upstream of the translation initiation signals. The sequence is numbered relative to the first nucleotide in the initiation codon. Panel C represents the primary structures of YwcE and of the S protein of phage λ. The lines below the amino acid sequence (in single-letter code) represent the locations of predicted transmembrane segments. Basic (+) and acidic (−) amino acids are indicated. Asterisks indicate the highly charged C-terminal regions of both proteins.
The B. subtilis genome appears to code for several holin-like proteins (35). For this study, we analyzed the expression and function of the ywcE gene of B. subtilis (35), because its predicted product is structurally related to class I holins (see Fig. 1C) and because ywcE is expressed during sporulation. We found that transcription of ywcE takes place from a σA-type promoter that is repressed during growth and induced at the onset of sporulation in a Spo0A-dependent manner. A functional YwcE-green fluorescent protein (GFP) fusion protein localizes to both the cell and spore membranes. ywcE mutants form spores with reduced levels of pyridine-2,6-dicarboxylic acid (DPA) that have an outer coat layer that lacks the normal striated pattern. Our results also indicate that ywcE expression is important for spore germination.
MATERIALS AND METHODS
Bacterial strains, media, and general techniques.
The bacterial strains used for this study are congenic derivatives of the wild-type strain MB24 and are listed in Table 1. The Escherichia coli strain DH5α (Gibco BRL) was used as the host for the construction and propagation of all plasmids described herein. Luria-Bertani (LB) medium was routinely used for the growth of both B. subtilis and E. coli strains at 37°C. Difco sporulation medium (DSM) was used to induce sporulation by nutrient exhaustion (10). Genetic manipulations of E. coli and B. subtilis and spore resistance or germination properties were assessed as previously described (10, 24). Other general methods were performed as previously described (10, 24, 25, 27). The levels of DPA were measured as previously described (10).
TABLE 1.
Bacterial strains used for this study
| Strain | Genotype/phenotype | Origin or reference |
|---|---|---|
| MB24 | trpC2 metC3; wild type | Laboratory stock |
| AH2357 | trpC2 metC3 ΔspoIIAC::em; Emr | Laboratory stock |
| AH38 | trpC2 metC3 spoIIGB55 | Laboratory stock |
| AH77 | trpC2 metC3 ΔsigK::erm; Emr | Laboratory stock |
| AH2544 | trpC2 metC3 ΔabrB::neo; Neor | Laboratory stock |
| AH3795 | trpC2 metC3 ΔspoIIIG | 51 |
| FB111 | ΔcwlJ::tet; Tetr | 42 |
| FB112 | ΔsleB::sp; Spr | 42 |
| FB113 | ΔcwlJ::tet ΔsleB::sp; Tetr Spr | 42 |
| AH630 | trpC2 metC3 ΔywcE::neo; Neor | This work |
| AH3113 | trpC2 metC3 Δspo0A::Pspac-spo0A; Cmr | This work |
| AH3362 | trpC2 metC3 Δspo0H::Pspac-spo0H; Cmr | This work |
| AH3279 | trpC2 metC3 ΔamyE::ywcE-gfpmut2; Neor | This work |
| AH3338 | trpC2 metC3 ΔamyE::PywcE-lacZ; Cmr | This work |
| AH3339 | trpC2 metC3 ΔspoIIAB::em ΔamyE::PywcE-lacZ; Cmr Emr | This work |
| AH3340 | trpC2 metC3 spoIIGB55 ΔamyE::PywcE-lacZ; Cmr | This work |
| AH3341 | trpC2 metC3 ΔspoIIIG ΔamyE::PywcE-lacZ; Cmr | This work |
| AH3342 | trpC2 metC3 ΔsigK::erm ΔamyE::PywcE-lacZ; Cmr Emr | This work |
| AH3344 | trpC2 metC3 ΔthrC::PywcE-gfpmut2; Emr | This work |
| AH3382 | trpC2 metC3 ΔamyE::PywcE-lacZ; Spr | This work |
| AH3384 | trpC2 metC3 Δspo0A::Pspac-spo0A ΔamyE::PywcE-lacZ; Cmr Spr | This work |
| AH3385 | trpC2 metC3 Δspo0H::Pspac-spo0H ΔamyE::PywcE-lacZ; Cmr Spr | This work |
| AH3386 | trpC2 metC3 ΔabrB::neo ΔamyE::PywcE-lacZ; Neor Spr | This work |
| AH3387 | trpC2 metC3 Δspo0A ΔabrB::neo ΔamyE::PywcE-lacZ; Cmr Spr Neor | This work |
| AH3410a | trpC2 metC3 ywcE::neoΩpGR182; Cmr | This work |
| AH3414 | trpC2 metC3 ywcE::neoΩpGR182 amyE::PywcE-gfp; Neor Cmr | This work |
| AH3443 | trpC2 metC3ΔcwlJ::tet; Tetr | This work |
| AH3444 | trpC2 metC3ΔsleB::sp ΔywcE::neo; Spr Neor | This work |
| AH3445 | trpC2 metC3ΔcwlJ::tet ΔywcE::neo; Tetr Neor | This work |
The Ω symbol denotes the single reciprocal integration of the indicated plasmid at the ywcE locus.
Construction of a ywcE insertional mutant.
Plasmid pAH103 has been described before (26). It was created by inserting a 1,755-bp EcoRI-EcoRV DNA fragment isolated from pλsacT2 between the EcoRI and HincII sites of plasmid pUS19 (26). The insert in pAH103 encompasses the 3′ end of the qoxD gene, the complete ywcE gene, and the 5′ end of the rodA gene (not represented in Fig. 1). After plasmid pAH103 was digested with HindII, a neomycin resistance (Nmr) determinant released from pBEST501 (32) with SmaI was inserted in the same orientation as the ywcE gene, thus producing pAH105. To transfer the Nmr determinant to the B. subtilis chromosome, pAH105 was linearized with ScaI and used to transform competent MB24 cells. A transformant, verified by PCR to result from a double-crossover event at the ywcE region, was named AH630 (Table 1).
Construction of ywcE-lacZ and ywcE-gfpmut2 transcriptional fusions.
A 386-bp fragment carrying DNA upstream from the ywcE start codon was PCR amplified with primers ywcE-392D(EcoRI) and ywcE-6R(BglII) (Table 2), simultaneously digested with EcoRI and BglII, and cloned between the EcoRI and BamHI sites of pSN32 (39), yielding pGR172 (Fig. 1). PstI-linearized pGR172 was used to transfer the ywcE-lacZ fusion to the amyE locus of wild-type strain MB24 to produce the Cmr AmyE− strain AH3338 (Fig. 1; Table 1). The chloramphenicol (Cm) resistance determinant was subsequently changed to a spectinomycin (Spec) resistance determinant by the transformation of AH3338 with plasmid pJL62 (36), producing the Cms Specr AmyE− strain AH3382 (Table 1). To construct a ywcE-gfpmut2 transcriptional fusion, we first constructed a vector for transcriptional fusions to gfp to be integrated at the thrC locus of the chromosome. Briefly, a 973-bp fragment carrying the spoIIGA promoter region, the spoVG Shine-Dalgarno sequence, and the gfp coding region was amplified from pMS201 (a gift from Monica Serrano) with primers spoIIG-131D(EcoRI) and gfpR(BglII) (Table 2) and introduced into EcoRI-BamHI-digested pDG1664 (20), yielding pGR168. The 398-bp PCR product amplified with primers ywcE-392D(EcoRI) and ywcE-6R(BglII) (see above) was digested with EcoRI and BglII and introduced between the EcoRI and BamHI sites of pLITMUS29 (New England Biolabs), yielding pGR174. A 412-bp EcoRI-HindIII fragment obtained from pGR174 was then introduced into EcoRI-HindIII-digested pGR168, yielding pGR176. This plasmid was used to transfer the ywcE-gfp transcriptional fusion to the thrC locus of wild-type strain MB24, creating the Ermr Specs strain AH3344 (Fig. 1; Table 1).
TABLE 2.
Oligonucleotides used for this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| ywcE-461R(gfpmut2) | GTGAAAAGTTCTTCTCCTTTACTGTTGTTGTTGTTGCTGCTTCGCTTTCTGCGAAGATACGGAAGGCCG |
| ywcE-356R | CAAATAAAGAGAGGCGTTGC |
| ywcE-392D(HindIII) | GCGGCGCTTTTTAAGCTTCACGAAATATGACG |
| ywcE-392D(EcoRI) | GCGGCGCTTTTTGAATTCCACGAAATATGACG |
| ywcE-6R(BgIII) | CCCAGATCTCCTTTCATTTTGTGATAACG |
| gfpR | GGCGAATTCTTATTTGTATAGTTCATCCATGC |
| spoIIG-131D(EcoRI) | GGAATTCATCGTCGCAGATGATTATGG |
| gfpR(BgIII) | GAAGATCTTCTTATTTGTATAGTTCATCCATGCG |
| gfp-30D | AGTAAAGGAGAAGAACTTTTCACTGGAG |
| spo0A-563D(HindIII) | GGTGAAGCTTGTTAACTACATTTGGGGAGG |
| spo0A-1064R(SpeI) | CCGATTTCATGGATAATACTAGTGATGCTC |
Native or engineered restriction sites are underlined.
Construction of a ywcE-gfp translational fusion.
A C-terminal fusion between ywcE and gfpmut2 was obtained using the splicing-by-overlap-extension technique (30). Briefly, a 653-bp PCR fragment including the ywcE gene and its promoter region was amplified from the chromosome of B. subtilis strain MB24 using primers ywcE-392D(HindIII) and ywcE-461R(gfpmut2) (Table 2). The gfpmut2 gene (710 bp) was PCR amplified using pEIA18 (46) as a template and primers gfp-30D and gfpR (Table 2). A linker encoding four asparagines was engineered in the ywcE-461R(gfpmut2) primer (17). The ywcE-gfpmut2 fusion was amplified with primers ywcE-392D(HindIII) and gfpR using an equimolar mix of both the ywcE and gfpmut2 PCR products as the template. The resulting fragment of 1,363 bp was digested with EcoRI and HindIII and inserted between the same sites in pMLK83 (33), generating pGR155. Strain AH3279 (Neor) resulted from the integration of pGR155 into the amyE locus of wild-type MB24 (Table 1). pGR182 was constructed by amplifying an internal fragment of a neomycin gene from pBEST501 (32) using the primers NeoD (NotI) (5′-GTCGTCAGACTGATGCGGCCGCTATTCGG-3′) and NeoR (XbaI) (5′-CTGCCATTGCTACCTCTAGAGTCAAGGATG-3′). The amplified product was digested with NotI and EcoRI and inserted between the same sites of pMS38 (60). The resulting plasmid, pGR182, was used to transform AH3319 to Cmr. A Neos transformant, AH3410, was then transformed with pGR155 to yield the AmyE− strain AH3414.
Conditional allele of spo0A.
A 502-bp DNA fragment corresponding to 470 bp of the 5′ coding sequence of spo0A was amplified using primers spo0A-563D and spo0A-1064R (Table 2), digested with HindIII and SpeI, and inserted between the HindIII and XbaI sites of pDH88 (23), yielding pGR96. Plasmid pGR96 was integrated into the host chromosome by means of a single-crossover (Campbell-type) recombinational event that occurred in the region of homology, giving strain AH3113 (Table 1).
RNA purification and primer extension analysis.
Total B. subtilis RNA was prepared at the indicated times from cells grown in DSM, as previously described (25). Primer extension reactions were carried out using primer ywcE-356R (Table 2). Sequencing and primer extension reactions were done as described previously (25). Appropriate double-stranded templates were used so that the initiation nucleotide in the mRNA species could be directly determined from the autoradiographs.
Purification of RNA polymerase.
The σA form of RNA polymerase (EσA) was isolated from exponentially growing cells of the wild-type strain B. subtilis MB24 (Table 1) grown in LB medium. The EσA holoenzyme was purified using a low-pressure affinity chromatography step on a heparin column, followed by anion-exchange fast protein liquid chromatography, as previously described (7, 57, 58).
In vitro transcription.
Plasmid pAH103 was cut with either BamHI or HpaI (Fig. 1) and used as a template for in vitro transcription reactions. RNA polymerase (0.04 μM) and the cut plasmid (5 nM) were preincubated at 37°C for 10 min in 50 μl (final volume) of a buffer containing 33 mM Tris acetate (pH 7.9), 10 mM magnesium acetate, 0.5 mM dithiothreitol, 0.15 mg bovine serum albumin/ml, and 66 mM potassium acetate. Ribonucleotides were added at the following concentrations: a 500 μM concentration (each) of ATP, CTP, and GTP (Boehringer Mannheim) and 10 μCi of [α-32P]UTP (800 Ci/mmol; Amersham). The mixture was incubated for 1 min before reinitiation was stopped by the addition of 10 μg of heparin (Sigma). Five minutes later, unlabeled UTP (Boehringer Mannheim) was added to a final concentration of 500 μM, and the mixture was incubated for another 5 min. The nucleic acids were then precipitated by the addition of sodium acetate (final concentration, 0.3 M) and ethanol and resuspended in 10 μl of a sequencing formamide dye. The samples were heated at 95°C for 3 min and then loaded onto a 5% polyacrylamide-7 M urea sequencing gel. Transcripts were localized by autoradiography on a PhosphorImager 445 SI (Molecular Dynamics).
Spore purification and extraction and analysis of spore coat proteins.
Spores were purified from 24-h DSM cultures on Renocal gradients as described previously (60). Extraction and analysis of the spore coat proteins and immunoblot analysis of CotG were performed as described previously (60).
Microscopy.
Samples for the visualization of fluorescence were prepared as previously described (47). For the visualization of membranes, the membrane dye FM4-64 (Molecular Probes) was added to a final concentration of 10 μg ml−1 before mounting the cells on agarose-coated slides. All samples were observed with a 63× objective lens. Phase-contrast or fluorescence images were acquired with a CoolSNAP HQ Photometrics camera (Roper Scientific, Tucson, Arizona), recorded, and processed for publication using Adobe Photoshop, version 4.0
Electron microscopy.
Spores were purified from DSM cultures of strain MB24 (wild type) and its congenic derivative AH630 (ywcE::neo) 24 h after the onset of sporulation, as formerly described (27). The spores were fixed and embedded as described before (27). Electron microscopy analysis and photography were conducted on a Phillips EM301 electron microscope operating at 80 keV.
RESULTS
The ywcE gene is expressed during sporulation from a σA-type promoter.
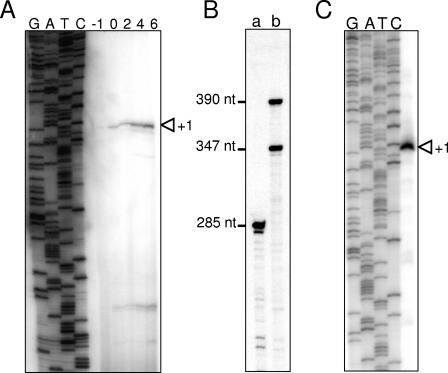
In a first step towards the functional analysis of the ywcE gene, we wanted to determine whether ywcE was transcribed during sporulation in DSM. We used an oligonucleotide complementary to the ywcE transcribed strand to prime cDNA synthesis of total RNA prepared during growth and sporulation of a wild-type strain in DSM. The results in Fig. 2A show that a single primer extension product was detected from the onset of sporulation (or time zero) until at least hour 6 after the initiation of the process. The presumed initiation nucleotide was an “A” located 76 bp upstream of the first nucleotide in the ywcE start codon (Fig. 1A and B). An inspection of the sequence upstream of this position did not reveal sequences similar to those known to be utilized by any of the four compartment-specific σ factors that control sporulation (21). Instead, the putative −10 sequence was identical to the consensus for σA-type promoters (TATAAT) (Fig. 1A and B). In contrast, the predicted −35 sequence (GTATCA), located 17 bp upstream, did not conform as well to the consensus (TTGACA) (21). However, note that the −10 region was preceded by the TG dinucleotide characteristic of extended −10 promoters (22). The results suggest that ywcE is transcribed during sporulation from a σA-type promoter with an extended −10 region. Because no transcript was detected during the exponential growth phase (Fig. 2A, lane −1), the results also suggest that the ywcE promoter is repressed during the exponential phase of growth.
FIG. 2.
Transcriptional analysis of the ywcE gene. Panel A shows an autoradiograph of ywcE-specific primer extension products after electrophoretic resolution in denaturing polyacrylamide gels. An oligonucleotide complementary to the 5′ end of the nontranscribed strand of ywcE was used to direct the cDNA synthesis of total RNA purified from DSM cultures of a wild-type strain (MB24) at the indicated times (in hours) before or after time zero, defined as the onset of sporulation. Panel B is an autoradiograph of runoff transcription assays, in which HpaI-linearized (a) or BamHI-linearized (b) pAH103 (see Materials and Methods) was used as the template. The sizes of the runoff products (in nucleotides) are shown on the left. Panel C shows an autoradiograph of the ywcE-specific primer extension product generated using the RNA produced in vitro with purified RNA polymerase and HpaI-linearized pAH103 after resolution in denaturing acrylamide gels. For panels A and C, the oligonucleotide ywcE-356R (Table 2) was used to prime RNA synthesis. The same oligonucleotide used for the primer extension experiments was also used in parallel to prime sequencing reactions (labeled G, A, T, and C) from pAH103 so that the initiating nucleotide in the nontranscribed strand (+1) could be determined by direct inspection of the sequencing ladder.
We then tested whether purified σA-containing RNA polymerase could direct transcription from the ywcE promoter in vitro. The RNA polymerase σA holoenzyme was purified from exponentially growing cells harvested at late exponential phase in LB medium (7, 57, 58). For in vitro transcription reactions, RNA polymerase was incubated with plasmid pAH103 cut with either HpaI, which cuts within the ywcE coding sequence, or BamHI, which cuts downstream of ywcE (Fig. 1). We found a runoff product of about 285 nucleotides (nt) with HpaI-cut pAH103 (Fig. 2B, lane a), consistent with the transcription initiation site determined in vivo (Fig. 2A). However, we found two products, of about 347 and 390 nt, with BamHI-cut pAH103 (Fig. 2B, lane b). The 347-nt product is consistent with transcription initiating at the +1 position found in vivo but stopping at the putative transcriptional terminator located between the ywcE and qoxD genes (Fig. 1A). The 390-nt species has the expected size for a runoff product reaching the end of the BamHI-linearized template. To determine whether the transcription initiation nucleotide was the same in vivo and in vitro, the RNA generated in vitro with HpaI-cut pAH103 was subjected to a primer extension reaction with the same oligonucleotide primer used for the in vivo experiment depicted in Fig. 2A. The results in Fig. 2C show that transcription in vitro was initiated from an “A” nucleotide 76 bp upstream from the first base of the ywcE start codon. From these results, we infer that the transcription of ywcE in vitro with purified σA-containing RNA polymerase and in vivo during sporulation takes place from the same σA-recognized promoter. We further infer that transcription from this promoter is repressed during vegetative growth and that this repression is relieved at the onset of sporulation.
The ywcE gene is repressed by AbrB.
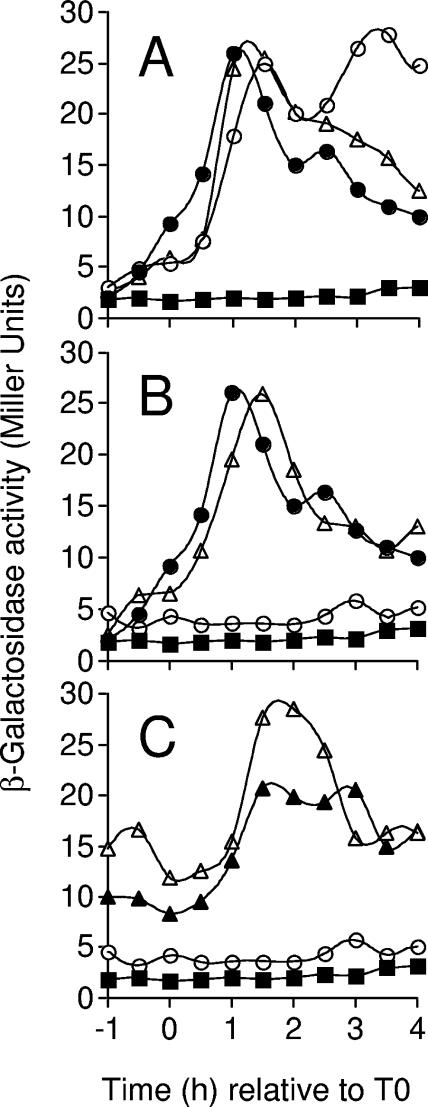
The dependence of ywcE expression on loci known to control the initiation of sporulation was examined by transferring a ywcE-lacZ fusion (see Materials and Methods) into a series of congenic mutants and by monitoring β-galactosidase production throughout growth and sporulation. In agreement with the primer extension analysis, the expression of ywcE-lacZ was not detected above background levels during vegetative growth but was induced around the onset of sporulation (Fig. 3A). Peak levels of β-galactosidase synthesis were reached between hours 1 and 2. The expression of ywcE-lacZ was independent of the expression of spo0H (Fig. 3A) but required the expression of spo0A from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter (Fig. 3B). Spo0A could be directly or indirectly involved in ywcE transcription. For example, Spo0A represses transcription of the abrB gene, which encodes the transition-state regulator AbrB, a repressor of several genes important for the initiation of sporulation (45, 61). The results in Fig. 3C show that mutation of the abrB gene leads to an increased expression of ywcE-lacZ during growth and at the onset of sporulation and that spo0A was no longer required for ywcE-lacZ expression in cells with an abrB mutation. These results suggest that AbrB represses transcription of the ywcE gene during growth and the initial stages of sporulation and that at the onset of sporulation, this repression is relieved in a Spo0A-dependent manner. In agreement with the results of the in vitro transcription studies described above, these results further suggest that Spo0A is not directly involved in the utilization of the ywcE promoter.
FIG. 3.
Analysis of ywcE-lacZ expression. The production of β-galactosidase from a ywcE-lacZ transcriptional fusion was monitored during growth and throughout sporulation in DSM cultures of a wild-type strain and congenic derivatives bearing mutations in loci known to control entry into sporulation. Panel A shows the expression of ywcE-lacZ in a wild-type background (AH3382) (•) and in a strain bearing an IPTG-inducible spo0H allele (AH3385) in the absence (▵) or presence (○) of 1 mM IPTG. (B) Effects of an IPTG-inducible allele of spo0A on the expression of ywcE-lacZ. The strains used are as follows: AH3382 (wild-type background) (•), AH3384 (Δspo0A::Pspac-spo0A; no inducer) (○), and AH3384 (Δspo0A::Pspac-spo0A; 1 mM IPTG) (▵). (C) Role of abrB on the expression of ywcE-lacZ with strains AH3351 (ΔabrB::neo) (▵), AH3352 (ΔabrB::neo Δspo0A::Pspac-spo0A; no inducer) (▴), and AH3384 (Δspo0A::Pspac-spo0A; no inducer) (○). For all panels, endogenous levels of activity were determined with strain MB24 (no fusion) (▪). The β-galactosidase activity is shown in Miller units. Time zero indicates the initiation of the stationary phase of growth, defined as the onset of sporulation.
A YwcE-GFP fusion localizes to the cell and spore membranes.
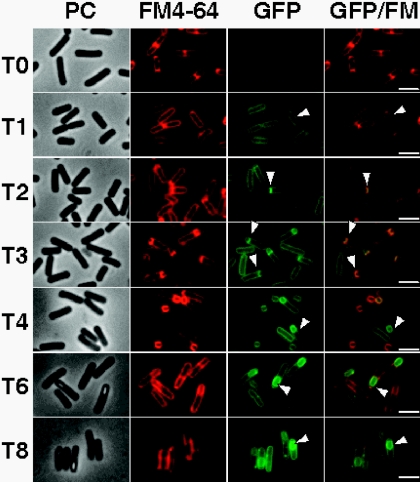
To investigate the localization of YwcE, we constructed a strain (AH3279) expressing a translational C-terminal YwcE-GFP fusion from the ywcE promoter at the nonessential amyE locus. Complementation of the ywcE phenotype (described below) by ectopic expression of ywcE-gfp showed that the GFP fusion protein was functional. The localization of YwcE-GFP was then investigated throughout sporulation by fluorescence microscopy. Samples of cultures grown in DSM were collected, and the cells were stained with FM4-64 to visualize the septal and engulfment membranes. In agreement with our analysis of ywcE transcription, GFP fluorescence was detected from hour 1 of sporulation onwards and was localized to the cell membrane (Fig. 4). At the onset of sporulation, the cell division protein FtsZ assembles into rings at two potential division sites near each cell pole of the sporangium (37). Only one of these two potential division sites is used to form the sporulation septum, while the other is aborted. At hour 2 of sporulation, many cells showed an asymmetric septum, and at this time, fluorescence from the YwcE-GFP fusion protein was found to colocalize with the polar septum, seen as a straight line across the short axis of the cell (Fig. 4). At later stages, the septal membranes migrate towards the proximal cell pole, during the process of engulfment. YwcE-GFP remained associated with the septal membranes during engulfment (Fig. 4). Once the engulfment process was complete, the signal from the lipophilic dye FM4-64 was lost (53), but the fusion protein remained associated with the spore membranes, completely encircling the developing spore (Fig. 4). YwcE-GFP remained associated with the spore during the subsequent stages of spore maturation when refractile spores developed and also after release from the mother cell (data not shown). In addition, the results show that YwcE-GFP remains associated with the mother cell membrane throughout sporulation. Since YwcE-GFP complemented the DPA phenotype of a ywcE mutant (see below) and since the pattern of YwcE-GFP localization in cells containing only the ywcE-gfp allele (not shown) did not differ in any discernible way from that described above for cells containing both the ywcE-gfp and wild-type alleles of ywcE, it seems likely that the localization of the fusion protein reflects that of untagged YwcE.
FIG. 4.
Localization of a functional YwcE-GFP fusion protein. Strain AH3279 was induced to sporulate in DSM, and samples were collected throughout sporulation. Cells were stained with the vital membrane stain FM4-64 (second column) and observed by fluorescence microscopy as described in Materials and Methods. Phase-contrast images are shown in the first column, the localization of YwcE-GFP is shown in the third column, and merges between the FM4-64 and GFP images are shown in the fourth column. Time zero (T0) represents the onset of sporulation, and the numbers for the other time points (shown on the left) indicate the time (in hours) after T0. Bars, 2 μm.
It is not known whether YwcE-GFP localizes to both the inner and outer membranes of the spore. However, in agreement with the temporal profile of ywcE-lacZ expression (Fig. 3), fluorescence resulting from GFP accumulation in a strain (AH3279) expressing a transcriptional fusion of the ywcE promoter to gfp was also detected from hour 1 (T1) after the onset of sporulation in cells that had not yet formed a prespore (not shown). The fusion protein YwcE-GFP also accumulated from T1 in cells that did not show any signs of asymmetric division, as assessed by staining with the membrane dye FM4-64 (Fig. 4). Since YwcE production is induced prior to the asymmetric division of the sporangium and decorates both the mother cell and the prespore membranes, we infer that YwcE is likely to be localized in both the inner and outer prespore membranes.
The ywcE gene is required for proper spore morphogenesis.
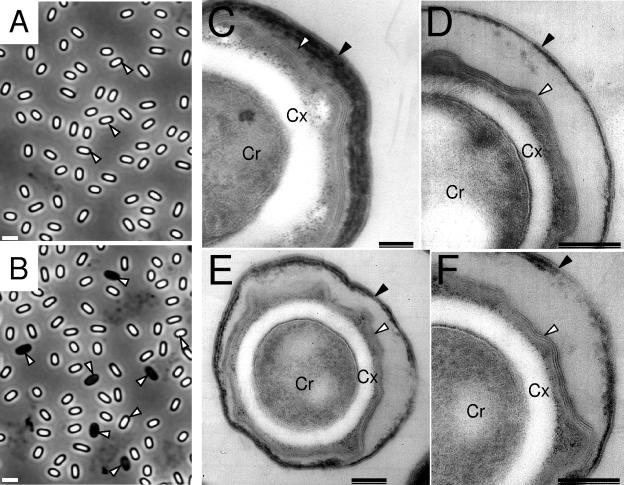
We examined whether ywcE expression was required for proper spore morphogenesis. In preliminary experiments, we observed spores purified from 24-h cultures of the ywcE mutant grown in DSM by phase-contrast microscopy. We found that almost 50% of the ywcE spores were phase dark, whereas <0.1% phase-dark spores were found for the wild-type strain (Fig. 5).
FIG. 5.
Structural alterations of ywcE mutant spores. Spores produced by MB24 (A and C) and by the ywcE insertional mutant AH630 (B, D, E, and F) were purified from 24-h DSM cultures of both strains and observed by phase-contrast microscopy (A and B) or transmission electron microscopy (C to F). The arrowheads in panel A point to phase-bright spores, whereas in panel B the arrowheads point to phase-dark spores. In panels C to F, black arrowheads point to the outer coat layer, and white arrowheads point to the inner coat structure. Cx, cortex; Cr, spore core. Bars, 1 μm in panels A and B and 0.2 μm in panels C to F.
Wild-type spores as well as ywcE spores were also observed by transmission electron microscopy. Wild-type spores exhibited a well-developed cortex layer and a coat that was closely apposed to it; the coat presented the normal pattern of structural differentiation into an inner lamellar layer and a thick, electron-dense, striated outer coat (Fig. 5). In contrast, ywcE spores showed several features that deviated from the wild-type pattern. The spore cores of ywcE mutant spores in the electron micrographs exhibited a punctate pattern, presumably due to the ribosomes (Fig. 5). The inner spore coat was adjacent to the cortex and did not appear reduced or otherwise compromised, but the outer coat formed a thin layer, essentially lacking the striated pattern of wild-type spores, and it did not adhere to the underlying inner coat (Fig. 5). Because the electron micrographs of ywcE spores revealed a reduced outer coat layer, we analyzed the coat polypeptide composition of ywcE spores by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). We observed only a few differences between the coat protein profiles of wild-type and ywcE mutant spores. We found that CotG (36 kDa) and CotB (64 kDa), both of which are outer coat proteins (50, 60), appeared to be more extractable from ywcE spores. This result is consistent with the observation that CotG is also more easily extractable (presumably less cross-linked) from sodA mutant spores. Moreover, mutations in sodA or cotG cause a structural alteration of the spore outer coat that is reminiscent of the phenotype described here for ywcE spores (27). The only other differences noted between the protein patterns of wild-type and ywcE spores were two minor unidentified proteins that were missing from ywcE spores. We also investigated the presence of SafA among the collection of spore coat polypeptides that could be extracted from the coats of ywcE spores by immunoblot analysis. However, we detected no difference in the patterns of assembly of SafA between wild-type spores and spores of the ywcE mutant (data not shown).
Because we found that expression of the σK-controlled gerE gene (9) occurred at the normal time and level in sporulating cells of the ywcE mutant (data not shown), the structural features seen for the ywcE mutant spores did not seem to arise from delayed sporulation. Therefore, the absence of the ywcE gene results in spores with structural deficiencies in the core and the outer coat layers. In spite of these alterations, ywcE spores showed wild-type levels of resistance to heat and lysozyme treatment (data not shown).
YwcE is required for normal DPA accumulation.
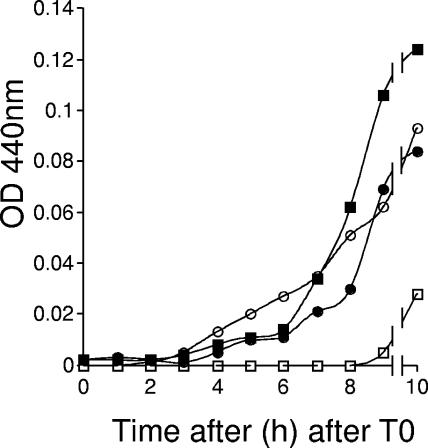
The presence of phase-dark spores in cultures of the ywcE mutant could be due to increased levels of core water. DPA-free spores show higher levels of core water and are less heat resistant than wild-type spores (42, 43). Since we found no impairment in the expression of at least one σK-governed gene (data not shown) and since the dipicolinate synthetase (SpoVFAB) is expressed under σK control (11), we assumed that the mutant cells were capable of producing DPA but could accumulate it in reduced amounts. We measured the levels of DPA in both the culture supernatants and developing spores of both the wild-type and ywcE mutant strains. DPA started to accumulate in wild-type sporulating cells between hours 4 and 5 of sporulation, and there was a marked increase from hour 6 onwards (Fig. 6). At least until hour 9, DPA remained undetectable in the supernatant of a wild-type culture. DPA accumulation also began between hours 4 and 5 of sporulation in the mutant, and it also increased from hour 6 onwards (Fig. 6). However, DPA accumulation in the mutant was slower than that in the wild type, and the peak level of DPA (at hour 16) in the mutant was about 40% lower than that in the wild type (Fig. 6). Also, in contrast to the case for the wild type, DPA accumulated in the culture medium in parallel with its accumulation in ywcE spores. These results indicate that the mutant produces DPA but loses about 40% of it to the medium. As a consequence, the levels of DPA in ywcE mutant spores are reduced relative to those in the wild type. The effect on DPA accumulation caused by the ywcE mutation was complemented by ectopic expression of ywcE-gfp (data not shown).
FIG. 6.
The ywcE mutation affects DPA accumulation. The graph shows DPA accumulation in sporulating cells and culture supernatants of various strains throughout sporulation in DSM. The strains used were as follows: MB24, wild-type cells (filled squares) and supernatant (open squares); and AH630, ywcE::neo cells (filled circles) and supernatant (open circles).
The ywcE gene is involved in spore germination.
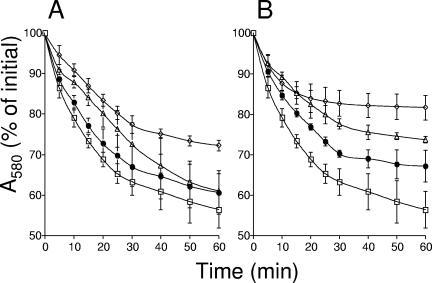
Hydrolysis of the spore cortex, which takes place during germination, results from the activity of two cortex-lytic enzymes, SleB and CwlJ, which have redundant roles (1, 6, 8). It is not clear what activates SleB (reviewed in reference 52). However, since wild-type but not cwlJ spores are induced to germinate by the addition of exogenous Ca2+-DPA, it seems that this compound directly or indirectly activates CwlJ (42-44). Since ywcE spores accumulated reduced levels of DPA, we reasoned that the germination of ywcE spores would be less dependent on CwlJ than on SleB. To test this idea, we examined the nutrient-induced germination of spores from strains mutated singly or doubly in cwlJ, sleB, and/or ywcE. ywcE spores germinated at a slightly lower rate than wild-type spores in response to l-alanine and attained a slightly lower final level of germination (Fig. 7). In agreement with previous observations (1, 6, 8), sleB and cwlJ spores were somewhat affected in both their initial rates of germination with l-alanine and their final levels of germination, and the double mutant was severely affected (Fig. 7). The introduction of a ywcE mutation into sleB cells did not greatly affect the phenotype of the sleB single mutant. However, spores of the cwlJ ywcE double mutant were much more affected in their response to the germinant l-alanine than were spores of either single mutant. Thus, in contrast to our expectations, these results suggest that ywcE affects germination independently of CwlJ function. The ywcE mutation had no effect on spore germination in response to a mixture of asparagine, glucose, fructose, and potassium and did not change the germination phenotype of sleB or cwlJ spores in response to this mix.
FIG. 7.
The ywcE mutation affects spore germination. The graphs show the change in optical density associated with germination in response to l-alanine of spores produced by the following strains: for panel A, MB24 (wild type) (open squares), AH630 (ywcE::neo) (filled circles), AH3443 (ΔcwlJ::tet) (open triangles), and AH3445 (ΔywcE::neo ΔcwlJ::tet) (open diamonds); for panel B, MB24 (wild type) (open squares), AH3442 (ΔsleB::sp) (filled circles), AH3444 (ΔywcE::neo ΔsleB::sp) (open triangles), and AH3433 (ΔsleB::sp ΔcwlJ::tet) (open diamonds). Germination was monitored for 60 min, and samples were collected every 5 or 10 min for absorbance readings at 580 nm.
DISCUSSION
The ywcE gene is expressed at the onset of sporulation from a σA-type promoter. We base this conclusion primarily on the consensus −10 motif in the ywcE promoter and on the observation that σA RNA polymerase utilized the ywcE promoter in vitro. The −35 region of the ywcE promoter deviated significantly from the consensus for σA-type promoters. However, the ywcE promoter appears to have an extended −10 region, which would presumably compensate for a weaker interaction of the polymerase with the −35 promoter element (22). Transcription of the ywcE gene is repressed during growth by the transition-state regulator AbrB (29, 54), and Spo0A-dependent repression of abrB at the onset of sporulation results in the activation of ywcE transcription in predivisional cells.
ywcE mutant spores were found to accumulate reduced levels of DPA, part of which was lost to the medium. DPA-free spores show reduced heat resistance (43), but ywcE spores were heat resistant. We infer that the decrease in DPA accumulation is not sufficient to affect the development of spore heat resistance. The reduced accumulation of DPA in the mutant is intriguing. One possibility is that YwcE is required to facilitate the localization to the cortex region of a factor (or factors) required for normal formation of the spore cortex and that in its absence the altered cortex structure is not able to promote the normal uptake or retention of DPA in the spore core. We speculate that the factors postulated to be controlled by YwcE could be proteins that are involved in both cortex and coat assembly because ywcE spores also show a defective coat structure. There are several proteins that associate with the coat layers that appear to have roles in spore cortex synthesis or degradation during spore germination. For instance, CwlJ, one of the two major cortex-lytic enzymes, and YaaH, which shares sequence similarity with CwlJ and other cortex-lytic enzymes, are both coat components (2, 34). It is not currently known whether the assembly or function of these or other proteins is controlled by YwcE. CwlJ and SleB have redundant roles in cortex hydrolysis during spore germination (1, 6, 8). CwlJ can be activated by either endogenous DPA, released from the spore core during the initial stages of germination, or exogenous DPA. However, a mutation in the ywcE gene exacerbated the germination phenotype of a cwlJ mutant specifically in response to l-alanine. It is not known what triggers SleB activity, although it has been suggested that the enzyme could only be active on peptidoglycan with a higher degree of stress than that found in dormant spores (52). The expression of ywcE was found to be important for germination with l-alanine in the absence of CwlJ, suggesting that ywcE may be required for the localization or function of SleB or that ywcE somehow affects the structure of the peptidoglycan that serves as a substrate for SleB. However, we cannot exclude the possibility that other alterations in the spore morphology of the ywcE mutant, such as the abnormal coat structure, affect SleB function.
The B. subtilis ywcE gene encodes a protein with features of the class I holins, of which the S protein from phage λ is a prototype member (18). YwcE is an 83-residue protein with three transmembrane domains and a highly charged C-terminal tail. Moreover, YwcE has a dual start motif, which plays a role in the regulation of class I or class II holins (3, 18, 59). However, YwcE differs from the λ S protein in two ways. First, the residue between the second and third methionines in YwcE is acidic (aspartate), whereas in the S protein the two methionines are separated by a basic (lysine in λ S) residue (Fig. 1C). However, the phage P1 holin LydA also contains an aspartate near its N terminus, and the holin Hol118 from the Listeria monocytogenes phage A118 contains an acidic residue between its two start codons (59). Second, YwcE is likely to have its N terminus on the outside of the membrane and its C terminus in the cytoplasm, whereas for λ S the N terminus is predicted to face the cytoplasm (18). We base our prediction for the membrane topology of YwcE on the observation that the fusion of the C-terminal region of YwcE to GFP results in a functional protein (at least when the accumulation of DPA is assessed as a functional criterion [see below]) which decorates both the mother cell and prespore membranes (Fig. 5). GFP does not fold properly when attached to periplasmic domains of inner membrane proteins in E. coli and hence has been used to discriminate between cytoplasmic and periplasmic domains (12). Because YwcE has only three predicted transmembrane domains, the only way for the C-terminally fused GFP moiety to remain in the cytoplasm is by assuming an N terminus-out, C terminus-in topology (Fig. 1C). We suspect that the different topology of YwcE from that of the λ S protein may be related to the particular arrangement of the prespore membranes, but since the exact role of YwcE in sporulation is currently unknown, this assumption remains to be tested. It is not known whether YwcE functions as a holin. We have constructed a fusion of the IPTG-dependent Pspac promoter to the ywcE gene but found no IPTG-dependent cell lysis during growth or sporulation (not shown). In addition, we found no delay in lysis of the mother cell during the late stages of sporulation for the ywcE mutant (not shown). The activity of holins is usually inhibited by the presence of a homologous antiholin (59). In the λ system, the antiholin, which inhibits holin activity, is the longer product of the S gene (S107), whereas the shorter protein (S105), which is the protein that initiates at the second methionine of the dual start motif, is the holin (reviewed in reference 59). However, there are reports of holins and their cognate antiholins being encoded by different chromosomal loci. For example, the lrgA gene of Staphylococcus aureus functions in a manner analogous to an antiholin, presumably controlling the activity of a holin encoded by the cidA gene (19, 48). Because of the similarity between holins and antiholins, we cannot exclude the possibility that the ywcE gene encodes an antiholin for which the cognate holin is unknown. If ywcE is an antiholin, then the phenotypes reported herein are due to uncontrolled activity of an as yet unknown holin. Alternatively, it is possible that the structural similarity of YwcE to holins and their antiholins is fortuitous and that the role of YwcE in sporulation is unrelated to a pore-forming capacity.
Acknowledgments
We thank David Popham for the gift of strains and Anne Moir and D. Popham for advice.
This work was supported by grant POCTI/BCI/48647/2002 from Fundação para a Ciência e a Tecnologia (F.C.T.) to A.O.H. and by NIH grant GM54395 to C. P. Moran, Jr. G.R. was the recipient of a Ph.D. fellowship (PRAXIS XXI/BD/21560/99) from the F.C.T.
REFERENCES
- 1.Atrih, A., and S. J. Foster. 2001. In vivo roles of the germination-specific lytic enzymes of Bacillus subtilis 168. Microbiology 147:2925-2932. [DOI] [PubMed] [Google Scholar]
- 2.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenboim, M., C. Y. Chang, F. dib Hajj, and R. Young. 1999. Characterization of the dual start motif of a class II holin gene. Mol. Microbiol. 32:715-727. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 5.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boland, F. M., A. Atrih, H. Chirakkal, S. J. Foster, and A. Moir. 2000. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 146:57-64. [DOI] [PubMed] [Google Scholar]
- 7.Buckner, C. M., G. Schyns, and C. P. Moran, Jr. 1998. A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation. J. Bacteriol. 180:3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirakkal, H., M. O'Rourke, A. Atrih, S. J. Foster, and A. Moir. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383-2392. [DOI] [PubMed] [Google Scholar]
- 9.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 10.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley and Sons, Ltd., New York, N.Y.
- 11.Daniel, R. A., and J. Errington. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J. Mol. Biol. 232:468-483. [DOI] [PubMed] [Google Scholar]
- 12.Drew, D., D. Sjostrand, J. Nilsson, T. Urbig, C. N. Chin, J. W. de Gier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 15.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 16.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-42. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 17.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 18.Graschopf, A., and U. Blasi. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 19.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 21.Helmann, J. D., and C. P. Moran, Jr. 2001. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 22.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henner, D. J. 1990. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 185:223-228. [DOI] [PubMed] [Google Scholar]
- 24.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques, A. O., E. M. Bryan, B. W. Beall, and C. P. Moran, Jr. 1997. cse15, cse60, and csk22 are new members of mother-cell-specific sporulation regulons in Bacillus subtilis. J. Bacteriol. 179:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 27.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 29.Hoch, J. A. 1995. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction system, p. 129-144. In T. J. Silhavy and J. A. Hoch (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 30.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 31.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 34.Kodama, T., H. Takamatsu, K. Asai, K. Kobayashi, N. Ogasawara, and K. Watabe. 1999. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J. Bacteriol. 181:4584-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 36.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478-488. [DOI] [PubMed] [Google Scholar]
- 38.Moriyama, R., H. Fukuoka, S. Miyata, S. Kudoh, A. Hattori, S. Kozuka, Y. Yasuda, K. Tochikubo, and S. Makino. 1999. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J. Bacteriol. 181:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mota, L. J., L. M. Sarmento, and I. de Sa-Nogueira. 2001. Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping. J. Bacteriol. 183:4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 183:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paidhungat, M., and P. Setlow. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 46.Quisel, J. D., D. C. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 47.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55:349-367. [DOI] [PubMed] [Google Scholar]
- 48.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano, M., A. Neves, C. M. Soares, C. P. Moran, Jr., and A. O. Henriques. 2004. Role of the anti-sigma factor SpoIIAB in regulation of sigmaG during Bacillus subtilis sporulation. J. Bacteriol. 186:4000-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 53.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, C. M., R. Daniel, N. Illing, and J. Errington. 1992. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatti, K. M., and C. P. Moran, Jr. 1996. RNA polymerase sigma factors of Bacillus subtilis: purification and characterization. Methods Enzymol. 273:149-162. [DOI] [PubMed] [Google Scholar]
- 58.Tatti, K. M., and C. P. Moran, Jr. 1995. Sigma E changed to sigma B specificity by amino acid substitutions in its −10 binding region. J. Bacteriol. 177:6506-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 60.Zilhao, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]