Abstract
The apparatus responsible for translocation of proteins across bacterial membranes is the conserved SecY complex, consisting of SecY, SecE, and SecG. Prior genetic analysis provided insight into the mechanisms of protein export, as well as the interactions between the component proteins. In particular, the prl suppressor alleles of secE and secY, which allow export of secretory proteins with defective signal sequences, have proven particularly useful. Here, we report the isolation of novel mutations in secE and secY, as well as the phenotypic effects of combinations of prl mutations. These new alleles, as well as previously characterized prl mutations, were analyzed in light of the recently published crystal structure of the archaeal SecY complex. Our results support and expand a model of Prl suppressor activity that proposes that all of the prlA and prlG alleles either destabilize the closed state of the channel or stabilize the open form. These mutants thus allow channel opening to occur without the triggering event of signal sequence binding that is required in a wild-type complex.
Transport of proteins across lipid bilayers to extracytoplasmic destinations is essential for growth of all organisms. In gram-negative bacteria, such as Escherichia coli, the most generally employed mechanism for translocating proteins across the inner membrane is the Sec pathway. Sec-dependent secretory proteins are synthesized in the cytoplasm with cleavable amino-terminal signal sequences. These precursor proteins often require binding by the export-specific chaperone SecB to maintain the precursor in a loosely folded conformation suitable for export. SecA interacts with both the precursor protein and SecB to form a ternary complex that is directed to the membranous translocation machinery. The core of the translocation complex consists of SecY, SecE, and SecG, all integral membrane proteins; these three proteins interact to form the SecY complex that physically transports proteins across the membrane (for reviews, see references 9, 12, 23, and 29). Understanding the functions of the Sec proteins, the interactions among them, and the structure of the translocation complex is vital to fully elucidating the process of protein translocation.
The SecY complex is conserved throughout evolution (7). The largest subunit is the SecY homolog (SecY in eubacteria and archaea, Sec61α in mammals, and Sec61p in yeast), which forms the channel core (19, 45). The SecE subunits (SecE in eubacteria and archaea, Sec61γ in mammals, and Sss1p in yeast) are smaller proteins, and although E. coli SecE has three membrane-spanning domains, its homologs in most other organisms consist of a single transmembrane segment (19). The nonessential SecG subunit does not show sequence conservation, but all of the Sec complexes contain a third small protein; thus, it is thought that SecG (eubacteria), Secβ (archaea), Sec61β (mammals), and Sbh (yeast) fulfill analogous roles (19, 25).
Most of the Sec proteins were originally identified via elegant genetic screens (for reviews, see references 4, 9, and 34). The sec alleles were defined as conditional-lethal mutations that conferred generalized protein export defects; such mutations have been found in secA, secD, secE, secF, and secY. In contrast, the prl alleles were isolated as suppressors that allow export of signal sequence-defective precursors and encode dominant mutations. Originally, prl alleles were identified in secA (prlD), secE (prlG), and secY (prlA). More recently, prlH alleles of secG have been characterized as well (6). It is critical to recognize that the sec and prl alleles are fundamentally different types of mutations. The sec alleles result in nonfunctional protein products under restrictive conditions, while the prl protein products not only retain function, but expand the repertoire of substrate secretory proteins to include those with mutant signal sequences or, indeed, with no signal sequence at all (10, 14). The prl alleles are not promiscuous in allowing nonsecretory proteins to be exported (27); however, this may be a secondary effect attributable to lack of targeting of these proteins to the SecY complex.
DNA sequence analysis of the sec and prl mutants, combined with predictions of secondary structure and membrane topology (1, 11, 15, 20, 24), led to initial rudimentary analyses of the topological location of each mutation (Table 1 and references therein). The secY mutations are scattered throughout the gene, consistent with the loss-of-function defect of these alleles. The secE mutations fall primarily in the region encoding the ribosome binding site or initial codons of the gene and most, possibly all, exert their effects by decreasing expression of secE rather than causing structural alterations to the protein (35). In contrast, it was observed that the prl mutations are more localized. The prlA mutations are found primarily in three domains of SecY: the 1st periplasmic loop (P1), the 7th transmembrane domain (TM7), and the 10th transmembrane helix (TM10) (24). Likewise, the prlG alleles of secE are localized to the third transmembrane region (TM3) and the second periplasmic domain (P2) (35).
TABLE 1.
Mutant alleles of secY and secEa
| Allele name | Mutation (E. coli) | Amino acid (M. jannaschii) | Domain | Reference |
|---|---|---|---|---|
| SecY | ||||
| secY24 | G240D | M229 | C4 | 37 |
| secY39 | R357H | A355 | C5 | 2 |
| secY40 | A363S | K364 | C5 | 2 |
| secY100 | P40S, A46V, G167E | D44, A50, G150 | TM1, P1, TM4 | 21 |
| secY104 | G175D | D158 | C3 | 42 |
| secY110 | R357C | R360 | C5 | 42 |
| secY115 | A363T | K364 | C5 | 42 |
| secY117 | G184D | G167 | TM5 | 42 |
| secY119 | P388S | L388 | TM9 | 42 |
| secY121 | I290T | W272 | TM7 | 31 |
| secY122 | G359R | S362 | C5 | 42 |
| secY124 | P84L | P87 | TM2 | 42 |
| secY125 | S76F | T69 | P1/TM2 | 42 |
| secY129 | C385Y | V385 | TM9 | 42 |
| secY161 | P287L | I269 | TM7 | 31 |
| secY205 | Y429D | L426 | C6 | 42 |
| prlA1 | V274G | N256 | TM7 | 13, 24 |
| prlA3 | F67C | I62 | P1b | 13, 32 |
| prlA4 | I408N (F286Y) | L406 | TM10 (TM7) | 13, 32 |
| prlA6 | I408N (S188L) | L406 | TM10 (TM5) | 13, 24 |
| prlA7 | L407R (A277E) | L405 | TM10 (TM7) | 13, 24 |
| prlA9 | G69D | A64 | P1b | 13, 24 |
| prlA11 | L407R (V411G) | L405 | TM10 (TM10) | 13, 24 |
| prlA200 | I191S | A174 | TM5 | 16, 24 |
| prlA202 | I278S | I260 | TM7 | 16, 24 |
| prlA205 | G69C | A64 | P1b | 16, 24 |
| prlA208 | I278N | I260 | TM7 | 16, 24 |
| prlA300 | F64C | W59 | P1b | 24 |
| prlA301 | L407R | L405 | TM10 | 24 |
| prlA302 | A71D | R66 | P1 | 24 |
| prlA303 | I278T | I260 | TM7 | 24 |
| prlA304 | I90N | I83 | TM2 | 24 |
| prlA306 | ΔS73 | P1 | 24 | |
| prlA401 | S282R | A264 | TM7 | 3, 32 |
| prlA666 | F67S | I62 | P1b | 28 |
| prlA726 | S68P | T63 | P1b | 14 |
| prlA799 | S68L | T63 | P1b | 15 |
| prlA8911 | S37F | G41 | TM1 | 10, 15 |
| prlA8913 | S68F | T63 | P1b | 10, 15 |
| prlA8914 | N65Y | Q60 | P1b | 10, 15 |
| secY(P42L) | P42L | Y46 | TM1 | This work |
| secY(F154C) | F154C | P137 | P2 | This work |
| SecE | ||||
| secE11 | N4Y | Start | 35, 36 | |
| secE12 | R12L | C1 | 35, 36 | |
| secE13 | N4N (T to C) | Start | 35, 36 | |
| secE15 | RBSc (G to A) | 5′ | 35 | |
| secE501 | −1 T to G | 5′ | 30 | |
| prlG1 | L108R | L48 | TM3 | 35, 41 |
| prlG2 | S105P | A45 | TM3 | 35, 41 |
| prlG3 | S120F | T60 | P2 | 35, 41 |
| prlG8 | ΔV116-R117 | H56-V57 | P2 | 14 |
| secE(T123P) | T123P | K63 | P2 | This work |
| secE(D112Y) | D112Y | G52 | TM3/P2 | This work |
All of the published and well-characterized alleles of secY and secE are shown, with amino acid alteration and topological location based on the original structural predictions. The amino acid alignment of E. coli and M. jannaschii sequences was based on supplemental Fig. 1 from van den Berg et al. (45). Domains are indicated as cytoplasmic (C), periplasmic (P), or transmembrane (TM), according to original topological predictions. For alleles that were originally isolated as double mutants (prlA4, prlA6, prlA7, and prlA11), both mutations are indicated, with the secondary mutation (non-prl mutation) in parentheses.
P1 mutations now known to be in the plug (TM2a).
RBS, ribosome binding site.
Originally, it was thought that prl mutations were located in domains of SecE and SecY that interact with the signal sequence and that they exerted their effects through altered protein-protein interactions (13). This hypothesis was abandoned following two significant observations. First, prl-mediated suppression of signal sequence mutations displays no allele specificity: with a few unusual exceptions, every prlA and every prlG allele is able to export any secretory protein with any signal sequence mutation (10, 14, 24). Second, all of the prlA and prlG suppressors promote export of secretory proteins that completely lack signal sequences, again suggesting that suppression is not due to altered interactions between the signal sequence and the Sec apparatus (10, 14).
These findings led to the hypothesis that wild-type SecE and SecY provide a proofreading mechanism whereby secretory proteins that are delivered to the SecY complex are exported only if they have a functional signal sequence (14, 24). A Prl suppressor of SecY or SecE would no longer perform this proofreading function, thereby allowing export of any secretory protein delivered to the SecY complex, even those with nonfunctional signal sequences. This hypothesis is supported by the observation that all prlA- and prlG-based suppression is dependent on SecA and SecB, suggesting that targeting to the SecY complex is a critical step for export of a mutant secretory protein (10, 14, 44). Even proteins that normally are secreted independently of SecB become SecB dependent when exported via the Prl pathway (10).
Understanding the mechanism of Prl suppression is intimately connected to discernment of the structure and function of the wild-type Sec apparatus. One approach to deciphering interactions between domains of SecE and SecY was to combine pairs of prl alleles and examine those combinations for altered, or synthetic, phenotypes (5, 15, 24). Out of 113 combinations of prlA and prlG alleles, 7 demonstrated a lethal phenotype. The pairs that showed synthetic lethality were extremely allele specific and topologically correlated, leading to the hypothesis that these alleles mapped to domains of interaction between the two proteins. Specifically, it was predicted that TM10 of SecY and SecE(TM3) interact, that SecY(TM10) also associates with SecY(TM7), and that the first periplasmic domain (P1) of SecY and SecE(P2) interact. Further, it was suggested that SecY(TM7) is the primary domain responsible for signal sequence recognition (15, 24).
Many of these predictions based on genetic analyses were corroborated by the recent elucidation of the crystal structure of the SecY complex from the archaeon Methanococcus jannaschii (45). In general, the early topological predictions (1, 11, 20) were accurate; the major exception was that the domain formerly predicted to be periplasmic loop 1 of SecY was found instead to be folded back into the channel. It is predicted that this region (now called TM2a) constitutes a “plug” that closes the translocation channel and must be displaced for export to occur. In addition to the general topology, the major SecE-SecY and SecY-SecY interactions predicted from synthetic lethality were substantiated by the solution of the SecY complex.
As screening for synthetic lethality was so useful in understanding SecY-SecE interactions, in this study, we sought suppressors of synthetic lethality in an attempt to further our knowledge both of the SecY complex structure and of Prl suppression. Analysis of these new mutations, as well as combinatorial analysis of previously characterized alleles, was merged with the recently released structural information to further our understanding of the mechanism of action of the prl suppressors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains used were derivatives of E. coli K-12 strain MC4100 (8) and are listed in Table 2. Strains were constructed using standard genetic techniques (38). The plasmids used are described in Tables 3 and 4, and all are derivatives of pBAD18 (17). Plasmids pAF26, pAF27, pAF28, and pAF29 have been described previously (15); plasmids pAF65, pAF66, and pAF67 were constructed by PCR amplification of the secY gene from chromosomal DNA isolated from strains AF230 (secY+), AF249 (prlA726), and AF233 (prlA4), respectively (15). Plasmids containing other mutant alleles were constructed by site-directed mutagenesis of one of the above-mentioned plasmids using the QuickChange protocol of Stratagene. All secY and secE sequences (including mutant alleles and original clones) were verified by DNA sequencing performed by MWG Biotech, Inc. (High Point, NC), or by the Colorado State University Sequencing Facility (Ft. Collins, CO). Sequences of oligonucleotides (Midland Certified Reagents) used for cloning, site-directed mutagenesis, and sequencing can be obtained by request. Preparation of competent cells and transformation were performed according to standard protocols (33, 38). Bacteria were grown in Luria-Bertani (LB), MacConkey, or M63 minimal medium supplemented with ampicillin (125 mg/liter), kanamycin (25 mg/liter), or tetracycline (25 mg/liter) when appropriate (38). Maltodextrin was prepared as described previously (43). Induction of plasmid-borne alleles was achieved using 0.2% arabinose.
TABLE 2.
Strains used in this studya
| Strain | Genotype | Source |
|---|---|---|
| MC4100 | F− λ−araD139 Δ(argF-lac)169 rpsL150 relA1 flhD5301 deoC1 fruA25 rbsR22 | 8 |
| AF230 | MC4100 Ara+lamB14D | 15 |
| AF232 | AF230 prlA3 | 15 |
| AF233 | AF230 prlA4 | 15 |
| AF249 | AF230 prlA726 | 15 |
| AF295 | AF249 secE15 zij::Tn5 recA::cam | 15 |
| AF314 | AF232 prlG3 zij::Tn10 | 15 |
| AF680 | MC4100 Δara714 secE15 zij::Tn5 recA::cam | This study |
| AF681 | AF680 lamBΔ78 | This study |
| AF682 | AF680 lamBΔ60 | This study |
| AF683 | AF680 lamB14D | This study |
| AF686 | AF680 lamBΔ111 | This study |
| MS28 | MC4100 secY39 | This study |
| MS29 | MS28 lamBΔ78 | This study |
| MS30 | MS28 lamBΔ60 | This study |
| MS31 | MS28 lamB14D | This study |
| MS32 | MS28 lamBΔ111 | This study |
| MS33 | AF314 slyD::kan | This study |
All strains are derivatives of E. coli K-12 strain MC4100 and were constructed by standard genetic techniques.
TABLE 3.
Phenotypes of mutant secE allelesa
| Line no | Plasmid | Allele | Complementation | Prl phenotype |
|---|---|---|---|---|
| 1 | pAF26 | Wild type | + | − |
| 2 | pAF27 | prlG1 (L108R) | + | + |
| 3 | pAF28 | prlG2 (S105P) | + | + |
| 4 | pAF29 | prlG3 (S120F) | + | + |
| 5 | pMAS15 | secE (T123P) | + | − |
| 6 | pAF71 | secE (D112Y) | + | + |
| 7 | pCH3 | prlG1 prlG2 (L108R, S105P) | + | ++ |
| 8 | pCH1 | prlG1 prlG3 (L108R, S120F) | + | ++ |
| 9 | pCH5 | prlG2 prlG3 (S105P, S120F) | + | ++ |
| 10 | pCH7 | prlG1 prlG2 prlG3 (L108R, S105P, S120F) | + | +++ |
| 11 | pMAS16 | prlG1 secE(T123P) (L108R, T123P) | + | + |
| 12 | pMAS14 | prlG1 secE(D112Y) (L108R, D112Y) | + | ++ |
| 13 | pMAS17 | prlG3 secE(T123P) (S120F, T123P) | − (DN) | NA |
| 14 | pAF72 | prlG3 secE(D112Y) (S120F, D112Y) | + | ++ |
| 15 | pMAS26 | secE(T123P), secE(D112Y) | + | + |
Complementation was measured as the ability of plasmid-borne alleles to rescue the cold-sensitive growth of strain AF680, carrying the chromosomal secE15 allele. Growth at 26°C resulted in a +, no growth was given a −, and DN is a dominant-negative allele. The Prl phenotype was determined by a compilation of dextrin utilization and λ sensitivity in strains containing lamB signal sequence mutations (AF681, AF682, AF683, and AF686). No detectable suppression of mutant lamB received a score of −, while suppression was qualitatively ranked in increasing + marks. Strains that produced a pink color on dextrin MacConkey and detectable λ sensitivity received one +, while red coloration and full λ sensitivity resulted in ++. The triple mutant was darker red than the double mutants, giving a +++ designation. NA indicates not applicable.
TABLE 4.
Phenotypes of mutant secY allelesa
| Line no. | Plasmid | Allele | Complementation | Prl phenotype |
|---|---|---|---|---|
| 1 | pAF65 | Wild type | + | − |
| 2 | pCH8 | prlA3 (F67C) | + | +++ |
| 3 | pAF67 | prlA4 (I408N) | + | +++ |
| 4 | pCH12 | prlA301 (L407R) | + | +++ |
| 5 | pAF66 | prlA726 (S68P) | + | +++ |
| 6 | pMAS6 | secY(P42L) (P42L) | + | − |
| 7 | pMAS19 | secY(F154C) (F154C) | + | − |
| 8 | pCH14 | prlA3 prlA4 (F67C, I408N) | − | NA |
| 9 | pCH10 | prlA3 prlA301 (F67C, L407R) | + | +++ |
| 10 | pCH6 | prlA3 prlA726 (F67C, S68P) | + | +++ |
| 11 | pCH4 | prlA4 prlA301 (I408N, L407R) | − | NA |
| 12 | pCH2 | prlA4 prlA726 (I408N, S68P) | − (DN) | NA |
| 13 | pMAS7 | prlA3 secY(P42L) (F67C, P42L) | + | +++ |
| 14 | pMAS8 | prlA4 secY(P42L) (I408N, P42L) | − | NA |
| 15 | pMAS9 | prlA726 secY(P42L) (S68P, P42L) | + | +++ |
| 16 | pMAS20 | prlA3 secY(F154C) (F67C, F154C) | + | +++ |
| 17 | pMAS21 | prlA4 secY(F154C) (I408N, F154C) | + | +++ |
| 18 | pMAS22 | prlA726 secY(F154C) (S68P, F154C) | + | +++ |
| 19 | pMAS25 | secY(P42L), secY(F154C) | + | − |
Complementation was measured as the ability of plasmid-borne alleles to rescue the cold-sensitive growth of strain MS28, carrying the chromosomal secY39 allele. Growth at 20°C resulted in a +, no growth was given a −, and DN is a dominant-negative allele. The Prl phenotype was determined by a compilation of dextrin utilization and lambda sensitivity in strains containing lamB signal sequence mutations (MS29, MS30, MS31, and MS32). No detectable suppression of mutant lamB received a score of −, while suppression was qualitatively ranked in increasing + marks as described in Table 3, note a. NA indicates not applicable.
Isolation of suppressors of synthetic lethality.
Suppressors of the cold-sensitive (Cs) phenotype conferred by the combination of prlA3 and prlG3 were isolated by plating aliquots of an overnight culture of strain AF314, grown at 37°C, onto LB plates and incubating them at 26°C. Individual colonies that arose were restreaked on LB plates and incubated at 37°C. These purified colonies then were streaked onto LB plates and incubated at 26°C; growth at 26°C was compared to that of the parental strain to verify the cold-resistant phenotype. Bacteriophage P1 mapping (38) was used to determine whether suppressors mapped to the vicinity of either prlG3 or prlA3. We did not find any suppressors that mapped to locations other than prlA or prlG. Following localization of the suppressor mutation, the prlA or prlG gene was amplified by PCR and the DNA sequence was determined.
To isolate suppressors of prlA726-prlG3 arabinose sensitivity, the prlG3 gene from plasmid pAF29 was amplified by mutagenic PCR (39), digested with EcoRI and BamHI, and ligated into pBAD18. Ligation products were transformed into AF295 and plated on LB-ampicillin-arabinose medium at 26°C to identify plasmids that conferred both cold resistance and arabinose resistance. This approach ensured that the plasmid expressed a functional gene product (to complement the cold sensitivity of secE15); thus, mutations that eliminated expression of the prlG3 allele were not selected. Those plasmids that allowed growth were isolated and retransformed into AF295 to verify the cold- and arabinose-resistant phenotypes, and then the prlG portion of the plasmid was subjected to DNA sequence analysis.
Characterization of mutant alleles.
Newly constructed mutant alleles of secE and secY were characterized for complementation ability by transforming strains AF680 (secE15) or MS28 (secY39) with the plasmids and assessing growth at the restrictive temperature of 26°C or 20°C, respectively. Prl suppressor activity was assessed phenotypically by transformation of strains containing lamB signal sequence mutations (AF681, AF682, AF683, and AF686 for secE mutants or MS29, MS30, MS31, and MS32 for secY alleles), followed by streaking colonies on dextrin MacConkey agar supplemented with 125 mg/liter ampicillin and 0.2% arabinose and incubation at either 37°C or the restrictive temperature. Additionally, suppression was assayed by cross-streaking the same plasmid-containing strains against λvir to assess sensitivity to λ infection.
Immunoblot analysis.
Strains containing plasmids expressing various secE suppressor alleles were assayed for steady-state levels of precursor and mature LamB14D as an indication of export. Plasmids were introduced into AF683 (secE15 lamB14D). Overnight cultures were grown at 37°C in LB-ampicillin medium and then subcultured into LB-ampicillin-arabinose medium at 26°C. At an A600 of ∼0.2, maltose was added to 0.2% to induce lamB expression. After 60 min, samples were removed and prepared for polyacrylamide gel electrophoresis by trichloroacetic acid precipitation on ice for 20 min. Following pelleting of the proteins, samples were resuspended in loading buffer, boiled, and analyzed on 7.5% polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose, and immunoblotting was performed using polyclonal antibody directed against LamB (14).
RESULTS
Suppressors of synthetic lethality.
Because synthetic-lethality studies had contributed to our understanding of SecYE structure, we predicted that suppressors of the lethal phenotypes would likewise expand our interpretation of the interactions between SecE and SecY. We took advantage of the unique features of two synthetic-lethal pairs to isolate such suppressors. Previous studies demonstrated that seven pairs of prlA (secY) and prlG (secE) alleles exhibit synthetic phenotypes (15); however, the severities of the combinatorial defects vary, resulting in different phenotypes. For example, the cold sensitivity conferred by the prlA3-prlG3 combination is recessive to wild-type alleles of either secY or secE, while the arabinose-sensitive phenotype of the prlA726-prlG3 pairing (due to the prlG3 allele expressed from an arabinose-inducible promoter) is dominant to both wild-type alleles. As they both provide selectable phenotypes, we sought suppressors of the lethality conferred by each of these pairs of alleles.
(i) Suppressors of prlA3 and prlG3.
As demonstrated previously (15), a strain containing both prlA3 (SecY F67C) and prlG3 (SecE S120F) on the chromosome is viable but exhibits a cold-sensitive phenotype, with poor growth at temperatures below 30°C. We isolated suppressors of the synthetic defect by selecting for spontaneous mutants that grew well at 26°C. Bacteriophage P1 mapping was used to determine whether any of the suppressor mutations mapped in proximity to either secY or secE.
Twelve mutants were identified that fulfilled these criteria; one contained a suppressor that mapped near or in secE; the other 11 suppressors cotransduced with secY. The secE or secY gene, as appropriate, was PCR amplified from the cold-resistant suppressor strains, and the DNA sequence was determined. Each isolate retained the original prlG3 or prlA3 mutation, and each had an additional novel mutation in either secE or secY. The single secE suppressor mutation resulted in an alteration of amino acid 123 from threonine to proline. The secY suppressors changed either amino acid 42 from proline to leucine or amino acid 154 from phenylalanine to cysteine.
(ii) Suppressors of prlA726 and prlG3.
The combination of prlA726 (SecY S68P) and prlG3 (SecE S120F) is particularly detrimental and confers sensitivity to arabinose induction of plasmid-borne prlG3 at any temperature, even in the presence of chromosomal prlG+ (secE+) (15). We initially sought spontaneous suppressors of this lethality by plating strain AF295 (prlA726) carrying plasmid pAF29 (prlG3 under arabinose regulation) at 26°C on LB-ampicillin plates containing 0.2% arabinose and selecting for arabinose-resistant mutants. Every isolate that we identified by this approach had one of two alterations to the secY sequence: either reversion of the prlA726 allele to wild type or a second mutation to the same codon, resulting in a leucine residue. This particular change, known as prlA799, has been observed previously (15).
To increase our chances of isolating new mutations, we performed mutagenic PCR using plasmid-borne prlG3 as the template, recloned the PCR product into pBAD18, transformed the resultant plasmids into strain AF295 (prlA726), and selected arabinose-resistant colonies at 26°C. Such colonies were verified by restreaking them on arabinose, and then the plasmids were isolated and retransformed into AF295 (prlA726) to confirm that the suppressor was carried on the plasmid. Only one such suppressor was identified; the plasmid DNA from this suppressor was isolated and sequenced. The original prlG3 mutation was still present, along with a suppressor mutation that altered codon 112 of secE from aspartate to tyrosine.
(iii) Characterization of suppressors of synthetic lethality.
Thus, in our search for suppressors of synthetic lethality, we obtained four new mutations, two each in secE and secY. To distinguish these suppressors from prl suppressors, they will be referred to as ssl (suppressor of synthetic lethality) alleles but for clarity will retain the sec nomenclature for allele names. To characterize these new ssl mutations, we used site-directed mutagenesis to introduce the new mutations into the corresponding wild-type plasmid-borne gene to create alleles that contained only the new mutations. The resulting alleles were tested for the ability to complement cold-sensitive mutations of either secE or secY and also to function as suppressors of signal sequence mutations (the Prl phenotype).
We thought it possible that synthetic lethality could result from such a drastic perturbation of SecY complex structure that suppressors would cause a significant but compensatory alteration, and it was conceivable, therefore, that the suppressors would be functional only in combination with the original prl mutation. Therefore, complementation of a chromosomal cold-sensitive allele was used to assess the functionality of mutants containing the single ssl mutation. Both secE(T123P) and secE(D112Y) complemented the secE15(Cs) mutation (Table 3, lines 5 and 6), while secY(P42L) and secY(F154C) both promoted growth of a secY39(Cs) strain (Table 4, lines 6 and 7). These results indicated that none of the new mutations interfered with production of a functional protein product and that these mutations did not adversely affect viability.
Similarly, we considered that the structural alteration required to rescue a synthetic phenotype might itself cause a Prl phenotype. When tested against a variety of lamB signal sequence mutations, only secE(D112Y) demonstrated the ability to promote export of signal sequence-defective preproteins, i.e., a Prl phenotype (Table 3, lines 5 and 6, and Table 4, lines 6 and 7). Therefore, although synthetic lethality is due to the combination of two prl suppressor alleles, the structural alterations required to rescue the lethality do not require generation of a Prl translocase. These new ssl alleles differ from all other previously characterized mutations of secE or secY in that they neither destroy function of the protein (sec alleles) nor are necessarily suppressors of signal sequence-defective precursors (prl alleles).
Construction of multiply mutant alleles.
An ongoing question has been whether all prlA and prlG alleles function by the same mechanism to facilitate export of defective preproteins. To partially address this issue, we sought to determine if prl alleles conferred additive or synergistic phenotypes or perhaps were even antagonistic to one another. To test this, we constructed plasmid-borne alleles of either secE or secY that contained two or more previously characterized mutations within the same gene. To start, the mutations chosen were some of those that had previously been identified as partners in synthetic-lethal pairs. After isolation of suppressors of synthetic lethality, we also included those new alleles in these multiple-mutation analyses.
(i) Multiple mutations in secE.
We combined the prlG1 (L108R), prlG2 (S105P), and prlG3 (S120F) mutations into multiply mutant alleles in all pairwise combinations and also combined all three mutations at once. All combinations complemented a chromosomally encoded Cs secE15 allele (Table 3, lines 7 to 10), indicating that multiple prlG mutations in a single gene did not adversely affect the integrity of the protein product.
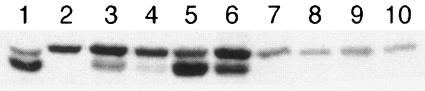
Each multiple mutant then was screened for Prl suppressor activity with a variety of lamB signal sequence mutations. As shown in Table 3 (lines 7 to 9), every double prlG mutant (i.e., combinations of prlG1, prlG2, and prlG3) promoted export of the mutant LamB molecules to a greater extent than did either parent allele (lines 2 to 4), and the triple mutant was a more effective suppressor than any single or double mutant based on these phenotypic assays (line 10). In addition, we performed immunoblot analyses to assess levels of precursor and mature LamB14D in strains carrying each of the plasmids with multiple prlG alleles. This provides an indication of the steady-state levels of mature, and therefore exported, protein. Consistent with the phenotypic assays, each multiple mutant resulted in a greater proportion of mature LamB14D than did any of the single-mutant strains (Fig. 1). Surprisingly, however, by this assay, the triple mutant did not appear to be a stronger suppressor than the double mutants, although it was more effective than single mutants. These results indicate that the alterations to SecYEG translocase caused by the prlG alleles are additive or synergistic in nature.
FIG. 1.
Immunoblot detecting precursor and mature forms of LamB14D. Lane 1, strain AF680 (wild-type LamB). Lane 2, strain AF683 (lamB14D). Lanes 3 to 10, AF683 with plasmids: lane 3, pCH1 (prlG1 prlG3); lane 4, pCH3 (prlG1 prlG2); lane 5, pCH5 (prlG2 prlG3); lane 6, pCH7 (prlG1 prlG2 prlG3); lane 7, pAF26 (prlG1+); lane 8, pAF27 (prlG1); lane 9, pAF71 (secE D112Y); lane 10 pMAS14 (prlG1 secE D112Y). Equivalent A600 units were loaded into each lane.
To address the effects of the newly isolated suppressors of synthetic lethality, we constructed combinations of the prlG1 and prlG3 mutations with the secE ssl alleles, as well as the two ssl alleles together. All except the prlG3-secE(T123P) pair rescued the cold-sensitive phenotype of secE15 (Table 3, lines 11 to 15), indicating that a functional protein product was produced. The unusual combination, prlG3 (S120F) plus secE(T123P) not only was unable to complement the cold-sensitive strain but exhibited a dominant-negative effect (line 13).
The prlG-ssl combinations were also analyzed for Prl activity. As described above, the secE(D112Y) mutation alone resulted in Prl suppressor activity, while secE(T123P) did not. Combinations that included a prlG mutation and either of these secE alleles all exhibited Prl activity, as did the combination of secE(T123P) and secE(D112Y), indicating that the Prl phenotype is dominant within a single molecule (Table 3, lines 11 to 15). Moreover, this demonstrates that the new mutations do not suppress synthetic lethality by quenching the Prl phenotype.
(ii) Multiple mutations in secY.
As with the prlG alleles, we combined selected prlA alleles into multiply mutant genes and examined the phenotypes conferred by those multiple mutations. The prlA3 (F67C), prlA726 (S68P), prlA4 (I408N, F286Y), and prlA301 (L407R) mutations were paired in all possible combinations, except the prlA301-prlA726 combination, which we were unable to construct. We tested the multiple mutants for complementation of a chromosomal cold-sensitive secY39 allele (Table 4, lines 8 to 12). The combinations of prlA3-prlA726 (pCH6) and prlA3-prlA301 (pCH10) complemented the cold-sensitive defect, albeit poorly. This indicates that the protein products produced by these genes were functional, although not as efficient as any single mutant. The prlA4-prlA3 (pCH14) and prlA4-prlA301 (pCH4) pairs were unable to complement secY39, indicating that these combinations resulted in nonfunctional or unstable proteins. The combination of prlA4 and prlA726 (pCH2) not only failed to complement secY39, but was a dominant-negative allele, as evidenced by arabinose sensitivity of wild-type cells carrying this multiple mutation, even at 37°C in the presence of a wild-type secY allele.
These results suggest that the individual alterations imposed by the prlA mutations disrupt the translocase structure and are increasingly detrimental. Thus, multiple prlA mutations can, in some pairs, result in defective complex formation. Indeed, combinations with prlA4 are particularly detrimental, as each of them was unable to complement.
As with the secE multiple mutants, we assessed the multiple secY alleles for Prl activity (Table 4, lines 8 to 12). We were unable to test those that did not complement secY39, leaving us with only two pairs to examine, prlA3-prlA301 and prlA3-prlA726. Both of these retained the capacity to suppress every lamB signal sequence defect tested. If the suppressor activity was greater than with any single mutation, it was not apparent by the phenotypic assays utilized.
Next, we combined three of these prlA mutations (prlA3, prlA4, and prlA726) with our newly isolated secY(P42L) and secY(F154C) alleles. All of the combinations except one were viable, as judged by their ability to complement the secY39 cold-sensitive strain (Table 4, lines 13 to 19). The exception again involved prlA4, the prlA4-secY(P42L) combination (pMAS8). When we examined these combinations for Prl activity, we found again that the Prl phenotype was dominant, as all combinations that contained a prl allele were able to suppress lamB signal sequence mutations.
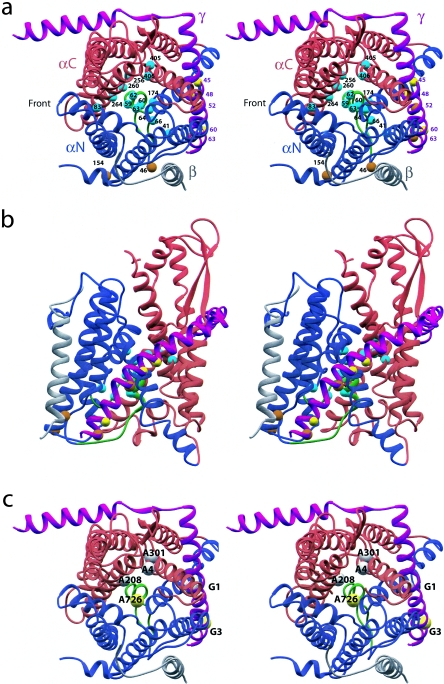
Localization of mutations on the SecY complex structure.
We used the amino acid alignments of van den Berg and coworkers (45) to localize the sec, prl, and ssl mutations from Table 1 onto the SecY complex structure (Fig. 2, prl and ssl mutations only). Although prl mutations have been isolated in secG, we cannot align those mutations with the structure, as there is no sequence similarity between the eubacterial SecG and the archaeal Secβ proteins.
FIG. 2.
Stereo views of the SecY structure represented as ribbons with functional mutations. SecY is drawn with TM1 to -5 in blue, TM6 to -10 in red, and the plug (TM2a) in green. SecE is in purple, and SecG is in gray. Locations of residues that are prlA (secY) mutations are shown as cyan spheres, prlG (secE) as yellow spheres, and suppressors of synthetic lethality (secY and secE) as orange spheres. (a) View from the cytoplasm. (b) View in the plane of the membrane from the “back.” The cytoplasmic side is at the top of the figure, and the periplasm is at the bottom. (c) View from the cytoplasm showing only alleles involved in synthetic-lethality pairs. The M. jannaschii SecY structure can be obtained as PDB 1RHZ (45).
The secY mutations are located throughout the gene, with no apparent pattern. This is not surprising, as these are conditional-lethal, loss-of-function mutations. Any mutation that disrupts the functional structure of SecY will result in a sec phenotype. It is noteworthy, however, that several mutations fall within the cytoplasmic domain that is predicted to act as a dock for cytoplasmic binding partners, which in the case of E. coli are the ribosome or SecA binding sites. It is possible that this is a hot spot for sec mutations, because disruption of SecA or ribosome binding would interfere with export. The secE mutations, as noted previously, are all located in domains predicted to affect expression levels. As it has been shown that SecE is required for SecY stability (22), these mutations probably act indirectly to inhibit export activity. As SecE is peripheral to the translocation channel, it may be less likely that alterations in the SecE protein affect pore structure sufficiently to result in a loss-of-function phenotype.
The distribution of the prlA alleles is very striking; all the prlA mutations from Table 1 fall inside the channel (Fig. 2). A large number of mutations are alterations to ring residues (prlA4, prlA6, prlA200, prlA202, prlA208, and prlA303), and many are located within the plug domain (prlA3, prlA9, prlA205, prlA300, prlA666, prlA726, prlA799, prlA8913, and prlA8914). The remainder (prlA1, prlA7, prlA11, prlA301, prlA304, prlA306, prlA401, and prlA8911) lie within the channel interior.
Two of the prlG alleles, prlG1 and prlG3, are localized to the periplasmic region, while prlG1 and prlG2 are located in TM3. As predicted through synthetic-lethality experiments (15), this transmembrane domain of SecE is in proximity to TM10 and TM7 of SecY. The periplasmic domain is not close to the periplasmic domain of SecY with which it was predicted to interact; however, the model of plug movement does place these regions close together. All of the ssl alleles alter periplasmic residues and may affect the interaction of the plug with SecE.
DISCUSSION
Analysis of the structure-function relationships in the SecYEG translocase complex has reached an exciting and highly informative juncture at which we are able to correlate functional alterations due to mutation with the recently solved crystal structure of the archaeal SecY complex (45). The availability of a very large number of alleles with quite different phenotypes expands our ability to decipher interactions between the protein subunits and to understand the contribution of each component. In the present work, we isolated and characterized new mutations in secE and secY that are unlike any previously isolated alleles, analyzed the phenotypes of combinations of alleles, and localized current and previously characterized mutations in the crystal structure of secE and secY to correlate functional alterations with predicted structural changes. Our results expand and refine the previously proposed model of Prl function to reflect a more complete catalog of mutations and to accommodate phenotypes associated with multiple mutations.
SecY complex structure.
The M. jannaschii SecY crystal structure reveals that the complex forms a roughly rectangular shape, with SecY (α subunit) constituting the central channel formed by two domains in a clamshell arrangement (Fig. 2). SecE (γ subunit) and SecG (β subunit) are positioned around the perimeter of SecY, leaving only the mouth of the clamshell open to the lipid bilayer. Viewed from the cytoplasm, the SecY complex forms a funnel, which narrows to a constricting ring in the center. The ring is composed of six hydrophobic residues (all Ile in E. coli), and it was predicted that this ring forms a seal around translocating polypeptides to maintain the integrity of the membrane barrier. It was hypothesized that the pore is closed by TM2a of SecY (formerly thought to be periplasmic and referred to previously as P1), which is postulated to move from a position in which it forms a plug closing the channel to one in which this domain moves out of the pore, binding to the C-terminal end of SecE to open the channel. The binding of the signal sequence between TM2b (formerly TM2) and TM7 is thought to trigger plug displacement, forming the open channel. In addition, the central hydrophobic ring must open slightly to allow passage of polypeptide domains while maintaining the seal around the translocating protein (45).
The plug displacement model gained credence from an earlier study in which a cysteine substitution at position 120 of SecE, when combined with prlA3 (SecY F67C), resulted in disulfide bridge formation and lethality (18). These residues are located a long distance from each other in the closed state, and the only likely explanation for the observed phenotype is that TM2a must move to the proposed open position. Thus, as predicted previously (18), the disulfide bond resulted in a constitutively open phenotype.
It was noted that several prlA alleles are located within the central SecY channel, particularly in ring residues. Based on these observations, it was proposed that at least some of the prlA alleles exert their effects either by destabilizing the closed state of the channel or by stabilizing the open state (45). We have extended this analysis to include most, if not all, of the published, characterized sec and prl mutations in secE and secY, as well as our newly isolated alleles and combinatorial mutants. Although the locations of the sec mutations were predictable, the prl alleles were enlightening. Our present findings are consistent with and expand upon the model proposed to explain how prl mutations may bypass signal sequence recognition.
The prlA alleles.
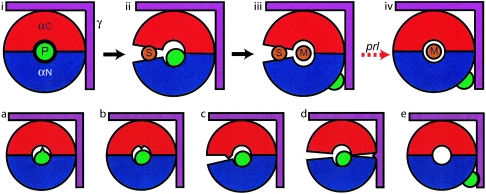
The prlA alleles all localize to ring residues, plug residues, or the channel interior. While the previous analysis (45) noted that a few prlA alleles lie within the channel interior, the present work expands on that observation and fills out the framework of the proposed model. We now suggest that the mechanism of action of all of the characterized prlA alleles can be interpreted as follows (Fig. 3 and Table 5). We propose that alterations to ring residues destabilize the ring in the absence of signal sequence binding (class B), while mutations in the plug allow displacement without a requirement for signal sequence binding (class A). Alterations to the channel interior may alter either property of the complex through effects on adjacent residues. In addition, a few mutations alter residues in the signal sequence binding domain and may thereby destabilize the closed state (class C). Therefore, through one of these mechanisms, the prlA mutations may bypass the requirement for triggering of the translocase via signal sequence binding.
FIG. 3.
Model of proposed Prl action. The SecY complex is colored as in Fig. 2, with SecY depicted as a blue-and-red donut, the plug in green, and SecE shown in purple. The top row illustrates the proposed normal model for plug opening. (i) The plug (P) starts in the closed position at the center of SecY. (ii) Binding of the signal sequence (S) destabilizes the domain interface, leading to a slight opening of the clamshell, which interrupts the interaction with the plug. (iii) Subsequent translocation of mature protein leads to displacement of the plug and full opening of the channel. (iv) Prl mutations remove the requirement for binding of the signal sequence and allow translocation of proteins with incorrect or absent signal sequences. In Prl complexes (bottom row), different types of mutations lead to similar effects. For prlA mutations, the plug may be partially displaced either by plug mutations (a) or by destabilization of the ring (b). Alternately, the closed state of the clamshell may be destabilized by mutations in the signal sequence binding domain (c). PrlG complexes may either destabilize the closed state through an indirect effect on ring stability (d) or stabilize the open plug state (e).
TABLE 5.
Proposed classes of prl suppressor mutationsa
| Alleles in class:
| ||||
|---|---|---|---|---|
| A (open-plug stabilization) | B (ring destabilization) | C (closed-state destabilization) | D (ring destabilization) (indirect) | E (open-plug stabilization) (indirect) |
| prlA3 | prlA4 | prlA1 | prlG1 | prlG3 |
| prlA9 | prlA6 | prA304 | prlG2 | prlG8 |
| prlA205 | prlA7 | prlA401 | ||
| prlA300 | prlA11 | |||
| prlA302 | prlA200 | |||
| prlA306 | prlA202 | |||
| prlA666 | prlA208 | |||
| prlA726 | prlA301 | |||
| prlA799 | prlA303 | |||
| prlA8913 | ||||
| prlA8914 | ||||
The prlA and prlG alleles can be categorized with respect to mechanism, as illustrated in Fig. 3. The class A alleles stabilize the open-plug state, class B mutations destabilize the ring, and class C mutations destabilize the closed state through mutations in the signal sequence binding domain. The prlG alleles act indirectly, either by destabilization of the ring (class D) or by stabilization of the open plug state (class E).
The prlG alleles.
The localization of the prlG alleles reflects alterations to the dynamic role of either the ring or the plug (Fig. 3 and Table 5). Two alleles, prlG3 and prlG8, are located in the periplasmic region, P2 (Fig. 2). This region is postulated to interact with the plug in the open conformation (45), and these alleles likely exert their effects through stabilization of the open state, reducing the need for the signal sequence triggering event (class E). It seems likely that the other prlG alleles, prlG1 and prlG2, exert an indirect effect on ring stability (class D). These SecE residues (S105 and L108) are in proximity to the rear of SecY at the interface of TM5 and TM10, the hinge of the clamshell. Alterations to these amino acids probably relax the clamping role of SecE. It is perhaps important that this region of SecE is situated directly next to helices that contain ring residues. The structural alterations caused by either of these mutations are likely to destabilize the integrity of the ring and permit opening of the channel without signal sequence binding.
It has been recognized that prlG alleles are, in general, less efficient Prl suppressors than are prlA mutations (14, 41), and the structural information now provides an explanation. The prlG1 and prlG2 alleles are not as effective as the prlA alleles because they affect the ring only indirectly, while the prlG3 and prlG8 mutations may not be as efficient because the SecY plug is not destabilized in the closed state; the PrlG effect is only to stabilize the open state once the plug has been displaced.
Synthetic lethality.
Several pairs of prlA and prlG alleles resulted in synthetic-lethal phenotypes when combined (15). Those combinations were both allele specific and topologically specific, leading to predictions of interactive domains between SecY and SecE. In particular, it was proposed that SecY(TM10) interacts both with SecY(TM7) and with SecE(TM3) and that two periplasmic domains interact, SecY(P1) and SecE(P2). Examination of the crystal structure validates these predictions (Fig. 2).
The prlA4-2 mutation is at one of the ring residues (I408), suggesting that this mutation destabilizes the ring structure. Mutations to the adjacent residue (L407R) introduce a positive charge (prlA301), potentially altering the conformation of the helix and pulling the neighboring I408 out of the ring, suggesting that a neighboring residue can affect ring stability. We predict that prlG1 (L108R) affects the integrity of the ring because it introduces a charge at the TM5-TM10 interface, destabilizing the closed state of the clamshell. Therefore, combining prlG1 with any of the prlA alleles that affect this ring residue (prlA4-2, prlA6-1, prlA7-1, prlA11-1, or prlA301) will compound the effect, leading to lethality. In fact, it was observed previously that each of these prlA alleles produced a synthetic-lethal phenotype when combined with prlG1 (15). In addition, prlA208 and prlG1 also resulted in a synthetic-lethal phenotype. As prlA208 (I278N) also alters the ring structure, we suspect that the lethality observed between prlA208 and prlG1 is also due to an additive effect on ring stability.
The remaining synthetic-lethal combinations were pairings between prlG3 and either prlA3 or prlA726. The prlG3 mutation alters Ser120 to Phe in the second periplasmic domain of SecE (P2), while prlA3 (F67C) and prlA726 (S68P) both alter residues in TM2a (SecY). The structure of the SecY complex led to the proposal that SecE(P2) interacts with SecY(TM2a) to bind the plug in the open position. Therefore, the synthetic-lethal pairs are predicted to favor the open plug position, resulting in an open channel and subsequent detrimental effects to the cell. The dominant phenotype of the prlA726-prlG3 combination is consistent with this hypothesis, while prlA3-prlG3 may be recessive due to assembly defects or because the shift to an open conformation is not as strong as with the prlA726-prlG3 pair. The model of plug movement and interaction with SecE is consistent with synthetic lethality in periplasmic domains.
Suppressors of synthetic lethality.
Of our newly isolated suppressors of synthetic lethality, we found that the two secY mutations affected periplasmic domains near the prlG3 residue. Because we predict that the lethality conferred by prlA726-prlG3 or by prlA3-prlG3 is the result of a stabilized open plug state, these new suppressor mutations are predicted to destabilize the open plug state. It is perhaps not surprising, then, that neither of these alleles exhibits a Prl phenotype. If they destabilize the plug-SecE interaction, the translocase would favor a closed state and Prl suppression would not occur. Again, the phenotypes observed are consistent with a dynamic structure in which both ring destabilization and plug displacement are necessary for translocation.
One of the suppressors of synthetic lethality found in secE, secE(T123P), is also located in the periplasm, only a few residues removed from prlG3. We predict that this mutation also alters the structure of the periplasmic domain to destabilize the open plug state. As mentioned above, and consistent with this prediction, secE(T123P) is not a prl allele. The secE(D112Y) allele falls within the membrane at the interface between SecY TM1 and TM5. Again, this mutation must shift the plug displacement activity to compensate for the synthetic lethality that it rescues, but this suppressor of lethality is also a Prl suppressor. We speculate that secE(D112Y) alters SecE P2, moving the prlG3 mutation to destabilize binding of the open plug while also disrupting the ring stability to create a Prl phenotype. We noted that secE(D112Y) is adjacent to several of the ring residues, particularly I174 located in TM5. We speculate that the mutation may disrupt the structure of TM5, resulting in a dislocation of I174 and destabilization of the ring, which is a Prl effect. If so, then it is possible to have a ring destabilization mutation, secE(D112Y), and a mutation that stabilizes the open plug (prlG3) in the same molecule without detrimental effects. Intriguingly, alterations to this same residue (D112) previously have been shown to result in either severe growth and secretion defects (D112P) or generation of a Prl suppressor phenotype (D112Q) (26), supporting our conclusion that D112 plays a critical role in SecE function.
Multiple-mutant analysis.
Combinations of prlG1, prlG2, and prlG3 were not deleterious and were additive (or synergistic) in their ability to suppress signal sequence defects. We speculated that prlG1 and prlG2 function indirectly to destabilize the ring and the closed state and that prlG3 stabilizes the open state. Therefore, as suggested above, these two effects can be present in the same translocase complex.
Combinations of any prlG allele with secE(D112Y) retained a Prl phenotype, indicating that the structural alteration imposed by secE(D112Y) to rescue the prlA726-prlG3 lethality does not counteract the structural alterations imposed by any of the prlG alleles (including prlG3). The secE(T123P) mutation is intriguing because it was isolated in combination with prlG3, as a suppressor of the prlA3-prlG3 combination, yet a prlG3-secE(T123P) double mutant expressed from a plasmid is lethal in some prlA backgrounds and, indeed, is detrimental even in a prlA wild-type strain. We do not fully understand this phenomenon and are continuing our studies of this combination.
Combinations of prlA alleles suggest that each single mutation is sufficiently disruptive to the structure of the SecY complex that combinations are likely to be deleterious. In particular, any combination involving prlA4 resulted in a nonfunctional complex. This is perhaps not surprising, because it has been thought that this allele is not completely innocuous. Notably, the prlA4 allele was originally isolated as a double mutant with one mutation in TM10 (I408N; prlA4-2) and a second alteration in TM7 (F286Y; prlA4-1). Subsequently, it was demonstrated that prlA4-2 (I408N) alone is sufficient to confer the Prl phenotype (32, 40), and therefore it is thought that the TM7 mutation relieves detrimental effects caused by the TM10 mutation. Additionally, the prlA6 allele contains the same suppressor mutation, I408N, and also has a second mutation, S188L, in TM5 (24), again suggesting that the I408N mutation requires a secondary mutation to produce a fully stable protein product. Importantly, all our combinations were constructed with the double prlA4 allele; that is, they contain the compensatory TM7 (F286Y) mutation in addition to the I408N alteration. In these combinations, apparently the TM7 mutation is not sufficient to alleviate negative effects imposed by I408N in combination with a second prlA allele. Although some of these multiple mutants may simply produce unstable protein products and thus fail to complement a cold-sensitive chromosomal allele, that is not the case with the prlA4-prlA726 combination. It is significant that prlA4-prlA726 is not only nonfunctional, but produces a dominant-negative phenotype. This implies both that the mutant protein is stable and either that it interacts with SecE and/or SecG or that the high level of such an abnormal membrane protein causes lethality. Thus, the multiple-mutant analysis demonstrates that mutations that destabilize the closed state and ones that stabilize the open plug can coexist in the same molecule. However, there is clearly a limit on the degree to which the open state can be tolerated without lethality.
Conclusions.
In summary, the correlation between genetic phenotypes and structural information has proved beneficial to understanding the SecY complex. It is gratifying to find that many predictions based on genetic analysis have been substantiated. In particular, analysis of prlA suppressors led to the prediction that SecY TM7 interacts with the signal sequence (24); the crystal structure also suggests that the signal sequence binds to TM7 and TM2b (45). Synthetic-lethality experiments generated predictions of interactive domains between SecE and SecY (15, 24); these were corroborated by the crystal structure (45). Genetic analysis led to a proofreading hypothesis that predicted that SecY and SecE were able to reject defective precursors from the export pathway while PrlA and PrlG allowed export (15, 24). This model is not completely validated by the structural analysis; instead, perhaps a “trigger-independent” model would be more accurate. According to this new model, PrlA and PrlG mutants allow export of defective preproteins independently of signal sequence binding, either by destabilization of the closed state or by stabilization of the open plug state of the translocase. The analyses presented here provide details to the model and suggest mechanistic actions for the prl suppressor alleles.
Acknowledgments
We are very grateful to Tom Hill, Tom Rapoport, Tom Silhavy, Kevin Young, and Nick Hand for critical reading and discussion. We also thank Kurt Cannon, Eran Or, and Andrew Osborne for reading the manuscript.
This work was supported by the National Science Foundation (A.M.F.) and a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation (W.M.C.).
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1987. Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J. 6:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., A. Jacq, E. Brickman, J. Beckwith, T. Taura, C. Ueguchi, Y. Akiyama, and K. Ito. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankaitis, V. A., and P. J. Bassford, Jr. 1985. Proper interaction between at least two components is required for efficient export of proteins to the Escherichia coli cell envelope. J. Bacteriol. 161:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieker, K. L., and T. J. Silhavy. 1990. The genetics of protein secretion in E. coli. Trends Genet. 6:329-334. [DOI] [PubMed] [Google Scholar]
- 5.Bieker, K. L., and T. J. Silhavy. 1990. PrlA (SecY) and PrlG (SecE) interact directly and function sequentially during protein translocation in E. coli. Cell 61:833-842. [DOI] [PubMed] [Google Scholar]
- 6.Bost, S., and D. Belin. 1997. prl mutations in the Escherichia coli secG gene. J. Biol. Chem. 272:4087-4093. [DOI] [PubMed] [Google Scholar]
- 7.Cao, T. B., and M. H. Saier. 2003. The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys. Acta 1609:115-125. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 10.Derman, A. I., J. W. Puziss, P. J. Bassford, Jr., and J. Beckwith. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 12:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downing, W. L., S. L. Sullivan, M. E. Gottesman, and P. P. Dennis. 1990. Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J. Bacteriol. 172:1621-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driessen, A. J., P. Fekkes, and J. P. W. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 13.Emr, S. D., S. Hanley-Way, and T. J. Silhavy. 1981. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23:79-88. [DOI] [PubMed] [Google Scholar]
- 14.Flower, A. M., R. C. Doebele, and T. J. Silhavy. 1994. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J. Bacteriol. 176:5607-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flower, A. M., R. S. Osborne, and T. J. Silhavy. 1995. The allele-specific synthetic lethality of prlA-prlG double mutants predicts interactive domains of SecY and SecE. EMBO J. 14:884-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gansheroff, L. J. 1988. B.A. thesis. Princeton University, Princeton, N.J.
- 17.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, C. R., and T. J. Silhavy. 1999. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 181:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann, E., T. Sommer, S. Prehn, D. Gorlich, S. Jentsch, and T. A. Rapoport. 1994. Evolutionary conservation of components of the protein translocation complex. Nature 367:654-657. [DOI] [PubMed] [Google Scholar]
- 20.Ito, K. 1990. Structure, function, and biogenesis of SecY, an integral membrane protein involved in protein export. J. Bioenerg. Biomembr. 22:353-367. [DOI] [PubMed] [Google Scholar]
- 21.Ito, K., Y. Hirota, and Y. Akiyama. 1989. Temperature-sensitive sec mutants of Escherichia coli: inhibition of protein export at the permissive temperature. J. Bacteriol. 171:1742-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama, S., J. Akimaru, and S. Mizushima. 1990. SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett. 269:96-100. [DOI] [PubMed] [Google Scholar]
- 23.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 24.Osborne, R. S., and T. J. Silhavy. 1993. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 12:3391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panzner, S., L. Dreier, E. Hartmann, S. Kostka, and T. A. Rapoport. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81:561-570. [DOI] [PubMed] [Google Scholar]
- 26.Pohlschroder, M., C. Murphy, and J. Beckwith. 1996. In vivo analyses of interactions between SecE and SecY, core components of the Escherichia coli protein translocation machinery. J. Biol. Chem. 271:19908-19914. [DOI] [PubMed] [Google Scholar]
- 27.Prinz, W. A., C. Spiess, M. Ehrmann, C. Schierle, and J. Beckwith. 1996. Targeting of signal sequenceless proteins for export in Escherichia coli with altered protein translocase. EMBO J. 15:5209-5217. [PMC free article] [PubMed] [Google Scholar]
- 28.Puziss, J. W., S. M. Strobel, and P. J. Bassford, Jr. 1992. Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations. J. Bacteriol. 174:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapoport, T. A., B. Jungnickel, and U. Kutay. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65:271-303. [DOI] [PubMed] [Google Scholar]
- 30.Riggs, P. D., A. I. Derman, and J. Beckwith. 1988. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics 118:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sako, T. 1991. Novel prlA alleles defective in supporting staphylokinase processing in Escherichia coli. J. Bacteriol. 173:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sako, T., and T. Iino. 1988. Distinct mutation sites in prlA suppressor mutant strains of Escherichia coli respond either to suppression of signal peptide mutations or to blockage of staphylokinase processing. J. Bacteriol. 170:5389-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schatz, P. J., and J. Beckwith. 1990. Genetic analysis of protein export in Escherichia coli. Annu. Rev. Genet. 24:215-248. [DOI] [PubMed] [Google Scholar]
- 35.Schatz, P. J., K. L. Bieker, K. M. Ottemann, T. J. Silhavy, and J. Beckwith. 1991. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10:1749-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schatz, P. J., P. D. Riggs, A. Jacq, M. J. Fath, and J. Beckwith. 1989. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 3:1035-1044. [DOI] [PubMed] [Google Scholar]
- 37.Shiba, K., K. Ito, T. Yura, and D. P. Cerretti. 1984. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 3:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Spee, J. H., W. M. de Vos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stader, J., S. A. Benson, and T. J. Silhavy. 1986. Kinetic analysis of lamB mutants suggests the signal sequence plays multiple roles in protein export. J. Biol. Chem. 261:15075-15080. [PubMed] [Google Scholar]
- 41.Stader, J., L. J. Gansheroff, and T. J. Silhavy. 1989. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev. 3:1045-1052. [DOI] [PubMed] [Google Scholar]
- 42.Taura, T., Y. Akiyama, and K. Ito. 1994. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol. Gen. Genet. 243:261-269. [DOI] [PubMed] [Google Scholar]
- 43.Trun, N. J., and T. J. Silhavy. 1987. Characterization and in vivo cloning of prlC, a suppressor of signal sequence mutations in Escherichia coli K12. Genetics 116:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trun, N. J., J. Stader, A. Lupas, C. Kumamoto, and T. J. Silhavy. 1988. Two cellular components, PrlA and SecB, that recognize different sequence determinants are required for efficient protein export. J. Bacteriol. 170:5928-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Berg, B., W. M. J. Clemons, I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]