Abstract
The phz operon of Pseudomonas fluorescens 2-79, which produces phenazine-1-carboxylate, is preceded by two genes, phzR and phzI, that are homologs of quorum-sensing gene pairs of the luxR-luxI family. Deleting phzR and phzI from strain 2-79 led to loss of production of the antibiotics, as well as a suite of six acyl-homoserine lactones (acyl-HSLs) that includes four 3-hydroxy- derivatives and two alkanoyl-HSLs. Strain 2-79 accumulates N-(3-hydroxy-hexanoyl)-l-HSL to levels 20 and 30 times those of N-(hexanoyl)-l-HSL and N-(3-hydroxy-octanoyl)-HSL, the next most abundant species produced by this isolate. Expression of a clone of phzI in Escherichia coli and P. fluorescens 1855 resulted in the synthesis of all six acyl-HSLs. Maximal activation of phzA and phzR fused to lacZ and uidA reporters, respectively, required PhzR and the acyl-HSL signals. PhzR-mediated expression of the phzA::lacZ fusion responded with highest sensitivity and greatest magnitude to pure N-(3-hydroxy-hexanoyl)-l-HSL. When exposed to organic extracts of culture supernatants containing the six acyl-HSLs at their normal levels, the reporter responded strongly to N-(3-hydroxy-hexanoyl)-l-HSL but did not respond to any of the other five acyl-HSLs. The transcriptional start sites for the divergently oriented phzA and phzR genes were mapped by primer extension analysis. An 18-bp almost perfect inverted repeat, the phz box, is located between the phzI and phzR promoters. Disrupting this repeat abolished PhzR-dependent activation of phzA and phzR. We conclude that PhzI of strain 2-79 synthesizes 3-OH acyl-HSLs and that P. fluorescens 2-79 uses N-(3-hydroxy-hexanoyl)-HSL as its quorum-sensing signal. We also conclude that PhzR, with its quormone, activates expression of phzA and phzR and that this activation requires an intact phz box sequence located in the divergent promoter region.
Many pseudomonads, including the generalist Pseudomonas aeruginosa and fluorescent soil forms, such as Pseudomonas fluorescens, Pseudomonas aureofaciens, and Pseudomonas chlororaphis, produce phenazine-1-carboxylic acid (PCA) or its derivatives (7, 11, 49). These secondary metabolites exhibit antimicrobial activity against a wide range of prokaryotic and eukaryotic microbes, and the synthesis and secretion of these compounds is thought to confer an advantage on the producer bacteria when in competition with other microflora in the soil habitat (29). Production of substituted phenazines is also required by these bacteria to fully protect plants against diseases caused by fungal and bacterial pathogens that are susceptible to the antibiotics (2, 7, 9, 33, 49). Such biocontrol properties are of considerable interest, since their application in the field could reduce reliance on the use of environmentally toxic antimicrobial chemicals. However, for such strategies to be successful, the antibiotics must be synthesized reliably in the appropriate environmental sites. Hence, it is important to identify the genetic determinants required for synthesis of the phenazines, as well as the mechanisms by which expression of these genes is regulated.
The phz cluster is organized as an operon of seven genes, the products of which catalyze the synthesis of chorismate and its conversion to PCA (9, 27, 28, 35). This parent compound can be substituted at several positions, depending upon the producer strain, and the genes coding for the enzymes involved in such substitutions are restricted to those strains producing the modified compounds. While gene nomenclature differs with the isolate (for an example, see reference 27), the order of the seven genes and the amino acid sequences of their products are conserved in all phz operons examined to date (9, 27, 28, 35).
Although regulation is complex and multifactorial, the phz operons of P. aureofaciens 30-84 and P. chlororaphis PCL1391 are controlled at the transcriptional level by a ligand-dependent transcription factor called PhzR (8, 34). This activator is a member of the LuxR family and, as such, requires a low-molecular-weight signal, an acyl-homoserine lactone (acyl-HSL), for activity (8, 56). Signal production in both pseudomonads is catalyzed by PhzI, an acyl-HSL synthase of the LuxI family (8, 56). The acyl-HSL most probably transits out of and back into the cell by diffusion and thus accumulates within the environment as the population size of the dividing bacteria increases. When the concentration of the signal reaches a critical level, which corresponds to a particular population size of the bacteria, it interacts with and activates PhzR, thus initiating expression of the phz operon.
In P. aureofaciens 30-84 and P. chlororaphis PCL1391, phzR and phzI are adjacent and convergently oriented, with phzR being located just upstream of and in opposite orientation to the phz operon (8, 56). Similarly, the phz operon of P. fluorescens 2-79 is preceded by phzR and phzI homologs organized in identical manners (Fig. 1). Moreover, the three phzR genes and the three phzI genes, as well as their encoded protein products, are all strongly orthologous (8).
FIG. 1.
Gene structure of the phz locus from P. fluorescens 2-79 and its mutants. (A) The organizations of the phz operon and the two regulatory genes phzR and phzI are shown by arrows. Alterations in the three mutants—strain 2-79IR, in which phzR and phzI have been deleted and replaced with a cassette coding for resistance to tetracycline; strain 2-79Z, in which lacZ has been fused to phzD; and strain 2-79IZ, in which the phzI gene of mutant 2-79Z has been deleted and replaced with the tet cassette—are shown as triangles. (B) Production of phenazines by P. fluorescens 2-79 and 2-79IR. Ethyl acetate extracts of culture supernatants of strain 2-79 and its phzR-phzI deletion mutant, 2-79IR, each grown to late exponential phase in LB medium, were subjected to HPLC, and column eluates were monitored by spectrophotometry at 250 nm as described in Materials and Methods. The material in the peak eluting at 14.5 min was analyzed by recording UV spectrophotometry between 200 and 450 nm (insert). AU, absorbance units.
Strain 2-79 has served as a model for the genetics, biochemistry, and enzymology of phenazine biosynthesis. We previously reported that this isolate produces at least six acyl-HSLs, including the 3-hydroxy forms, N-(3-hydroxy-hexanoyl)-l-homoserine lactone (3-OH-C6-HSL), N-(3-hydroxy-octanoyl)-l-homoserine lactone (3-OH-C8-HSL), and N-(3-hydroxy-decanoyl)-l-homoserine lactone (3-OH-C10-HSL); the alkanoyl forms hexanoyl-homoserine lactone (C6-HSL) and octanoyl-homoserine lactone (C8-HSL); and an active signal compound of unknown structure (45). However, P. aureofaciens 30-84 and P. chlororaphis PCL1391 both have been reported to produce and utilize the alkanoyl signal, C6-HSL (8, 57). Moreover, the last two strains have not been reported to produce 3-hydroxy-acyl-HSLs. Given the very close amino acid sequence relatedness of the PhzI and PhzR proteins from these three strains, it became important to determine what acyl-HSL signals are produced by PhzI of P. fluorescens 2-79 and which of these signals activates PhzR of this strain. In this report, we show that PhzI catalyzes synthesis of all of the 3-OH- and alkanoyl-acyl-HSLs produced by P. fluorescens 2-79. We also show that 3-OH-C6 accumulates to the highest concentration in culture supernatants and that PhzR from strain 2-79 responds with greatest sensitivity to this signal. In addition, we show that activation of the phz operon by PhzR is dependent upon an intact inverted repeat, the phz box, located in the divergent phzA-phzR promoter region and that PhzR weakly autoactivates its own transcription, also in a phz box-dependent manner.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. All E. coli strains were grown in Luria-Bertani (LB) medium at 37°C unless stated otherwise. Pseudomonas fluorescens strains 2-79 and 1855 were grown in King's medium B (23), ABM minimal medium (6), and LB medium at 28°C. The bioreporter strains, Agrobacterium tumefaciens NTL4(pZLR4) (4) and Chromobacterium violaceum CV026blu (30), were grown at 28°C in ABM and LB media, respectively. The P. fluorescens 1855 indicator strain, which harbors pSF105 and pSF107 (Table 1; see also below) was grown in ABM medium at 28°C. When required, antibiotics were added to the following final concentrations, in μg per ml: carbenicillin, 100 or 200; streptomycin or spectinomycin, 100; kanamycin, 50; gentamicin, 15 or 50; rifampin, 75; and tetracycline, 12.5 or 25.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characterstics | Source or reference |
|---|---|---|
| Pseudomonas fluorescens | ||
| 2-79 | Phz+ Rifr wild-type strain | 49 |
| 2-79IR | Phz+ Rifr Tetr ΔphzIR | This study |
| 2-79Z | Phz− RifrphzD::lacZ | This study |
| 2-79IZ | Phz− RifrphzI::TetrphzD::lacZ | This study |
| 1855 | Wild-type strain lacking any known homologues of luxR and luxI | 4 |
| E. coli | ||
| S17-1 | Pro− Res− Mod+recA; integrated RP4-Tet::Mu-Kan::Tn7 Mob+ | 47 |
| S17-1(λpir) | Pro− Res− Mod+recA; integrated RP4-Tet::Mu-Kan::Tn7 Mob+ λpir | 47 |
| Others | ||
| A. tumefaciens NTL4(pZLQR) | Indicator strain for detection of acyl-HSLs | 4 |
| C. violeceum CV026blu | Indicator strain for detection of alkanoyl-acyl HSLs | 30 |
| Plasmids | ||
| pBluescript II SK | Co1E1 f1(+) Ampr, cloning vector | Stratagene |
| pUCP22 | Broad-host-range cloning vector; aac1 bla lacZ | 54 |
| pUCPIR | pUCP22 containing a 1.67-kb BglII fragment from pPHZ108A coding for phzI under the control of Plac | This study |
| pUCPRI | pUCP22 containing a 1.67-kb BglII fragment from pPHZ108A coding for phzR under the control of Plac | This study |
| pUCPR | pUCP22 containing phzR under control of Plac | This study |
| pZLQ | Kmr; pBBR1MCS-2-derived expression vector containing trc promoter and lacIq | 26 |
| pDLB4 | Kmr; pBBR1MCS-2-derived expression vector containing the BAD promoter of pBAD22 | 24 |
| pRG970b | Broad-host-range promoter selection vector; Strr Carbr | 51 |
| pMC1871 | Source of promoterless lacZ gene | Amersham |
| pALTER-Ex1 | ColE1 vector containing SP6, tac, and T7 promoters; Tetr | Promega |
| pNOT19 | ColE1 bla accessory plasmid | 42 |
| pMOB3 | Kanrcat oriT; source of sacBR cassette | 42 |
| pEX18Tc | TetroriT sacB; gene replacement vector | 20 |
| pEX18Gm | aac1 oriT sacB; gene replacement vector | 20 |
| pPHZ108A | pLAFR3 containing P. fluorescens 2-79 genomic DNA; Phz+ | 27 |
| pT7-5CDE | pT7-5 containing a 4.3-kb EcoRI-BglII fragment from pPHZ108A with phzCDE genes | 27 |
| pKS-Pst6.3 | pBluescript II KS containing an approx 6.3-kb PstI fragment from pPHZ108A with phzIRABCD genes | This study |
| pNOT-Pst6.3-Tcr | pNOT19 containing an approx 6.3-kb PstI fragment from pKS-Pst6.3 with tet gene inserted into BglII site | This study |
| pNOT-Pst6.3-Tcr-MOB | pNOT-Pst6.3-Tcr ligated with sacBR cassette from pMOB3 | This study |
| pEX18Tc-CDE | pEX18Tc containing a 4.3-kb EcoRI-BglII fragment from pT7-5CDE with the phzCDE genes | This study |
| pEX18Tc-CDE-lacZ | pEX18Tc-CDE containing a phzD::lacZ fusion | This study |
| pEX18Gm-I | pEX18Gm containing a 515-bp PCR fragment of the 5′ end of phzI | This study |
| pEX18Gm-I-Bam | pEX18Gm-I containing a 2.8-kb fragment with the phzIRAB genes | This study |
| pEX18Gm-I-Tcr | pEX18Gm-I-Bam containing the tet cassette inserted into BamHI site | This study |
| pSF101 | PCR-amplified 0.7-kb phzR gene cloned in the BamHI site of pBluescript II SK | This study |
| pSF102 | 0.6-kb phzI gene cloned in the EcoRI site of pBluescript II SK | This study |
| pSF103 | pBluescript II SK containing the 0.38-kb phzR-phzA intergenic region | This study |
| pSF105 | pZLQ containing a 0.7-kb BamHI-NdeI fragment coding for phzR from pSF101 | This study |
| pSF106 | pDLB4 containing a 0.6-kb EcoRI-HindIII fragment coding for phzI from pSF102 | This study |
| pSF107 | pRG970b containing the 0.38-kb phzR-phzA intergenic region from pSF103 | This study |
| pSF108 | pRG970b containing the 0.38-kb phzR-phzA intergenic region with a disrupted phz box sequence | This study |
DNA manipulations and analysis.
Standard methods were used for plasmid DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation (3). Plasmids were mobilized into P. fluorescens isolates using E. coli strains S17-1 and S17-1λpir as donors (Table 1). DNA was sequenced by using an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA), and sequence data were analyzed with the OMIGA 2.0 software package (Accelrys, San Diego, CA).
Plasmid constructions.
A 1.67-kb BglII fragment containing the phzI and phzR genes of P. fluorescens 2-7 in their native configurations was subcloned from pKS-BIIB (D. V. Mavrodi, unpublished data) into the BamHI site of pUCP22, a Pseudomonas-E. coli shuttle vector (Table 1), in both orientations, giving pUCPIR and pUCPRI. The phzR gene alone was cloned as a 789-bp PCR amplicon containing the entire activator gene and its ribosome binding site from pPHZ108A (27) (Table 1) between the SmaI and BamHI sites (restriction sites were part of the PCR primers) of pUCP22 to yield pUCPR. The two regulatory genes were also cloned separately into broad-host-range expression vectors as follows. Amplicons of phzR and phzI, produced by PCR from pPHZ108A, were cloned in pBluescript II SK (Stratagene, La Jolla, CA) to give plasmids pSF101 and pSF102, respectively. The phzR gene was excised from pSF101 and cloned between the NdeI and BamHI sites (sites were generated during PCR amplification) in the broad-host-range expression vector pZLQ (26), yielding pSF105. In this plasmid, which also codes for lacIq, phzR is under the control of the trc promoter. The phzI gene from pSF102 was excised by digestion with EcoRI and HindIII and ligated into pDLB4 (24) to yield pSF106. In this vector, phzI is under the control of PBAD, and its expression is inducible with arabinose. All constructs were confirmed by DNA sequence analysis.
The PhzR-dependent promoter-reporter plasmid pSF107 was constructed by amplifying the intergenic region between the divergently oriented phzR and phzA genes of strain 2-79 using as a template pPHZ108A. The resultant 379-bp fragment, with a BamHI site at the phzA end and EcoRI and SmaI sites at the phzR end, was cloned into pBluescript II SK, producing pSF103. The BamHI-SmaI fragment was excised from pSF103 and ligated into the promoter selection vector pRG970b (51) (Table 1), yielding pSF107. In this construct, transcription toward phzA drives expression of lacZ while transcription toward phzR drives expression of uidA.
The lux box-like sequence located in the divergent promoter region was disrupted by digesting pSF103 with BglII, which cuts in the middle of the box. The ends were filled in with Klenow fragment. Following self-ligation, plasmids isolated from transformants were tested for the loss of the BglII site by restriction digestion. The disruption was confirmed by sequence analysis, and the mutated bidirectional promoter was cloned into pRG970b, giving rise to pSF108.
Gene replacement mutagenesis.
Mutants of P. fluorescens 2-79 were constructed using the gene replacement strategy previously described by Schweizer (42). To generate strain 2-79IR, which lacks the phzI and phzR genes (Fig. 1A), a 6.3-kb PstI fragment of pPHZ108A, which contains the phz operon together with the phzI and phzR genes of strain 2-79 (Fig. 1A), was subcloned into pBluescript II KS, yielding pKS-Pst6.3. The plasmid was digested with BglII, thereby removing a 1.67-kb fragment containing the phzI and phzR genes, and the ends were polished with the Klenow fragment. The 1.67-kb fragment was replaced with a 2.5-kb PvuII fragment encoding a tetracycline resistance gene from pALTER-Ex1 (Promega Corp., Madison, WI). The interrupted insert was subcloned into pNOT19 (42), yielding pNOT-Pst6.3-Tcr, which was digested with NotI and ligated with a 5.3-kb sacB cassette from pMOB3 (Table 1). The resulting plasmid, pNOT-Pst6.3-Tcr-MOB, was mobilized into P. fluorescens 2-79, and transconjugants were selected on LB agar supplemented with rifampin and tetracycline. Following selection for double crossovers on LB agar supplemented with sucrose (28), isolates were screened phenotypically for the absence of the plasmid-borne bla and sacB markers and the presence of tetA. The deletion replacement event was confirmed by PCR analysis.
To construct strain 2-79Z containing lacZ fused to phzD (Fig. 1), a 4.3-kb fragment containing the phzCDE interval was subcloned from pT7-5CDE (27) into pEX18Tc (20) (Table 1), yielding pEX18Tc-CDE. The plasmid was treated sequentially with NcoI, Klenow fragment, and PstI and ligated with a 3.1-kb fragment from pMC1871 containing a promoterless lacZ gene, giving pEX18Tc-CDE-lacZ. This construct contains a translational fusion of lacZ to phzD. pEX18Tc-CDE-lacZ was introduced into strain 2-79 by electroporation, and transformants were selected on LB agar supplemented with tetracycline. Double crossovers were selected, and mutations were verified by PCR as described above.
To construct strain 2-79IZ, a phzI disruption mutant of strain 2-79Z (Fig. 1), a 515-bp 5′ segment of phzI was amplified by PCR and cloned as a KpnI-BamHI fragment (sites were part of the primers) into pEX18Gm (20) to give pEX18Gm-I. A second, ca. 2.4-kb fragment containing a 3′ segment of phzI, as well as phzR, phzA, and phzB, was amplified from pPHZ108A by PCR and cloned into pEX18Gm-I between the BamHI and XbaI sites, yielding pEX18Gm-I-Bam. These manipulations resulted in introduction of a unique BamHI site within the phzI gene. A 2.5-kb PvuII fragment containing the tet cassette from pALTER-Ex1 was cloned into the BamHI site of phzI in pEX18Gm-I-Bam, yielding pEX18Gm-I-Tcr. This plasmid was introduced into strain 2-79Z by electroporation, and double-crossover events were selected for and confirmed as described above.
Primer extension analysis.
Transcription start sites were mapped by primer extension essentially as described by Ausubel et al. (3). Total RNA was isolated from cultures of strain 2-79 using an RNeasy Midi Kit (QIAGEN). The transcriptional start site of phzA was determined using primer PHSPA (5′-CGC GGT TTT TTC GGC GAA GTT C-3′ [positions 1872 through 1893 in GenBank entry L48616]). The transcriptional start site of phzR was determined using primer PHSPR (5′-CTC CTC CAT GTT CAT GGC TTG TGC-3′ [positions 1368 through 1391 in GenBank entry L48616]) (27). Oligonucleotides end labeled with [γ-32P]ATP were mixed with 15 μg of RNA at 65°C in a buffer containing 50 mM Tris-HCl, pH 8.3, and 45 mM KCl and annealed by slowly cooling them to room temperature. Primer extension was performed at 42°C with SuperScript II RNase H− reverse transcriptase (Life Technologies, Inc.). Extension products were separated on denaturing 6% polyacrylamide-8 M urea gels next to sequencing ladders generated from pKS-Pst6.3 (Table 1) with the same primers using Sequenase version 2.0 (Amersham Biosciences Corp.).
Analysis of phenazines.
Cultures of P. fluorescens 2-79 and its mutants were grown for 24 h at 28°C in LB medium supplemented with 0.2% glucose and antibiotics as appropriate. The cells were removed by centrifugation, and the culture supernatants were acidified with trifluoroacetic acid and extracted twice with ethyl acetate. Organic phases were pooled and evaporated to dryness, and residues were redissolved in 35% acetonitrile-0.1% trifluoroacetic acid. PCA was resolved by high-performance liquid chromatography (HPLC) on a NOVA-PAK C18 reverse-phase Radial-PAK cartridge (4 μm; 8 by 100 mm) (Waters Corp., Milford, MA) with a linear gradient (8 to 100%) of acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml per min using a Waters HPLC system. Elution profiles were monitored at the absorbance maxima (248 nm and 366 nm) characteristic of PCA in the solvent system using a Waters 990 photodiode array detector. Reference samples of PCA were purchased from Color Your Enzyme (Bath, Ontario, Canada).
Extraction of acyl-homoserine lactones from culture supernatants.
Cultures of strain 2-79 and its mutants were grown in King's medium B for 48 h at 28°C. Cultures of Escherichia coli and P. fluorescens 1855 expressing the arabinose-inducible phzI gene of strain 2-79 in pSF106 were grown in LB and ABM media, respectively, to an optical density at 600 nm of 0.4. Arabinose was added to a final concentration of 0.3%, and incubation was continued for another 15 to 18 h at 28°C. The bacteria were removed by centrifugation, the supernatants were extracted twice with equal volumes of ethyl acetate or dichloromethane, the organic phases were collected and pooled, and residual water was removed by addition of anhydrous sodium sulfate. The extracts were taken to dryness under vacuum, and the residues were redissolved in volumes of HPLC grade ethyl acetate or dichloromethane sufficient to yield concentration factors of 100 to 1,000.
Synthesis of acyl-homoserine lactone standards.
The N-(3-hydroxy-hexanoyl)-l-HSL and N-(3-hydroxy-octanoyl)-l-HSL standards were synthesized by the Reformatsky reaction of hexanal or butanal with ethyl bromoacetate in the presence of activated zinc (16, 46). This reaction results in the formation of ethyl 3-hydroxy-octanoate (with hexanal) or ethyl 3-hydroxy-hexanoate (with butanal), each of which was subsequently hydrolyzed by lithium hydroxide in tetrahydrofuran-water (1:1 [vol/vol]) and condensed with l-homoserine lactone (Sigma-Aldrich) using 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride-dimethylaminopyridine. The structures of the synthetic acyl-HSLs were confirmed by analytical thin-layer chromatography (TLC) and by 1H nuclear magnetic resonance on a Varian Gemini 300-, Inova 400-, or Inova 500-MHz spectrometer. 1H nuclear magnetic resonance shifts were referenced to tetramethylsilane (0.0 ppm). N-(Hexanoyl)-l-HSL was purchased from Sigma-Aldrich.
Separation and visualization of acyl-homoserine lactones.
Two- to 5-μl volumes of samples to be analyzed were applied to C18 reverse-phase TLC plates and developed in methanol-water (60:40) as described previously (4, 45). The plates were dried, overlaid with appropriate indicator strains, incubated, and examined as described previously (15, 45).
Quantification of acyl-HSLs in culture supernatants.
Serial dilutions of ethyl acetate extracts of culture supernatants from strain 2-79 were chromatographed on TLC plates alongside dilution series of standard acyl-HSLs of known concentrations. Following development, the TLC plates were overlaid with cultures of the A. tumefaciens or C. violaceum indicator strain and incubated at 28°C. The diameters of the spots of different AHLs present in the mixture were measured and compared with the spots of standard acyl-HSLs of known concentrations chromatographed on the same plate as previously described (15, 45).
β-Galactosidase and β-glucuronidase assays.
β-Galactosidase or β-glucuronidase activities were assessed qualitatively by patching cultures onto solid ABM or LB media containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) or 5-bromo-4-chloro-3-indolyl-β-d-glucuronoside, respectively. Colony color and intensity were visually assessed after 18 to 36 h of growth at 28°C. β-Galactosidase activity was quantified according to the method of Miller (31), while β-glucuronidase activity was quantified as described by Jefferson et al. (22).
RESULTS
Production of PCA by P. fluorescens 2-79 requires PhzR and PhzI.
The phz operon of P. fluorescens 2-79 is preceded by two genes, phzR and phzI (Fig. 1A), the organization and sequence of which are highly conserved relative to homologs forming a quorum-sensing regulatory pair that activates expression of the phz operon in other soil pseudomonads (8, 34, 56). To determine if phzI and phzR are required for expression of the phz operon in strain 2-79, we replaced the two genes with a tet cassette (Fig. 1A). Whereas the parent strain produced PCA as detected by biological activity and chemical analysis, the deletion mutant, 2-79IR, failed to produce any detectable PCA (Fig. 1B and data not shown). Production of PCA was partially restored to strain 2-79IR by complementation with either pUCPRI or pUCPIR (Table 1), both of which carry phzI and phzR (data not shown).
When pUCPR, expressing only phzR (Table 1), was introduced into strain 2-79IR, the mutant failed to produce detectable levels of PCA (data not shown). However, when the culture was supplemented with an ethyl acetate extract of a culture supernatant of strain 2-79Z, the complemented mutant produced small but detectable amounts of the antibiotic (data not shown). Addition of the ethyl acetate extract to cultures of strain 2-79IR lacking the phzR clone did not restore production of PCA (data not shown). Combined, the data suggest that production of PCA by P. fluorescens 2-79 is controlled by PhzR and that this LuxR homolog requires an organic-soluble autoinducer likely produced by PhzI.
The phzI gene of strain 2-79 codes for the synthesis of 3-hydroxy- and alkanoyl-acyl-homoserine lactones.
The phz quorum-sensing systems of P. aureofaciens 30-84 and P. chlororaphis PCL1391 respond to C6-HSL (8, 57). On the other hand, we previously reported that strain 2-79 produces six signals: C6- and C8-HSL and three 3-OH derivatives with side chains of 6, 8, and 10 carbons (45). By mass spectrometry and chemical synthesis, we have since identified the sixth signal as the rare seven-carbon species N-(3-hydroxy-heptanoyl)-l-HSL (Fig. 2) (reference 12 and data not shown).
FIG. 2.
Deletion of phzI results in the loss of production of all acyl-HSLs normally synthesized by P. fluorescens 2-79. Ethyl acetate extracts of supernatants from cultures of wild-type P. fluorescens 2-79 (lanes 1), the phzD::lacZ mutant, 2-79Z (lanes 2), and the phzI deletion mutant, 2-79ZI (lanes 3), each grown in King's medium B to late exponential phase, were subjected to thin-layer chromatography, and the developed plates were overlaid with cultures of the A. tumefaciens (A) or C. violaceum (B) bioreporter, all as described in Materials and Methods. The position of migration of each active acyl-HSL component, identified as described previously (45), is indicated at the margin of each panel.
We assessed the role of the phzI gene in the production of these signals by examining culture supernatants of P. fluorescens 2-79IZ, in which phzI is deleted, for quormone content. As assessed by TLC, extracts from supernatants of late-exponential-phase cultures of strain 2-79IZ did not contain any signals detectable using either the A. tumefaciens or C. violaceum reporter (Fig. 2A and B, lanes 3).
It is possible that PhzI produces C6-HSL and that this signal, interacting with PhzR, activates another quorum-sensing system in strain 2-79 responsible for production of the 3-hydroxy-HSL signals. To test this hypothesis, we analyzed the acyl-HSL complement produced by heterologous bacteria expressing the phzI gene of strain 2-79 cloned in pSF106 (Table 1). Whether expressed in E. coli DH5α or in P. fluorescens 1855, neither of which produce their own acyl-HSLs (4, 45), the cloned phzI gene directed the synthesis of all six of the 3-hydroxy- and alkanoyl-acyl-HSLs characteristically produced by strain 2-79 (Fig. 3A and B). Strains harboring the pDLB4 vector alone did not produce detectable amounts of any active compound.
FIG. 3.
The phzI gene of P. fluorescens 2-79 confers production of all six acyl-HSLs. Ethyl acetate extracts of culture supernatants of strains harboring the cloned phzI gene in pSF106 or the vector pDLB4 were chromatographed, and the plates were overlaid with the A. tumefaciens (A) or C. violaceum (B) reporter strain as described in Materials and Methods. The lanes contain extracts from cultures of (1) P. fluorescens 2-79, (2) E. coli DH5α(pDLB4), (3) DH5α(pSF106), (4) P. fluorescens 1855(pDLB4), and (5) 1855(pSF106). Lane 6 contains a sample of pure N-hexanoyl-HSL standard. The locations and identities of the six acyl-HSLs are indicated at the margin of each panel.
We quantified the amounts of the major 3-OH and alkanoyl forms present in culture supernatants of strain 2-79 as described in Materials and Methods. In one such sample taken from a culture grown in minimal medium to late exponential phase, 3-OH-C6-HSL, at a concentration of 1.3 μM, dominated the mixture, while C6-HSL accumulated to 0.075 μM. 3-OH-C8-HSL, present at 0.04 μM, was the least abundant of the three species examined. Although the absolute amounts of each species varied as a function of culture phase and medium, the relative amounts of each of the tested species remained the same in all samples tested (data not shown). Moreover, the relative amounts of each species did not differ in samples prepared by extraction with ethyl acetate or dichloromethane (data not shown).
PhzR recognizes 3-OH-C6-HSL with the highest sensitivity.
To assess how PhzR-dependent regulation in strain 2-79 responds to the different signals, we constructed a reporter system composed of the cloned phzR gene on pSF105 and the phzR-phzA divergent dual promoter region fused between the oppositely oriented uidA and lacZ reporters on pSF107 (Table 1 and Fig. 4A). The two plasmids were introduced into P. fluorescens 1855, which neither synthesizes detectable acyl-HSLs nor produces detectable amounts of β-galactosidase or β-glucuronidase (data not shown). When grown in AB minimal medium to late exponential phase, the lacZ and the uidA reporter fusions were expressed at low but detectable levels (Table 2). However when grown in medium supplemented with an ethyl acetate extract of a culture supernatant of strain 2-79, the phzA reporter was induced almost 30-fold compared to cells grown without extract (Table 2). The phzR reporter was induced about twofold by the ethyl acetate extract (Table 2). Although low, this extract-dependent autoactivation of phzR by PhzR was observed consistently. In qualitative assays on medium containing X-Gal, induction of both activities required PhzR; strains lacking pSF105 exhibited no significant reporter activity even when cultured with the acyl-HSL (data not shown).
FIG. 4.
PhzR responds with greatest sensitivity to N-(3-OH-hexanoyl)-l-homoserine lactone. The intergenic region between phzA and phzI was cloned in the promoter vector pRG970b to form pSF107 as described in Materials and Methods. In this vector, the divergent promoter region forms reporter fusions to lacZ and uidA (A). P. fluorescens 1855(pSF107), also harboring pSF105, which codes for PhzR, was grown for 4 hours in ABM medium containing different concentrations of (□) N-(3-OH-hexanoyl)-HSL, (⋄) N-hexanoyl-HSL, or (○) N-(3-OH-octanoyl)-HSL. The cells were assayed for β-galactosidase activity from the phzA::lacZ reporter, expressed as Miller units (B), as described in Materials and Methods. The experiment was repeated twice, and a representative data set is shown. AI, acyl-HSL autoinducer.
TABLE 2.
Induction of phzA::lacZ and phzR::uidA requires acyl-HSLsa
| Reporter fusion | Enzyme activity
|
Inductionc | |
|---|---|---|---|
| − Acyl-HSL | + Acyl-HSLb | ||
| phzA::lacZd | 102 | 2,853 | 27 |
| phzR::uidAe | 35 | 101 | 1.9 |
Tested in P. fluorescens 1855(pSF105, pSF107) grown in ABM minimal medium.
Cultures were supplemented with an ethyl acetate extract containing the acyl-HSLs synthesized by strain 2-79.
Calculated as [(+AI) − (−AI)]/(−AI) (n-fold).
Measured in Miller units.
Measured in modified Miller units (22).
When tested with increasing concentrations of pure samples, the phzA::lacZ reporter of pSF107 exhibited the greatest sensitivity to and was induced to the highest levels by 3-OH-C6-HSL (Fig. 4B). The reporter also responded to 3-OH-C8-HSL and to C6-HSL but required concentrations of these two acyl-HSLs almost 10-fold higher than that of the 3-OH-C6 signal. Moreover, even when grown with saturating concentrations of C6-HSL or 3-OH-C8-HSL, the phzA::lacZ reporter yielded levels of maximum activation only about half that observed in cultures grown with 3-OH-C6-HSL (Fig. 4B).
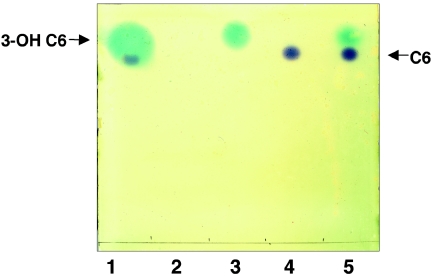
To assess the response of PhzR to the mixture of acyl-HSLs produced by strain 2-79, we chromatographed a sample of an ethyl acetate extract of a culture supernatant from the wild-type phenazine producer and overlaid the developed TLC plate with a mixture of the C. violaceum and P. fluorescens 1855 phz reporter strains (Fig. 5). In this mixed indicator system, the C. violaceum reporter, which recognizes C4 and C6 alkanoyl-HSLs (4, 30, 45), produces a purple pigment while the P. fluorescens phzA::lacZ indicator produces indol from hydrolysis of X-Gal, each in response to the acyl-HSLs to which it can react. The C. violaceum reporter in the mixture clearly responded to the C6-HSL, both in pure form (Fig. 5, lanes 4 and 5), and that present in the ethyl acetate extract (Fig. 5, lane 1). Moreover, of the six acyl-HSLs present in the sample, only C6-HSL was detectable by C. violaceum. The P. fluorescens phzA::lacZ reporter responded to only one standard, 3-OH-C6-HSL, and only to the 3-OH-C6 species present in the extract from the culture supernatant (Fig. 5, lanes 1, 3, and 5). This reporter did not respond to the alkanoyl quormone standard (Fig. 5, lane 4) or to the amount of C6-HSL present in the extract of the culture supernatant from strain 2-79 (Fig. 5, lane 1), nor did the P. fluorescens reporter respond to any of the other 3-OH- or alkanoyl-acyl-HSLs present in the culture extracts (Fig. 5, lane 1).
FIG. 5.
PhzR responds only to the 3-OH-hexanoyl-HSL present in culture supernatants of P. fluorescens 2-79. An ethyl acetate extract of a culture supernatant of P. fluorescens 2-79 grown to late exponential phase in King's medium B, as well as several standard compounds, was chromatographed, and the TLC plate was overlaid with a mixture of P. fluorescens 1855(pSF105, pSF107) and C. violaceum CV026blu as described in Materials and Methods. The blue spots represent acyl-HSLs detected by the P. fluorescens phzA::lacZ reporter, while the purple spots correspond to species detected by C. violaceum. Lanes contain samples of (1) extract from a culture supernatant of P. fluorescens 2-79 and pure standards of (2) N-(3-OH-octanoyl)-HSL, (3) N-(3-OH-hexanoyl)-HSL, (4) N-hexanoyl-HSL, and (5) a mixture of N-(3-OH-hexanoyl)-HSL and N-hexanoyl-HSL.
Other acyl-HSLs do not significantly influence activation of the phzA lacZ reporter by PhzR in response to 3-OH-C6-HSL.
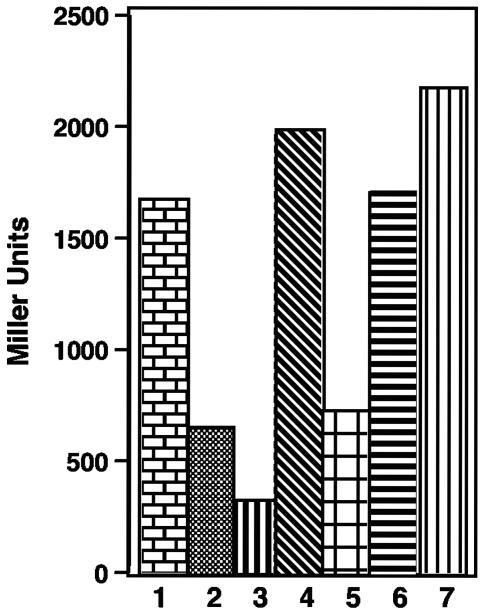
We considered the possibility that one or more of the acyl-HSLs produced by P. fluorescens 2-79 exert a modulating activity on PhzR-mediated gene activation. When grown in medium supplemented with only one of the three most abundant acyl-HSLs produced by strain 2-79, the P. fluorescens 1855 phzA::lacZ reporter responded with highest sensitivity to the 3-OH-C6 species and with less sensitivity to C6-HSL and 3-OH-C8-HSL (Fig. 6). Growth in medium supplemented with mixtures containing equal molar amounts of C6-HSL or 3-OH-C8-HSL, along with 3-OH-C6-HSL, gave slightly higher levels of response than cultures grown with only 3-OH-C6-HSL. A culture supplemented with equal molar amounts of all three acyl-HSLs induced the phzA::lacZ reporter to levels 30 to 40% higher than that seen in cultures incubated with 3-OH-C6-HSL alone or with mixtures of 3-OH-C6-HSL and one of the other two components (Fig. 6). Cultures incubated with a mixture of C6-HSL and 3-OH-C8-HSL but containing no 3-OH-C6-HSL induced the reporter to low levels similar to that observed in cultures grown with C6-HSL or 3-OH-C8-HSL (Fig. 6).
FIG. 6.
Noncognate acyl-HSLs have little effect on activation of PhzR by N-(3-OH-hexanoyl)-HSL. Individual cultures of P. fluorescens 1855(pSF105, pSF107), grown for 4 h in ABM medium with a single acyl-HSL or with mixtures of component acyl-HSLs, all at final concentrations of 500 nM, were assayed for β-galactosidase activities from phzA::lacZ as described in Materials and Methods. The cultures were supplemented with (1) N-(3-OH-hexanoyl)-HSL, (2) N-hexanoyl-HSL, (3) N-(3-OH-octanoyl)-HSL, (4) N-(3-OH-hexanoyl)-HSL and N-hexanoyl-HSL, (5) N-hexanoyl-HSL and N-(3-OH-octanoyl)-HSL, (6) N-(3-OH-hexanoyl)-HSL and N-(3-OH-octanoyl)-HSL, or (7) N-(3-OH-hexanoyl)-HSL, N-hexanoyl-HSL, and N-(3-OH-octanoyl)-HSL.
Transcriptional start sites for phzA and phzR.
Using primer extension analysis, we located the transcriptional start site for the phz operon to a G residue positioned 111 bp upstream of the translational initiation codon of phzA (Fig. 7B). This site is located 10 bp downstream from the center of a hexamer, 5′-TATAAA-3′, with a good sequence match to that of the consensus −10 promoter element (Fig. 7A). We also located the transcriptional start site of the divergently oriented phzR gene to a G residue 63 bp upstream of the putative translational start codon (Fig. 7C). This site is located downstream of properly spaced hexamers showing only weak relatedness to consensus −10 and −35 elements (Fig. 7A).
FIG. 7.
The divergent promoter region between phzR and the phz operon of P. fluorescens 2-79 contains cis-acting sites required for PhzR-mediated activation. The nucleotide sequence of the 379-bp region between phzR and phzA (A) contains putative −10 and −35 elements and ribosomal binding sites (RBS) as marked. The transcriptional start sites, shown as arrows labeled +1, were determined by primer extension analysis for phzA (B) and phzR (C) as described in Materials and Methods. The region also contains an 18-bp almost perfect inverted-repeat (phz box) that is related to the lux box sequence of Vibrio fischeri and also to inverted repeats located in the promoter regions of the phz operons of P. aureofaciens 30-84 and P. chlororaphis PCL1391 (D).
An intact phz box is required for PhzR-mediated activation of the phzA and phzR genes.
The intergenic promoter region also contains an 18-bp almost-perfect inverted repeat that resembles the lux box family of transcription factor binding sites (Fig. 7A and D). This repeat is situated upstream and immediately adjacent to the probable −35 element of the phzA promoter and approximately 120 bp upstream from the putative −35 element of the phzR promoter (Fig. 7A). We tested the requirement of this site for transcription by constructing a 4-bp insertion that duplicates the central GATC of the putative phz box sequence present in our standard reporter plasmid pSF107 (Table 1). Cultures of the strains harboring the wild-type and mutant reporter clones grown in the absence of an acyl-HSL signal expressed the phzA::lacZ and phzR::uidA fusions at low basal levels (Table 3). As expected, the strain harboring the wild-type promoter segment strongly expressed the lacZ reporter when grown with the acyl-HSL signal. However, in the reporter harboring the insertion mutant, expression of the phzA::lacZ fusion remained at basal levels even when the strain was grown with 3-OH-C6-HSL (Table 3). Similarly, the phzR::uidA fusion was induced almost twofold in response to the 3-OH-C6 signal in the strain harboring pSF107, but expression of this reporter remained at basal levels in the strain harboring the mutant phz box sequence in pSF108 (Table 3).
TABLE 3.
An intact phz box is required for phzR-dependent activation of phzA and phzRa
| Fusion | phz box | Enzyme activity
|
Inductionc | |
|---|---|---|---|---|
| − Acyl-HSL | + Acyl-HSLb | |||
| phzA::lacZd | Wte | 110 | 2,641 | 23 |
| Mutf | 95 | 97 | ||
| phzR::uidAg | Wte | 38 | 97 | 1.6 |
| Mutf | 26 | 28 | ||
Tested in P. fluorescens 1855(pSF105) grown in ABM minimal medium.
Cultures were supplemented with an ethyl acetate extract containing the acyl-HSLs synthesized by strain 2-79.
Calculated as [(+AI) − (−AI)]/(−AI) (n-fold).
Measured as Miller units.
Expressed from pSF107, which contains the wild-type (Wt) phz box sequence.
Expressed from pSF108, which contains a 4-bp insertion in the middle of the phz box sequence. Mut, mutant.
Measured as modified Miller units (22).
DISCUSSION
Expression of the phz operon of P. fluorescens 2-79 clearly is controlled by an autoinducer-type regulatory system composed of the LuxR homolog PhzR and soluble and diffusible acyl-HSL signals produced by the LuxI homolog PhzI. Moreover, the fact that a mutant lacking the two genes expresses the phz operon at its low basal level suggests that PhzR functions as a transcriptional activator. The regulatory ensemble resembles the systems that control phz gene expression in P. aureofaciens 30-84 (34, 56, 57) and P. chlororaphis PCL1391 (8). However, whereas these two pseudomonads are reported to produce and respond to the alkanoyl signal C6-HSL (8, 57), we confirm our previous report that P. fluorescens 2-79 produces at least six detectable acyl-HSLs, four of which are 3-OH derivatives (Fig. 3) (45). Moreover, the quantitatively dominant signal produced by strain 2-79 is 3-OH-C6-HSL, not the alkanoyl-C6 species.
From analyses of the acyl-HSL complement produced by strains of E. coli and P. fluorescens expressing the cloned phzI gene from P. fluorescens 2-79, we conclude that the enzyme coded for by this gene catalyzes synthesis of all six of the derivatives produced by wild-type strain 2-79 (Fig. 3). Consistent with this conclusion, the phzI mutant of strain 2-79 does not produce any acyl-HSLs detectable by the A. tumefaciens or C. violaceum bioreporters. Strain 2-79 therefore differs from P. aureofaciens 30-84, which produces additional signals via a second acyl-HSL synthase, CsaI (60). Our attempts to detect a csaI homolog in the genome of strain 2-79 by low-stringency hybridization with a csaI probe derived from strain 30-84 were unsuccessful (data not shown). Similarly, we did not detect a PCR product following amplification of genomic DNA from strain 2-79 using primers based on the sequence of the csaI gene from strain 30-84 (data not shown).
Several lines of evidence indicate that PhzR of strain 2-79 recognizes 3-OH-C6-HSL as its cognate signal. First, this hydroxy derivative is the most abundant of the six acyl-HSLs detected, accumulating in culture supernatants to levels almost 20 times higher than that of C6-HSL, the next most abundant species. Second, when used in the TLC assay, the P. fluorescens reporter strain expressing phzR and the phzA::lacZ of strain 2-79 responded to only the 3-OH-C6 species among the six acyl-HSLs present in extracts of supernatants from cultures of strain 2-79 (Fig. 5). While the reporter may have detected one or more of the other five acyl-HSLs had we chromatographed a more concentrated sample, these results show that PhzR clearly can be activated by 3-OH-C6-HSL under conditions in which the other acyl-HSLs, present in the mixture at their physiological ratios, did not elicit a detectable response. Finally, the phzA::lacZ reporter responded to pure 3-OH-C6-HSL at concentrations more than 10-fold lower than the concentrations of C6-HSL and 3-OH-C8-HSL standards required for activation of the reporter to similar levels (Fig. 4). Moreover, at saturating levels of each of the signals, PhzR from cells grown with 3-OH-C6-HSL activated the reporter to levels almost twice those observed in cells grown with the pure forms of the other tested acyl-HSLs (Fig. 4).
Given that the additional, less active acyl-HSLs synthesized by PhzI accumulate to levels much lower than 3-OH-C6-HSL, the cognate signal, it is not likely that they play a major role in modulating, either in a positive or negative manner, the activity of PhzR. Consistent with this hypothesis, even when provided at concentrations equal to that of the 3-OH-C6 signal, neither C6-HSL nor 3-OH-C8-HSL significantly affected PhzR-mediated activation of the phzA::lacZ reporter (Fig. 6). We suspect that the production of these additional acyl-HSLs represents the relaxed substrate specificity of PhzI, a characteristic that is common among the LuxI family of acyl-HSL synthases (21, 32, 40, 58). However, while these alternative signals play little role in controlling expression of the phz operon in strain 2-79, they may influence the acyl-HSL-dependent regulatory systems of other bacteria occupying the same habitat (57).
Consistent with the model that PhzR regulates production of phenazines by strain 2-79, the promoter region of the phz operon contains an inverted repeat similar in sequence to the lux box of Vibrio fischeri (Fig. 7A and D). Such boxes function as binding sites for LuxR-type activators (1, 13, 14, 18, 25, 37, 39, 41, 50, 52, 59). The sequence is identical to those of inverted repeats located in similar positions upstream of the phz operon of P. aureofaciens 30-84 (GenBank entry AF 007801) and of the phzI genes of strain 30-84 (56) and P. chlororaphis PCL1391 (8) (Fig. 7D), although there is no experimental evidence to show that these elements play a role in controlling expression of these gene sets in either bacterium. Our analysis of the 4-bp insertion in the inverted repeat of pSF108 indicates that this sequence must be properly intact in order for PhzR to activate transcription of the phz operon in vivo (Table 3). Similar disruptions block the in vivo and in vitro binding of TraR to the analogous tra box sequence located upstream of TraR-dependent promoters (25, 37).
Given the position of the transcriptional start site for the phz operon of strain 2-79, the upstream untranslated sequence contains good matches for −10 and −35 promoter elements (Fig. 7A). The inverted repeat, which our in vivo experiments suggest is a bona fide phz box, is located just upstream of the −35 element (Fig. 7A). Such a location for cognate activator binding sites is strongly conserved in other promoters dependent upon positively acting LuxR-like transcription factors, including TraR (18), LuxR (10, 14), and LasR (39, 41). The site placement also suggests that the cis-active sequences upstream of the phz operon constitute a class II promoter and that PhzR functions at this promoter as an ambidextrous activator in a manner similar to that proposed for LuxR, TraR, and LasR (14, 38, 41, 48, 52, 55, 59).
PhzR also activates expression of its own gene some 2-fold (Table 2), a level comparable to the 3.5-fold autoactivation of phzR by PhzR reported in strain 30-84 (34). Such relatively weak autoregulation is common among the luxR gene family (17, 36, 43, 44). Autoactivation of phzR in strain 2-79 requires an intact phz box (Table 3), suggesting that PhzR must bind to this site to activate expression of its own gene. However, the phz box is located more than 100 bp upstream of the putative −35 element of the phzR promoter, a position quite different from that of the phzA promoter. This difference in spacing suggests that PhzR promotes expression of phzR by a mechanism different from that by which it activates expression of the divergently oriented phz operon. This hypothesis, in turn, is consistent with a recent report showing that TraR can activate a gene from a tra box located well upstream of the promoter region (55) and suggests that both LuxR homologs can activate transcription at class I-type promoters.
PhzR from P. fluorescens 2-79 is 88% and 87% identical to the PhzR proteins of P. aureofaciens 30-84 and P. chlororaphis PCL1391, respectively, whereas the PhzR proteins of strains 30-84 and PCL1391 are 93% identical. While the PhzR proteins of strains 30-84 and PCL1381 apparently sense C6-HSL as their cognate signal (8, 57), PhzR of 2-79 is activated by 3-OH-C6-HSL. Sequence differences between PhzR from 2-79 and the other two pseudomonads are distributed over the entire lengths of the proteins. However, several variant residues are located in the N-terminal region, which in TraR and LuxR is required for acyl-HSL binding and ligand specificity (5, 19, 26, 52, 59). It is possible that one or more of these N-terminal substitutions is responsible for the apparent difference in signal specificity exhibited by PhzR from strain 2-79. Similarly, the PhzI protein from strain 2-79 is 87% and 86% identical to the PhzI proteins from strains 30-84 and PCL1391, respectively, which themselves are 96% identical. It is conceivable that one or more of the variant residues is responsible for the reported differences in the profiles of acyl-HSL signals produced by strains 30-84 and PCL1391 compared to strain 2-79. However, the three PhzI proteins share 100% sequence identity in the region between residues 121 and 150, a segment of this family of synthases suggested to be important for specifying the nature of the substitution at the C-3 position of the acyl-HSL product (53).
Clearly, the phz operons of the three rhizosphere pseudomonads all are controlled by a PhzR-PhzI regulatory system, the pairs of genetic components of which themselves are very closely related among the three bacteria. What is not clear is why the regulatory system of strain 2-79 would have evolved a signal specificity different from that of the other two pseudomonads.
Acknowledgments
Portions of this work were supported by grant no. R01 GM52465 from the NIH and C-FAR grant no. IDACF 01I-3-3 CS from the State of Illinois to S.K.F. and by grant no. 03-35319-13800 from the USDA NRI Grants Program to L.S.T.
We thank James Mitchell of the USDA-ARS Root Disease and Biocontrol Unit, Pullman, WA, for help with phenazine analyses.
REFERENCES
- 1.Anderson, R. M., C. A. Zimprich, and L. Rust. 1999. A second operator is involved in Pseudomonas aeruginosa elastase (lasB) activation. J. Bacteriol. 181:6264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anjaiah, V., N. Koedam, B. Nowak-Thompson, J. E. Loper, M. Hofte, J. T. Tambong, and P. Cornelis. 1998. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythium spp. Mol. Plant-Microbe. Interact. 11:847-854. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Short protocols in molecular biology. J. Wiley and Sons, New York, N.Y.
- 4.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe. Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 5.Chai, Y., and S. C. Winans. 2004. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Mol. Microbiol. 51:765-776. [DOI] [PubMed] [Google Scholar]
- 6.Chilton, M. D., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. USA 71:3672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin-A-Woeng, T. F. C., G. V. Bloemberg, A. J. Van der Bij, K. M. G. M. Van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. A. H. M. Bakker, H.-V. Tichy, F. J. De Bruijn, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 1998. Biocontrol by phenazine-1-carboxamide producing bacterium Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersic. Mol. Plant-Microbe. Interact. 11:1069-1077. [Google Scholar]
- 8.Chin-A-Woeng, T. F. C., D. van den Broek, G. de Voer, K. M. van der Drift, S. Tuinman, J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2001. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol. Plant-Microbe. Interact. 14:969-979. [DOI] [PubMed] [Google Scholar]
- 9.Chin-A-Woeng, T. F. C., J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2001. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe. Interact. 14:1006-1015. [DOI] [PubMed] [Google Scholar]
- 10.Choi, S. H., and E. P. Greenberg. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J. Bacteriol. 174:4064-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway. H. F., W. C. Haynes, R. W. Jackson, J. M. Locke, T. G. Pridham, V. E. Sohns, and F. H. Stodola. 1956. Pseudomonas aureofaciens Kluyver and phenazine α-carboxylic acid, its characteristic pigment. J. Bacteriol. 72:412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly, S. L. 1992. Characterization of bacterial autoinducers by mass spectrometry. M.S. thesis. University of Illinois at Urbana-Champaign, Urbana.
- 13.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. USA 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 15.Farrand, S. K., Y. Qin, and P. Oger. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358:452-484. [DOI] [PubMed] [Google Scholar]
- 16.Frankenfeld, J. W., and J. J. Werner. 1969. Improved procedure for the Reformatsky reaction of aliphatic aldehydes and ethyl bromoacetate. J. Org. Chem. 34:3689-3691. [Google Scholar]
- 17.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuqua, W. C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Hwang, I., P. L. Li, L. Zhang, K. R. Piper, D. M. Cook, M. E. Tate, and S. K. Farrand. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA 91:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 24.Luo, Z. Q., and S. K. Farrand. 1999. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J. Bacteriol. 181:618-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, Z. Q., and S. K. Farrand. 2001. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. USA 96:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, Z. Q., A. J. Smyth, P. Gao, Y. Qin, and S. K. Farrand. 2003. Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J. Biol. Chem. 278:13173-13182. [DOI] [PubMed] [Google Scholar]
- 27.Mavrodi, D. V., V. N. Ksenzenko, R. F. Bonsall, R. J. Cook, A. M. Boronin, and L. S. Thomashow. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzola, M., R. J. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson III. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierson, L. S., III, and L. S. Thomashow. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant-Microbe. Interact. 5:330-339. [DOI] [PubMed] [Google Scholar]
- 34.Pierson, L. S., III, V. D. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierson, L. S., III, T. Gaffney, S. Lam, and F. Gong. 1995. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30-84. FEMS Microbiol. Lett. 134:299-307. [DOI] [PubMed] [Google Scholar]
- 36.Piper, K. R., S. Beck Von Bodman, I. Hwang, and S. K. Farrand. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 32:1077-1089. [DOI] [PubMed] [Google Scholar]
- 37.Qin, Y., Z. Q. Luo, A. J. Smyth, P. Gao, S. Beck von Bodman, and S. K. Farrand. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin, Y., Z. Q. Luo, and S. K. Farrand. 2004. Domains formed within the N-terminal region of the quorum-sensing activator TraR are required for transcriptional activation and direct interaction with RpoA from Agrobacterium. J. Biol. Chem. 279:40844-40851. [DOI] [PubMed] [Google Scholar]
- 39.Rust, L., E. C. Pesci, and B. H. Iglewski. 1996. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J. Bacteriol. 178:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 43.Shadel, G. S., and T. O. Baldwin. 1992. Identification of a distantly located regulatory element in the luxD gene required for negative autoregulation of the Vibrio fischeri luxR gene. J. Biol. Chem. 267:7690-7695. [PubMed] [Google Scholar]
- 44.Shadel, G. S., and T. O. Baldwin. 1992. Positive autoregulation of the Vibrio fischeri luxR gene. LuxR and autoinducer activate cAMP-catabolite gene activator protein complex-independent and -dependent luxR transcription. J. Biol. Chem. 267:7696-7702. [PubMed] [Google Scholar]
- 45.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shriner, R. L. 1942. Reformatsky reaction. Org. Reactions 1:1-37. [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering. Transposon mutagenesis in gram negative bacteria. Bio/Technol. 1:784-791. [Google Scholar]
- 48.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Eede, G., R. Deblaere, K. Goethals, M. Van Montagu, and M. Holsters. 1992. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol. Plant-Microbe. Interact. 5:228-234. [DOI] [PubMed] [Google Scholar]
- 52.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, W. T., T. D. Minogue, D. L. Val, S. Beck von Bodman, and M. E. A. Churchill. 2002. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell 9:685-694. [DOI] [PubMed] [Google Scholar]
- 54.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 55.White, C. E., and S. C. Winans. 2005. Identification of amino acid residues of the Agrobacterium tumefaciens quorum-sensing regulator TraR that are critical for positive control of transcription. Mol. Microbiol. 55:1473-1486. [DOI] [PubMed] [Google Scholar]
- 56.Wood, D. W., and L. S. Pierson III. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]
- 57.Wood, D. W., F. Gong, M. M. Daykin, P. Williams, and L. S. Pierson III. 1997. N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179:7663-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, L., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446-448. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Z., and L. S. Pierson III. 2001. A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 67:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]