Abstract
CS1 is the prototype of a class of pili of enterotoxigenic Escherichia coli (ETEC) associated with diarrheal disease in humans. The genes encoding this pilus are carried on a large plasmid, pCoo. We report the sequence of the complete 98,396-bp plasmid. Like many other virulence plasmids, pCoo is a mosaic consisting of regions derived from multiple sources. Complete and fragmented insertion sequences (IS) make up 24% of the total DNA and are scattered throughout the plasmid. The pCoo DNA between these IS elements has a wide range of G+C content (35 to 57%), suggesting that these regions have different ancestries. We find that the pCoo plasmid is a cointegrate of two functional replicons, related to R64 and R100, which are joined at a 1,953-bp direct repeat of IS100. Recombination between these repeats in the cointegrate generates the two smaller replicons which coexist with the cointegrate in the culture. Both of the smaller replicons have plasmid stability genes as well as genes that may be important in pathogenesis. Examination by PCR of 17 other unrelated CS1 ETEC strains with a variety of serotypes demonstrated that all contained at least parts of both replicons of pCoo and that strains of the O6 genotype appear to contain a cointegrate very similar to pCoo. The results suggest that this family of CS1-encoding plasmids is evolving rapidly.
Enterotoxigenic Escherichia coli (ETEC) is an important cause of diarrheal diseases in humans (1, 2, 14, 20). Although ETEC infections are usually self-limiting in adults, the disease results in death in infants and young children in developing countries. Following colonization of the small intestine, these bacteria produce the heat-labile (LT) and heat-stable (ST) toxins, which are responsible for the extensive fluid secretion from the intestine that characterizes this disease.
An important first step in ETEC infection, colonization of the host, requires attachment of the bacteria to cells of the small intestine. In gram-negative bacteria, such attachment is usually mediated by pili. In ETEC strains associated with human disease, at least 21 serologically distinct pili have been identified (12, 27). CS1, the best characterized of these, is the prototype of a family of ETEC pili that includes CS1, CS2, CS4, CS14, CS17, CS19, and colonization factor antigen I (CFA/I) (12), whose structural and assembly proteins have similar sequences.
For production of functional CS1 pili, only four genes (cooB, -A, -C, and -D) are necessary when cloned in E. coli K-12 (11, 26, 30). Like many other virulence genes, the coo genes are located on a plasmid, called pCoo (9, 26). In ETEC strains, a positive regulator, rns, is required for expression of the coo operon. Rns is encoded on another plasmid, which has IncF1 incompatibility (3, 5, 10, 22). In some CS1-producing strains, this plasmid also encodes the LT and ST and the CS3 colonization factor (22).
A partial sequence of the pCoo plasmid (36 kb) from the ETEC-derived strain C921b-1 revealed regions homologous to parts of the IncI1 plasmid R64 (9). This homology includes the R64 replication region, and, as expected, pCoo is incompatible with R64. In addition, this homology includes most of the genes required for synthesis of the R64 thin pilus, required for conjugation in liquid. However, because this locus is not complete, pCoo is not self-conjugative.
In this work, we report the complete sequence of the 98,369-bp pCooKm plasmid. Like many other virulence plasmids, pCoo is a mosaic made up of DNA from various sources. Surprisingly, we found that pCoo is a cointegrate with two functional replication regions (R64- and R100-like). The two smaller replicons are separated by 1,953-bp direct repeats of insertion sequence 100 (IS100) that serve as homologous recombination sites leading to resolution into the individual small plasmids. Each of the smaller replicons has genes important for stable plasmid inheritance. We also examined pCoo plasmids from 17 CS1-producing ETEC strains with a variety of serotypes and found that all contain portions of both replicons.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains used are listed in Table 1. Bacteria were grown in Luria-Bertani medium at 37°C with aeration. The plasmid pHSG576 is a pSC101 replicon (Cmr) (33). R64colK is a conjugative IncI1 Tcr plasmid (36) and will be referred to as R64 in the rest of this paper. Antibiotics used were kanamycin (50 μg/ml) and tetracycline (10 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant traitsa | Reference or sourceb |
|---|---|---|
| 1392-75-2A | ETEC O6:H16, biotype A, CS1+ CS3+ | Myron Levine |
| 294 | ETEC, CS1+ | Foodborne and Diarrheal Diseases Laboratory at the CDC |
| 295 | ETEC, CS1+ | Foodborne and Diarrheal Diseases Laboratory at the CDC |
| 296 | ETEC, CS1+ | Foodborne and Diarrheal Diseases Laboratory at the CDC |
| 60R75 | ETEC, CS1+ | Bernard Rowe via Eileen Barry |
| C921b-1 | ETEC O6:H16, biotype A, CS1+ CS3+ | 31 |
| E4377/0/A | ETEC O139:H8, CS1+ CS3+ | 29 |
| JEF100 | Derivative of C921b-1, CooB::ΩKm | 30 |
| K1250 | E. coli/R100 (Tcr) | E. Lederberg |
| EU2574 | JEF100/pHSG576/R64 (Kmr Cmr Tcr) | 9 |
| M424C1 | ETEC O6:H16, CS1+ CS3+ | 21 |
| TW03577 | ETEC O25, CS1+ | 15, 37 |
| TW03605 | ETEC O6:H16, CS1+ CS2+w CS3+ CS5+ | 15, 37 |
| TW03923 | ETEC O6, CS1+ CS2+ CS3+ CS6+ | 15, 37 |
| TW03875 | ETEC O6, CS1+ CS3+ | 15, 37 |
| TW04205 | ETEC O6, CS1+ CS3+ CS6+w | 15, 37 |
| TW04215 | ETEC O6, CS1+ CS3+ | 15, 37 |
| TW04237 | ETEC O27, CS1+ CS4+ CS6+ | 15, 37 |
| TW04144 | ETEC O−, CS1+ CS2+w CS4+ CS5+w CS6+ | 15, 37 |
| TW04316 | ETEC O148, CS1+ CS2+ CS6+ | 15, 37 |
| TW04234 | ETEC O148, CS1+ CS3+w CS6+ | 15, 37 |
| TW04244 | ETEC O−, CS1+ | 15, 37 |
| TW04211 | ETEC O159, CS1+ CS6+ | 15, 37 |
Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant; w, weakly positive.
CDC, Centers for Disease Control and Prevention.
DNA sequencing of pCoo.
About 36 kb of pCoo has been previously sequenced (9). Plasmid DNA isolated from JEF100 (Table 1) was used for sequencing the rest of the pCooKm plasmid. The DNA was fragmented by sonication and size fractionated before constructing libraries in pUC19. The sequence was generated from 821 paired end-reads from two pUC19 libraries with insert sizes of 1.4 to 2 kb and 713 paired end-reads from two pUC19 libraries with insert sizes of 2 to 4 kb. These 1,534 sequence reads were performed with ABI BigDye Terminator chemistry on ABI3730 sequencing machines and gave a total coverage for the plasmid of 9.94-fold. All identified repeats were bridged with read pairs or end-sequenced PCR products. Error checking and finishing of the sequence were performed to standard criteria, and the final sequence had a quality score of >30 at each base (equivalent to an estimated error rate of <1 bp per 1.73 Mb). The sequence was annotated using Artemis (28). Predicted coding sequences (CDSs) were identified manually with reference to positional base composition and amino acid usage plots. The entire sequence was searched in all six reading frames against the nonredundant TrEMBL database using BLASTX to ensure no genes were missed. Each CDS was searched against the nonredundant databases using FASTA and BLASTP and against the PFAM and Prosite databases of protein motifs. Transmembrane helices were identified with TMHMM (18) and signal sequences with SignalP (24). Repeats were identified using Dotter (32).
Nucleotide sequence accession number.
The nucleotide sequence of the pCoo plasmid has been deposited in EMBL/GenBank under accession no. CR942285.
RESULTS AND DISCUSSION
General structure of pCoo.
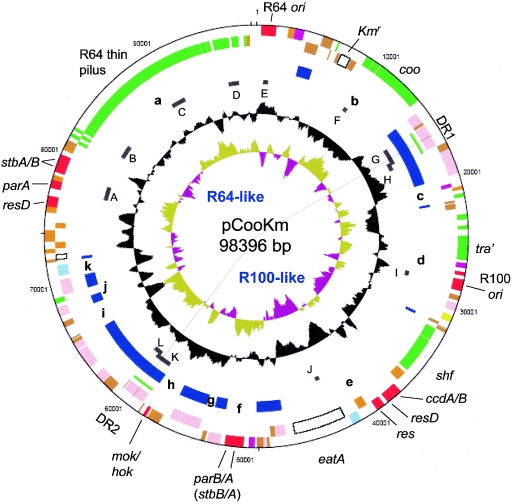
The pCoo plasmid with a kanamycin resistance gene inserted into cooB was obtained from the ETEC-derived strain JEF100 (Table 1) and its sequence determined. Analysis of the sequence of this 98,396-bp plasmid revealed 129 predicted CDSs, including the inserted Km gene (see Table S1 in the supplemental material for the locations and homologies of all CDSs of pCoo). Like many other virulence plasmids, pCoo appears to be a mosaic consisting of regions derived from multiple sources. DNA homologous to IS elements is scattered throughout the plasmid and comprises 24% of the total pCooKm DNA (Fig. 1). There are 11 regions between 195 and 7,612 bp in length that contain intact and partial IS elements. Many of these regions contain fragments of several different IS elements, which suggests that they are not the result of single insertion events, although some recent simple insertions were identified.
FIG. 1.
Circular representation of pCooKm. The concentric circles represent, from the outside in, the following: 1 + 2, coding sequences on the forward and reverse strands (red, plasmid functions [replication, partition, maintenance, etc.]; green, cell surface structures; pink, IS elements and phage proteins; white, virulence and drug resistance; yellow, metabolic proteins; blue, regulators; purple, unknown; orange, conserved hypothetical; brown, pseudogenes); 3, DR1 and DR2 shown as green arcs; 4, IS elements and group II introns shown as blue blocks, with the unique regions between, designated with lowercase italic letters a through k in the text and indicated in bold in the figure; 5, PCR-amplified regions, labeled in uppercase letters A through L; 6, G+C content (window size, 700 bp; step, 50 bp); 7, GC bias [(G-C)/(G+C)] (window size, 1,000 bp; step, 50 bp); values of >1 for khaki and <1 for magenta. The radial lines indicate the site of recombination between DR1 and DR2 that resolves pCoo into two smaller plasmids. The blue labels in the center indicate the R64-like and R100-like sections of pCoo.
The G+C content of the pCoo DNA sequences between these IS elements ranges from 57% to 35%. This wide range of G+C content is consistent with pCoo being derived by the amalgamation of DNA from a variety of sources. The pCoo sequences between the IS fragments are labeled with a bold lowercase letter in Fig. 1, and they have G+C contents of 49%, 40%, 49%, 46%, 42%, 45%, 53%, 57%, 36%, 35%, and 36% for regions a to k, respectively. These regions are described below.
A large contiguous part of pCoo region a (bp 83057 through 98396 and 1 through 2250) is 95% identical at the level of DNA to a contiguous region of plasmid R64 (bp 105487 through 120826; accession no. AP 005147) (9). Part of region a of pCoo (pCoo bp 72009 through 78749) is similar to the closely related plasmids R64 and ColIb-P9 (accession no. AP 005147 and NC 002122, respectively). Although these plasmids are in the same incompatibility group, there are DNA segments that differ in sequence between them. This part of pCoo appears to be a hybrid between these two plasmids because it has one segment (bp 72009 through 72759) similar to a unique region in ColIb-P9 and another (bp 75370 through 77265) similar to a unique region in R64. Within the part similar to both R64 and ColIb-P9, there are two genes with no similarities to genes of known function. The rest of the CDSs in this section have homologies to genes of known function and will be discussed later.
A homologue of the AraC family regulatory gene csvR (accession no. X60106) is located in a part of pCoo (bp 65421 through 69147; spanning regions i and j, the IS element between them, and portions of the flanking IS elements) that is 96% identical to a section of a plasmid from an ETEC strain of serotype O167 H5 (6). Although CsvR is 75% identical to Rns, the positive regulator for the coo operon present on another plasmid in the same ETEC strain, the pCoo copy of CsvR contains a deletion leading to a frameshift, so it is not expected to be expressed.
A number of conserved hypothetical genes in pCoo show similarity to genes from several other different gram-negative chromosomes and plasmids, contributing to the impression of a mosaic origin for pCoo.
Potential virulence genes encoded on pCoo.
In addition to the four coo genes (Fig. 1, region b) required for CS1 pilus production, several predicted gene products of pCoo resemble proteins implicated in pathogenesis in other bacteria, and these may be important for the virulence of an ETEC strain containing pCoo.
Region e of pCoo (Fig. 1, between bp 32036 and 36029) is 95% identical to the shf operon of the Shigella flexneri virulence plasmid pINV (accession no. AY206446 [19]). The two last proteins of the pCoo shf locus are 97% identical to their counterparts in pINV of Shigella flexneri, and these genes have been implicated in virulence. The first of these proteins, VirK (pCoo045), mediates intracellular spreading of Shigella flexneri by regulating the expression of VirG (IcsA), an outer membrane protein, and the last protein encoded in this locus, MsbB2 (pCoo044), is required for the full virulence of a septicemic E. coli strain (17, 23).
The protein encoded by pCoo056 (Fig. 1, region e) is 94% identical to the secreted autotransporter EatA of an ETEC strain that produces CFA/I pili, another member of the CS1 pilus family. EatA is a serine protease that increases fluid accumulation in a rabbit ileal loop model of ETEC infection (25). PCR analysis of 41 human clinical ETEC isolates showed that about 61% contain the eatA gene, suggesting that it may be important in human disease as well. Directly downstream of the pCoo eatA gene and transcribed convergently is a member of the AraC family of transcription regulators (Fig. 1, region e) (pCoo055), which, because of its proximity, might regulate the expression of eatA. The sequence of the corresponding region in the CFA/I plasmid is not yet available.
A second AraC family member is pCoo089, which is 70% identical to cafR, the regulator of the Yersinia pestis operon that encodes the f1 nonfimbrial adhesin (accession no. P26950). This protein is encoded in region k (Fig. 1). If it is expressed from pCoo, it might regulate other virulence factors on plasmids and/or the host chromosome.
Several CDSs of pCoo are homologous to genes whose functions do not suggest roles in pathogenesis. A segment homologous to the conjugative transfer region of the IncFV plasmid pED208 (TraM, TraJ, TraA, TraL, TraE, and a TraK/X fusion [pCoo28-33] are 65%, 27%, 83%, 86%, 92%, and 85%/73% identical, respectively; accession no. AF411480) is found at region d of pCoo. In pED208, there are 20 transfer genes between TraK and TraX which are absent from pCoo, suggesting that the fusion TraK/X protein was formed as a result of a deletion of a large portion of this transfer region.
Surprisingly, pCoo region d encodes a protein 98% identical to the E. coli peptide deformylase (pCoo040; accession no. P71251), which is essential for bacterial protein maturation. To our knowledge, this is the first report of this essential enzyme being encoded on a plasmid, although homologues of other essential chromosomal genes, often involved in DNA metabolism, have been found on other plasmids.
Two other predicted pCoo proteins (pCoo071 and pCoo072, region h) have homology to R100 proteins thought to contribute to the conjugative processing of DNA (accession no. P18148 and P28044). Both of these are only partial genes and probably represent remnants of genes from other plasmids since pCoo is not conjugative (9).
Plasmid pCoo has two functional origins of replication.
In addition to the functional replication region homologous to the conjugative IncI1 plasmid R64 (9), pCoo contains a second replication region (pCoo bp 27285 through 29452) (Fig. 1) that is closely related to that of the IncFII plasmid R100. This region contains all of the gene products and cis-acting sites required for the replication of R100, including the replication initiation protein RepA1 (pCoo036). This contiguous region of pCoo is organized identically to that of R100. The expression of repA1 is positively regulated by RepA6 (pCoo35a) and negatively regulated by RepA2 and by the antisense RNA, RNA-I (7). The incompatibility determinant, RNA-I, of pCoo, is 90% identical to that of R100. The RepA1 and RepA6 proteins of pCoo are 99% identical to the R100 RepA1 and RepA6 proteins, and pCoo RepA2 (pCoo0035) is 71% identical and 90% similar to the RepA2 of R100 (accession no. AP000342). The pCoo origin is 99% identical to the origin region, oriR, of R100. There are additional cis-acting sites downstream of the origin in R100 that are important for stable inheritance of the plasmid, and these are also present in pCoo (16). These homologies strongly suggest that the R100 origin in pCoo is functional (see below).
It seems likely that pCoo is a cointegrate formed by the recombination of two independent replicons because the two pCoo origins are separated by long (1,953-bp) direct repeats comprised of recent IS100 insertions. Analysis of the patterns of target site duplications, and the genes into which the IS100s have inserted, suggests that they transposed into separate genes and subsequently recombined (Fig. 1, direct repeat I [DRI] and DRII). Recombination to resolve the cointegrate is expected to be frequent because of the length of the repeated region. To detect the presence of the independent replicons that would be generated by recombination, we performed PCR analysis using primers that flank the direct repeats (Fig. 2). Primers 1 and 2, which flank DRII, and primers 3 and 4, which flank DRI, will amplify PCR products from unresolved intact pCoo but not from the two independent plasmids (Fig. 2). Primer pairs 2 and 3 and 1 and 4 will amplify PCR products from the separate replicons, R64 and R100, respectively, but not from the pCoo cointegrate. By using total plasmid DNA from the pCooKm-containing strain JEF100 as template, PCR products of the expected sizes were obtained with all four primer pairs (1 + 2, 3 + 4, 3 + 2, and 1 + 4) (data not shown). This demonstrates that the pCoo cointegrate plasmid coexists with the two separate replicons in a single culture.
FIG. 2.
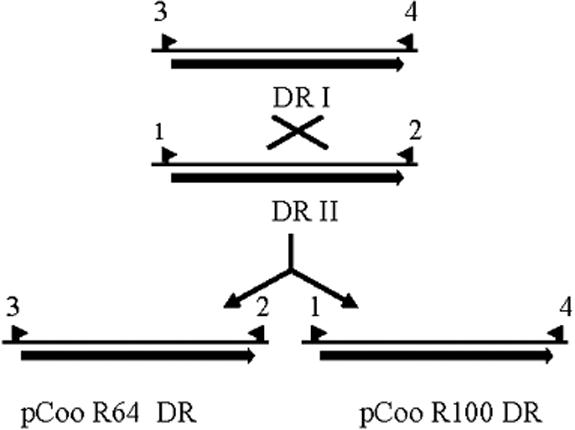
DNA region containing the direct repeats of whole pCoo and repeats after resolution of the pCoo cointegrate. The repeats (arrows) in whole pCoo are labeled DRI and DRII. The repeat regions contained within the products of recombination are labeled pCoo R64 DR and pCoo R100 DR. The start sites and directions of primers 1, 2, 3, and 4 which flank the repeated region are indicated by small arrows. See Table S2 in the supplemental material for primer sequences.
We have shown previously that the R64 origin is functional (9); however, it is possible that the R100 origin is not functional. The R100-related small plasmid may be continuously generated from the cointegrate replicon by recombination. Assuming that the pCoo origins have low copy numbers like both R100 and R64, a nonreplicating R100 plasmid would not be expected to reside in many cells containing the cointegrate because of incompatibility with the cointegrate. However, a nonreplicative R100 might be present transiently. To determine whether the R100-related plasmid is a functional replicon, we used incompatibility to isolate colonies that had lost the R64 portion of pCoo (9) and asked whether an R100 replicon was still present.
A culture of strain EU2574, which carries both R64Tc and pCooKm (the Km marker is in cooB, which is in the R64 part of the large plasmid) (Fig. 1), was grown for 24 generations without antibiotic selection, and colonies were scored for Km and Tc. We found that all 643 of the EU2574 colonies scored still contained the Tc marker of R64. In 24 generations, 80% of the colonies had lost the Km marker, indicating a loss of both pCooKm and its smaller R64Km derivative. If the R100-like plasmid could replicate independently, cells that had lost pCooKm should still contain DNA from the R100-like replicon. Plasmid DNA was isolated from 7 Kms colonies, and the presence of the R100-like replicon was assessed by PCR using primers 1 and 4, which flank the repeat in the R100-like replicon (Fig. 2, pCoo R100 DR). A PCR product of the expected size was amplified from all of the colonies tested, demonstrating the presence of the R100-like replicon (data not shown). As expected, fragment G within the R64 part of pCoo (Fig. 1) was not amplified from these Kms colonies. Thus, the R100-like replicon is stable in the absence of the R64 replicon of pCoo, indicating that the R100-like origin, like the R64 origin, is functional. It remains formally possible that the R100 (and R64) resolution products undergo chromosomal integration in such a way that the recombinant junctions are intact, since this could not be detected by our PCR analysis.
Since the incompatibility determinant, RNA-I, of the R100-like part of pCoo is not 100% (but only 90%) identical to that of R100, it seemed possible that the two plasmids might be compatible. We showed above that the pCoo cointegrate plasmid resolves to form the two separate replicons, and since introduction of an IncI1 plasmid leads to loss of the R64 replicon of pCoo (9), we applied the same test for R100 incompatibility. We investigated whether the introduction of R100 (Tcr) into a strain carrying pCooKm (Table 1, JEF100) leads to loss of the R100-like part of the pCoo cointegrate. After 25 generations in the absence of selection, 300 colonies were found to grow on plates containing both Km and Tc, indicating that both the R64 replicon of pCooKm (Kmr) and R100 (Tcr) were present. Plasmid DNA was isolated from 35 of these colonies and the presence of the R100-like replicon of pCooKm was assessed by PCR, using primers within the eatA gene and the araC homolog directly downstream of eatA (Fig. 1). A PCR product of the expected size was amplified from all colonies tested (data not shown). Using the same primers, no PCR product was amplified from plasmid isolated from the negative control (Table 1, K1250/R100). These results demonstrate that the R100-like pCoo replicon can coexist with R100 in the absence of selection, demonstrating that they are in different incompatibility groups.
Plasmid stability regions of pCoo.
Since both replicons of pCoo can function independently, they might be expected to be lost independently in the absence of known selection. The strain analyzed was selected for production of the CS1 pili encoded by the R64 homologue, but there was no obvious selection for the R100 homologue. Thus, the continued presence of both replicons suggests that they are both inherited very stably. Several different mechanisms are used by plasmids to ensure stable and equal distribution into daughter cells following replication (for a review, see reference 38). More than one of these mechanisms is often present in the same plasmid. Often plasmid replication results in the production of multimers that would be segregated as one molecule into the same daughter cell at division unless they were resolved. Thus, production by the plasmid of a site-specific recombinase to resolve these multimers into monomers is an important stability mechanism. A second mechanism is an active centromere-like partition process (encoded by par or stb genes), which precisely distributes copies of the plasmid to each daughter cell. Third, and perhaps most effective (4), is the postsegregation killing or plasmid addiction mechanism. In the last case, plasmid-free segregants are killed because of the production of a plasmid-encoded stable toxin that is counteracted in plasmid-containing cells by the production of an unstable antidote.
The plasmid stability region in the R64 portion of pCoo (Fig. 1, region a) contains genes for the first two types of stability mechanisms: site-specific recombination and active partition (9). The resolvase ResD (pCoo097; 91% identical to the Rsv protein required for maintenance of the F plasmid), ParA (pCoo98; 61% identical to a member of the ParA family from Xanthomonas citri), StbA (pCoo100; 66% identical to StbA of EPEC plasmid pB171), and StbB (pCoo101; 46% identical to StbB of pB171) are all likely to contribute to the stable inheritance of pCoo and the pCoo R64 replicon.
The R100 part of pCoo also has genes homologous to those that encode all three stability mechanisms described above. This region encodes two postsegregation killing systems: ccdA/B (Fig. 1, region e) and hok/sok/mok (Fig. 1, region h). CcdB (pCoo051) is the cytotoxic protein that kills plasmid-free segregants, and CcdA (pCoo050) is the antitoxin protein that protects the plasmid-containing cells (13). In plasmid pCoo, CcdA and -B are 100% identical to the CcdA and -B proteins found in E. coli plasmid pB171 (accession no. BAA84907 and BAA84908, respectively [35]). The pCoo Hok protein (pCoo075), which is the stable toxin that kills plasmidless cells, is predicted to be 74% identical to Hok of R100, and pCoo Mok (pCoo074), the modulator of Hok, is 51% identical to Mok of R100 (accession no. AP000342 [34]).
In addition to the postsegregation killing systems, the R100 half of pCoo encodes two proteins homologous to those of an active partitioning system. ParA (pCoo064; region f) is 87% identical to ParA from Citrobacter freundii (accession no. AF550415) and 53% identical to StbA from E. coli plasmid R100 (accession no. AP000342). ParB (pCoo065; region f) is 75% identical to ParB of Citrobacter freundii (accession no. AF 550415) and 39% identical to StbB of E. coli plasmid R100 (accession no. AP000342).
Both replicons of pCoo also encode putative resolvases. The R100 part encodes two resolvase homologues. One, Res (pCoo053; region e), is 94% identical to a Proteus vulgaris resolvase (accession no. AP004237), and the second, ResD (pCoo052; region e), is 96% identical to the Rsv protein of the F plasmid (8).
There is also a ResD homologue in the R64 part of pCoo (pCoo097; region a). The two ResD proteins are oriented in the same direction, and a 400-bp segment of the two CDSs is 95% identical. These direct repeats, like the repeats between the two replicons, could serve as a substrate for recombination. However, in this case, one recombinant would contain a replication origin and the other would not. That pCoo has not lost the region between the two ResD homologues is probably due to the presence of the mok/hok postsegregation killing system on the nonreplicating recombinant circle. The presence of the stable Hok protein in the culture would insure the killing of any cell lacking this region. Because so many plasmid stability genes are present, it is not surprising that both of the smaller replicons, as well as the pCoo cointegrate, are stably maintained.
Structure of pCoo plasmids from other CS1-expressing ETEC strains.
Although both parts of the pCoo cointegrate are likely to be stably inherited both because of stability functions and possibly because of genes providing a selective advantage, the advantage to maintaining them on one large cointegrate plasmid is not obvious. Therefore, one might expect to find some ETEC strains that have only the two smaller plasmids and have lost the direct repeat region required for recombination, leading to formation of the cointegrate.
To learn about the presence and configuration of the pCoo-related plasmids of other ETEC strains, we used PCR to probe unrelated clinical ETEC isolates that produce CS1 pili (Table 1). With the exception of strains 294, 295, and 296, which had not been studied as completely, these strains were chosen for the study because they differed in one or more known genetic traits, like serotype, drug resistance, or type of pilus (in addition to CS1) produced. Primer pairs (Table S2) were selected to amplify the pCoo regions shown as uppercase letters in Fig. 1. As expected from the presence of CS1 pili on their surfaces, region E, containing cooA, and region A, which is specific to pCoo and within the R64 replicon, were amplified from all of the strains (data summarized in Table 2).
TABLE 2.
PCR analysis of plasmid DNA isolated from CS1-positive ETEC strainsa
| Class and strain | Serotype | CS pili | Toxinsb
|
Amplified regionsc
|
Tcf | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LT | ST | pCoo specific | R64 pilus
|
R64 ori | Coo | DRI
|
R100 ori | EatA | DRII
|
||||||||
| A | B | C | D | E | F | G | H | I | J | K | L | ||||||
| Class A | |||||||||||||||||
| C921b-1 | O6H16 | 1, 3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | S |
| 1392-75-2A | O6H16 | 1, 3 | − | ? | + | + | + | ND | + | + | + | + | + | + | + | + | S |
| M424CI | O6H16 | 1, 3 | ? | ? | + | + | + | ND | + | + | + | + | + | + | + | + | S |
| TW03605 | O6H16 | 1, 2, 3, 4 | + | + | + | ND | + | + | + | + | + | + | + | + | + | + | S |
| TW03923 | O6H− | 1, 2, 3, 6 | + | + | + | ND | − | − | + | + | + | + | + | + | + | + | S |
| TW03875 | O6 | 1, 3 | + | + | + | ND | + | + | + | + | + | + | + | + | + | + | S |
| TW04215 | O6 | 1, 3 | + | + | + | ND | + | ND | + | + | + | + | + | + | + | + | S |
| TW04205 | O6 | 1, 2, 6 | + | + | + | ND | + | + | + | + | + | + | + | + | + | + | S |
| 60R70 | O6 | 1 | + | ? | + | − | − | + | + | + | + | + | + | + | + | + | S |
| 294 | 1 | ? | ? | + | + | + | ND | + | + | + | + | + | + | + | + | S | |
| 295 | 1 | ? | ? | + | + | + | ND | + | + | + | + | + | + | + | + | S | |
| 296 | 1 | ? | ? | + | − | + | ND | + | + | + | + | + | + | + | + | R | |
| Class B | |||||||||||||||||
| TW04244 | O− | 1 | + | + | + | ND | − | + | + | + | − | + | + | + | − | − | R |
| TW04144 | O− | 1, 2, 4, 5, 6 | − | + | + | ND | + | + | + | + | − | + | + | + | + | + | S |
| TW03677 | O25 | 1 | + | − | + | ND | − | − | + | + | +d | + | + | − | − | − | S |
| TW04237 | O27 | 1, 3, 4, 6 | − | + | + | ND | + | + | + | + | + | + | + | + | − | + | S |
| E24377/O/A | O139H8 | 1, 3 | + | + | + | ND | − | + | + | + | − | + | + | + | − | − | S |
| TW04316 | O148 | 1, 2, 6 | − | + | + | ND | − | + | + | + | − | + | + | + | − | − | S |
| TW04211 | O159 | 1, 6 | − | + | + | ND | − | + | + | + | + | +e | + | + | + | + | S |
The table is divided into two parts, classes A and B by O type and the presence of DRI and DRII PCR products identical to those for C921b-1.
?, unknown.
The amplified fragments G, H, K, and L each use one primer within the direct repeat and the second flanking the repeat. ND, not done.
PCR product larger than that for C921b-1.
PCR product smaller than that for C921b-1.
S, sensitive; R, resistant.
At least a part of the region encoding the R64 thin pilus (Fig. 1; Table 2, regions B, C, and D) was amplified from all but two of the strains (TW03923 and TW03677). From both of these, a product was obtained using primers within the R64 origin region (E), which is just upstream of the R64 thin pilus region. This suggests that most, but not all, of the R64 homology found in pCoo is absent from these strains. Deletions of parts of the thin pilus operon in some of the ETEC strains suggested by the inability to amplify regions B, C, and/or D indicates that this operon may not be essential for ETEC infection. This is supported by the nonfunctionality of this system in pCoo.
All of the ETEC strains have the R100-like origin region (Table 2, amplified region I), and primers within eatA (region J), which is also in the R100 part of pCoo, generated a PCR product from all but one of the strains (TW03677). Thus, all of the strains have some regions from both the R64 and R100 halves of pCoo.
To determine whether the cointegrate structure found in pCoo of C921b-1 is retained in the plasmids of the other strains, we used primers flanking the direct repeats (DRI and DRII) together with primers within the repeat. If DRI is present, amplified fragments G and H will be obtained, and if DRII is present, K and L should be seen. The presence or absence of DRI and DRII divides these strains into two classes.
The largest class, A, with 10 members (Table 2), contains both repeats and includes our reference strain, C921b-1. Where known, the serotype of all class A strains examined is O6. To determine whether the cointegrate plasmid is resolved during growth of these strains, we sought to amplify the expected junctions of the cointegrate and of the individual small plasmids from three of these strains (1392-75-2A, 60R70, and 296) as described above for C912b-1. As we had found for C921b-1, the two smaller replicons were present in the same culture, with the whole cointegrate plasmid in all three strains examined (data not shown).
The other class (Table 2, class B) of pCoo-related plasmids in these ETEC strains has eight members, all of which lack parts of one or both of the repeats or have repeats of sizes different from those in pCoo from strain C921b-1. The members of class B have a variety of serotypes and are probably more distantly related than the O6 group of strains. In summary, we find that several segments of pCoo, such as region A of R64, some portion of the R64 thin pilus, the R64 and R100 origin regions, and eatA, are common to the plasmids in most of the ETEC strains of all serotypes. However, only from the ETEC strains of serotype 06 could we amplify fragments with all the pCoo primer pairs used to identify both DRI and DRII. Thus, it appears that the pCoo cointegrate structure may be characteristic of CS1-producing strains of serotype O6. The traits that distinguish the O6 strains we examined include the production of LT and ST and production of additional pili (CS2, -3, -4, and/or -6), which are all plasmid encoded. Thus, these strains may be very close relatives that differ only in plasmid content. The strains with other O serotypes may be more distantly related, and thus their pCoo-related plasmids may have had a greater time to diverge from that in C921b-2.
Conclusions.
Comparison of the pCoo plasmids from the CS1-producing ETEC strains suggests that they were all derived from an ancestor plasmid that has diverged over time. They all contain both the R64- and R100-like origins of replication, the pCoo-specific region A (that encodes the CS1 pilus genes) in the R64 half of pCoo, and at least some portion of one of the direct repeats, DRI and DRII, that delimit the individual replicons in the cointegrate plasmid. The extent of the R64 thin pilus operon present in the ETEC strains varies; deletions in this region suggest that it is not essential. All ETEC strains that we examined with the O6 serotype have both of the direct repeats, but for the more distantly related strains with other serotypes, the primers we used did not always amplify all parts of the direct repeats. In some of the non-O6 strains, one of the direct repeats was present, suggesting that the cointegrate was formed. For the rest of the strains, we were unable to determine whether the two parts of pCoo formed a cointegrate and, if so, whether the cointegrate would resolve. It is not clear whether there is selective value to maintenance of the cointegrate plasmid over that of the two independent replicons which constitute it, but its continued presence suggests some type of advantage.
The presence of at least part of both the R100-like and R64-like halves of pCoo in all of the CS1-piliated ETEC strains suggests that there might be essential functions on both. On the R64 part, the coo genes are probably required for virulence. On the R100 portion of pCoo, candidates for functions important for pathogenesis include the shf operon, which is almost identical to that found on the pINV virulence plasmid of Shigella flexneri, and eatA, which encodes the secreted serine protease that enhances the virulence in a CFA/I-pilus-producing ETEC strain (25).
In the CS1-producing ETEC strains, replicons of three incompatibility groups are apparently necessary for pathogenesis. An IncFI plasmid contains the positive regulator, rns, required for expression of the coo genes, LT and ST, and, in some strains, the colonization factor CS3. The pCoo cointegrate plasmid is made up of IncI1 (R64) and an R100-like replicon, both of which may contain genes that may be important for virulence. A better understanding of the role of these genes and of contributions to virulence from the IncF1 plasmid and from the chromosome awaits further sequence analysis and studies on the pathogenesis of ETEC.
Supplementary Material
Acknowledgments
This work was supported by grant AI24870 from the National Institutes of Health and by the Wellcome Trust. We gratefully acknowledge the support of the Sanger Institute core sequencing and informatics teams.
We thank Myron Levine for ETEC strain 1391-75-2A; Eileen Barry for ETEC strains 60T75 and E4377/O/A; The Foodborne and Diarrheal Disease Laboratory at the CDC for ETEC strains 294, 295, 296; and Tom Whittam for ETEC strains TW03577, TW03604, TW03923, TW03875, TW04205, TW04215, TW04237, TW04144, TW04316, TW04244, and TW04211.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Black, R. E. 1993. Epidemiology of diarrhoeal disease: implications for control by vaccines. Vaccine 11:100-106. [DOI] [PubMed] [Google Scholar]
- 2.Black, R. E. 1990. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev. Infect. Dis. 12(Suppl. 1):S73-S79. [DOI] [PubMed] [Google Scholar]
- 3.Boylan, M., and C. J. Smyth. 1985. Mobilization of CS fimbriae-associated plasmids of enterotoxigenic Escherichia coli of serotype O6:K15:H16 or H-into various wild-type hosts. FEMS Microbiol. Lett. 29:83-89. [Google Scholar]
- 4.Brendler, T., L. Reaves, and S. Austin. 2004. Interplay between plasmid partition and postsegregational killing systems. J. Bacteriol. 186:2504-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan, L. A., G. A. Willshaw, B. A. van der Zeijst, and W. Gaastra. 1991. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol. Lett. 67:341-346. [DOI] [PubMed] [Google Scholar]
- 7.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Disque-Kochem, C., and R. Eichenlaub. 1993. Purification and DNA binding of the D protein, a putative resolvase of the F-factor of Escherichia coli. Mol. Gen. Genet. 237:206-214. [DOI] [PubMed] [Google Scholar]
- 9.Froehlich, B., E. Holtzapple, T. D. Read, and J. R. Scott. 2004. Horizontal transfer of CS1 pilin genes of enterotoxigenic Escherichia coli. J. Bacteriol. 186:3230-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froehlich, B., L. Husmann, J. Caron, and J. R. Scott. 1994. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 176:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froehlich, B. J., A. Karakashian, L. R. Melsen, J. C. Wakefield, and J. R. Scott. 1994. CooC and CooD are required for assembly of CS1 pili. Mol. Microbiol. 12:387-401. [DOI] [PubMed] [Google Scholar]
- 12.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 13.Hiraga, S., A. Jaffe, T. Ogura, H. Mori, and H. Takahashi. 1986. F plasmid ccd mechanism in Escherichia coli. J. Bacteriol. 166:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huilan, S., L. G. Zhen, M. M. Mathan, M. M. Mathew, J. Olarte, R. Espejo, U. Khin Maung, M. A. Ghafoor, M. A. Khan, Z. Sami, et al. 1991. Etiology of acute diarrhoea among children in developing countries: a multicentre study in five countries. Bull. W. H. O. 69:549-555. [PMC free article] [PubMed] [Google Scholar]
- 15.Hyams, K. C., A. L. Bourgeois, B. R. Merrell, P. Rozmajzl, J. Escamilla, S. A. Thornton, G. M. Wasserman, A. Burke, P. Echeverria, K. Y. Green, et al. 1991. Diarrheal disease during Operation Desert Shield. N. Engl. J. Med. 325:1423-1428. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, T., Y. N. Min, W. Liu, D. D. Womble, and R. H. Rownd. 1993. Insertion and deletion mutations in the repA4 region of the IncFII plasmid NR1 cause unstable inheritance. J. Bacteriol. 175:5350-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S. H., W. Jia, R. E. Bishop, and C. Gyles. 2004. An msbB homologue carried in plasmid pO157 encodes an acyltransferase involved in lipid A biosynthesis in Escherichia coli O157:H7. Infect. Immun. 72:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 19.Lan, R., G. Stevenson, and P. R. Reeves. 2003. Comparison of two major forms of the Shigella virulence plasmid pINV: positive selection is a major force driving the divergence. Infect. Immun. 71:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 21.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, et al. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullany, P., A. M. Field, M. M. McConnell, S. M. Scotland, H. R. Smith, and B. Rowe. 1983. Expression of plasmids coding for colonization factor antigen II (CFA/II) and enterotoxin production in Escherichia coli. J. Gen. Microbiol. 129:3591-3601. [DOI] [PubMed] [Google Scholar]
- 23.Nakata, N., C. Sasakawa, N. Okada, T. Tobe, I. Fukuda, T. Suzuki, K. Komatsu, and M. Yoshikawa. 1992. Identification and characterization of virK, a virulence-associated large plasmid gene essential for intercellular spreading of Shigella flexneri. Mol. Microbiol. 6:2387-2395. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Casal, J., J. S. Swartley, and J. R. Scott. 1990. Gene encoding the major subunit of CS1 pili of human enterotoxigenic Escherichia coli. Infect. Immun. 58:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichel, M., N. Binsztein, and G. Viboud. 2000. CS22, a novel human enterotoxigenic Escherichia coli adhesin, is related to CS15. Infect. Immun. 68:3280-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 29.Scotland, S. M., M. M. McConnell, G. A. Willshaw, B. Rowe, and A. M. Field. 1985. Properties of wild-type strains of enterotoxigenic Escherichia coli which produce colonization factor antigen II, and belong to serogroups other than O6. J. Gen. Microbiol. 131:2327-2333. [DOI] [PubMed] [Google Scholar]
- 30.Scott, J. R., J. C. Wakefield, P. W. Russell, P. E. Orndorff, and B. J. Froehlich. 1992. CooB is required for assembly but not transport of CS1 pilin. Mol. Microbiol. 6:293-300. [DOI] [PubMed] [Google Scholar]
- 31.Smyth, C. J. 1982. Two mannose-resistant haemagglutinins on enterotoxigenic Escherichia coli of serotype O6:K15:H16 or H-isolated from travellers' and infantile diarrhoea. J. Gen. Microbiol. 128:2081-2096. [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1-10. [DOI] [PubMed]
- 33.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 34.Thisted, T., N. S. Sorensen, E. G. Wagner, and K. Gerdes. 1994. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13:1960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren, G., and D. Sherratt. 1977. Complementation of transfer deficient ColE1 mutants. Mol. Gen. Genet. 151:197-201. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, M. K., D. N. Taylor, E. C. Boedeker, K. C. Hyams, D. R. Maneval, M. M. Levine, K. Tamura, R. A. Wilson, and P. Echeverria. 1993. Characterization of enterotoxigenic Escherichia coli isolated from U.S. troops deployed to the Middle East. J. Clin. Microbiol. 31:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.