Abstract
Complex gene regulatory circuits contain many features that are likely to contribute to their operation. It is unclear, however, whether all these features are necessary for proper circuit behavior or whether certain ones are refinements that make the circuit work better but are dispensable for qualitatively normal behavior. We have addressed this question using the phage λ regulatory circuit, which can persist in two stable states, the lytic state and the lysogenic state. In the lysogenic state, the CI repressor positively regulates its own expression by stimulating transcription from the PRM promoter. We tested whether this feature is an essential part of the regulatory circuitry. Several phages with a cI mutation preventing positive autoregulation and an up mutation in the PRM promoter showed near-normal behavior. We conclude that positive autoregulation is not necessary for proper operation of the λ circuitry and speculate that it serves a partially redundant function of stabilizing a bistable circuit, a form of redundancy we term “circuit-level redundancy.” We discuss our findings in the context of a two-stage model for evolution and elaboration of regulatory circuits from simpler to more complex forms.
Complex gene regulatory systems often contain a large number of interacting components whose actions are presumed to contribute to the overall behavior of the system. It is plausible that some or all of these features are advantageous and have been selected for during the course of evolution, since they are found in the natural system. However, this does not mean that all the features of a natural system are essential for its proper operation—instead, a given feature might be a refinement that confers a small but decisive selective advantage. If this is the case, certain features of contemporary circuits might be dispensable—either they could be removed without destroying qualitatively normal system behavior, or suppressors might compensate for their loss well enough to restore such behavior. In this work, we have removed a regulatory feature from a complex circuit both to test its importance in that circuit and in the hope that the behavior of the simplified circuit might provide insights into the evolution of complex circuits.
We have carried out this test in the well-characterized regulatory circuitry of phage λ. This virus can exist in two alternative stable regulatory states (8, 12, 34). An infected cell can follow the lytic pathway, in which a temporal program of lytic gene expression culminates in the production of about 100 new virions, followed by lysis of the cell. Alternatively, the infected cell can follow the lysogenic pathway, in which the virus establishes and maintains a stable association with the host. The phage genome becomes physically integrated into that of the host, and expression of viral genes is repressed by the action of the CI repressor. The initial choice between these two regulatory states involves several aspects of host physiology that are not well understood and will not be considered further here.
The lysogenic state is extremely stable, but it can break down as a result of the host SOS response, which is triggered by DNA damage or inhibition of DNA replication (26). Inducing treatments lead to activation of RecA protein; activated RecA mediates the specific cleavage of CI (25, 36), inactivating it. This leads to expression of lytic genes, excision of the viral genome from the host, and production of new virions. This process, termed prophage induction, exhibits threshold behavior—at low doses of DNA damage, little or no induction is observed, while at a particular dose, termed the set point, induction rather abruptly becomes efficient (e.g., see reference 27). At higher doses, essentially all the cells are induced.
The operation of this regulatory circuitry involves many complex features, several of which depend on the properties of a central regulatory protein, CI or λ repressor (34). CI has a diverse set of activities (Fig. 1B). It reversibly forms dimers. These dimers bind specifically to several operator sites. Pairs of dimers bind cooperatively to adjacent operator sites (17), and two cooperatively bound tetramers support the formation of a DNA loop (6, 35), another form of cooperativity. Depending on the context, bound CI can act as either a repressor or an activator of transcription. Finally, CI is capable of a specific self-cleavage reaction, stimulated by activated RecA, that results in its inactivation under some circumstances (not shown in Fig. 1). It is almost certain that most or all of these features of CI are selectively advantageous, since they have been found in the CI proteins of several related viruses, such as 434, P22, and HK022.
FIG. 1.
Diagram of OR region, and activities of CI. (A) Diagram of OR. Locations of PR and PRM promoters are shown. Cro and CI bind to OR1, OR2, and OR3, although the relative affinities differ; Cro binds most tightly to OR3, while CI binds most tightly to OR1. The proximal portions of the cI and cro genes are shown. The map is to scale. (B) Activities of CI. The N- and C-terminal domains of CI are indicated by the letters N and C, respectively. CI dimerizes with a dissociation constant of ∼10 nM. Dimers bind to three operators at OR and to three other sites in the OL region (depicted only at the bottom). Dimers bind cooperatively to adjacent operators, generally to OR1 and OR2 as shown. CI bound to OR2 stimulates its own expression from PRM. When CI is bound to two adjacent operators at OR and at OL (not shown for the unlooped form), the two tetramers can interact to form a loop as depicted. In a second step, two more dimers bind to the empty sites (depicted as OR3 and OL3); binding to OR3 represses PRM. Details of the contacts among CI dimers in the complex are not known; one likely possibility is depicted. Diagrams are not to scale.
Several of these activities are believed to be crucial to the maintenance of a stable lysogenic state. CI dimers can bind to six different operators (Fig. 1B), three apiece in the OL and OR regions. CI acts most directly to influence the choice of regulatory states by its action at OR. At moderate levels of CI, it binds to OR1 and OR2; when bound to OR2, it stimulates its own expression from a weak promoter, termed PRM, about 10-fold (31). Intrinsically, OR1 and OR2 are, respectively, strong and weak binding sites for CI; when OR1 is occupied, cooperativity favors binding to OR2. When CI is bound to either or both of these two sites, it represses the lytic promoter PR. At OL, CI binds strongly and cooperatively to OL1 and OL2, repressing the lytic PL promoter. In addition, two dimers of CI bound at OL engage in a higher-order cooperative interaction with two dimers at OR. Finally, at somewhat higher CI levels, two more dimers bind to sites OR3 and OL3; occupancy of OR3 shuts off the PRM promoter (6). The resulting negative autoregulation results in a relatively constant level of expression in a lysogen.
Are these properties of CI essential to the operation of the λ circuitry? We previously tested whether a subtle feature of CI action, its ability to bind preferentially to certain of its binding sites, was necessary for the operation of the circuit and found that it was dispensable (27). Here we test the effect of removing a different CI function, positive regulation of its own expression.
Positive autoregulation, or positive control, is likely to be an adaptive feature for λ, since it is also observed in other lambdoid phages; the most detailed analyses have been done for 434 and P22. P22 has a different relationship between PRM and OR2 than that seen in λ (34), suggesting that positive autoregulation might operate by different interactions between CI and RNA polymerase and might have evolved independently in each case.
Positive autoregulation of λ cI involves a protein-protein interaction between a CI dimer bound at OR2 and the σ70 subunit of RNA polymerase, which is involved in promoter recognition. Recent crystallographic evidence (using a fragment of σ70 from Thermus aquaticus) shows that this contact involves a small interface between the two proteins (16), an interface that includes electrostatic interactions between two residues on CI, Glu34 and Glu38, and particular Arg residues in σ70. Both CI residues have been shown genetically to be required for positive control (13); in addition, genetic evidence (20, 22) indicates that Glu38 interacts with Arg596 in σ70, as seen in the crystal structure, since the cI D38N mutation is suppressed by the σ70 mutation R596H. Accordingly, in this work we have examined the properties of phage mutants carrying either of two mutations in Glu38. We find that we can readily isolate variants of λ that lack positive autoregulation and yet exhibit essentially normal behavior. These findings demonstrate that positive autoregulation is not an essential feature of the λ circuitry.
MATERIALS AND METHODS
Reagents and media.
Restriction enzymes and T4 DNA ligase were from New England Biolabs (Beverly, MA), Roche, and Promega Corp. (Madison, WI). Taq DNA polymerase was from Roche. Polymyxin B was from Sigma. Oligonucleotides were from Midland Certified Reagent Corp. (Midland, TX) and from QIAGEN and are listed in Table S1 of the supplemental material. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was from Research Products International Corp. TMG was as previously described (27). TZ8 (6) was 100 mM Tris-HCl (pH 8.0)-1 mM MgSO4-10 mM KCl. LB and tryptone broth were as previously described (27) and supplemented with antibiotics as appropriate at the levels previously described (27). LBGM and LBMM were LB with 1 mM MgSO4 and either 0.2% glucose or 0.2% maltose, respectively (27). DNA sequencing was carried out as previously described (27). For each variant phage, DNA sequencing was done on a PCR product extending from the distal end of cro to the end of cI; sequencing was done from the end of cro through about the first third of cI, spanning the entire region that could recombine from the plasmid.
Bacterial and phage strains and plasmids.
A list of many of the strains and plasmids used is given in Table 1. All bacterial strains were derivatives of Escherichia coli K-12. Phages not listed include those carrying reporter gene fusions and most of those isolated in the presence of wild-type CI (see Table S2 of the supplemental material). Plasmids not described include those used as intermediates in the construction of reporter gene fusions and those used for backcrosses of PRM alleles.
TABLE 1.
Strains, phages, and plasmids employed in this study
| Strain, phage, or plasmid | Relevant genotype | Vector | Source or reference |
|---|---|---|---|
| Bacterial strains | |||
| JL468 | AB1157/F′ lacIq | 23 | |
| JL2497 | N99 lacZΔM15/F′ lacIqlacZΔM15::Tn9; used as wild type | 30 | |
| JL6039 | JL2497 hflKC | 32 | |
| JL6142 | JL2497 Δ(lacIPOZYA)F− | 1 | |
| JL6994 | JL6142/pJWL615/pJWL486 | This work | |
| JL6995 | JL6142/pJWL615/pA3B2 | This work | |
| JL6996 | JL6142/pJWL615/pJWL710 | This work | |
| JL6997 | JL6142/pJWL615/pJWL709 | This work | |
| Phages | |||
| λJL163 | λ+bor::kan | 27 | |
| λJL611 | ′amp-′lacZY′ imm21bor::kan vector for reporter gene fusions | 32 | |
| λJL628 | Same as λJL611 but with λ OL operators distal to lacZ | 32 | |
| λJL800 | λ cI D38R bor::kan | This work | |
| λJL801 | λ Δ(PRM) bor::kana | This work | |
| λJL802 | λ Δ(PRM) cI D38R bor::kana | This work | |
| λJL804 | λ Δ(PRM) cI D38N bor::kana | This work | |
| λJL815 | λ cI D38N bor::kan | This work | |
| Plasmids | |||
| pA3B2 | Δ−35 lacP::cI; provides low level of CI; Cmr | pACYC184 | 41 |
| pACYC184 | Origin of p15a; compatible with pGB2 and ColE1-derived plasmids; Cmr | pACYC184 | New England Biolabs |
| pBS(−) | Cloning vector with extensive polylinker; Ampr | ColE1 | Stratagene |
| pJWL334 | Derivative of pBS(−) with modified polylinker | pBS(−) | 32 |
| pJWL486 | pACYC184 derivative (control for pA3B2) | pACYC184 | 32 |
| pJWL615 | lacIq promoter driving lacI; compatible with pACYC184- and ColE1-derived plasmids; Spcr | pGB2 | 32 |
| pJWL701 | cI amino acid 80-BglII interval spanning OR region with cI D38Rb | pBS(−) | This work |
| pJWL702 | NsiI-BglII interval spanning OR region with Δ(PRM) | pBS(−) | This work |
| pJWL704 | cI amino acid 80-BglII interval spanning OR region with Δ(PRM) and cI D38Ra,b | pBS(−) | This work |
| pJWL706 | cI amino acid 80-BglII interval spanning OR region with cI D38Nb | pBS(−) | This work |
| pJWL707 | cI amino acid 80-BglII interval spanning OR region with Δ(PRM) and cI D38Na,b | pBS(−) | This work |
| pJWL709 | cI D38N version of pA3B2 | pACYC184 | This work |
| pJWL710 | cI D38R version of pA3B2 | pACYC184 | This work |
In this phage or plasmid, the deletion removes the first four codons of cI; hence, the cI gene is not intact, symbolized by ′cI in a phage. The deletion is termed Δ(PRM cI-K4) or Δ(PRM) for brevity.
This plasmid contains a segment of cI extending to residue 80, indicated by “cI amino acid 80,” beyond which a PstI site was introduced by PCR to allow cloning.
Creation of doped PRM pools.
Pools of DNA fragments was made in which eight positions of PRM were doped as previously described (32), with the following modifications. First, pools of fragments containing the cI D38N or D38R allele were made by PCR as described previously (32), with primers prmdope-10 and cIright (Table S1) and λJL815 and λJL800 templates, respectively, to make the fragment extending from PRM beyond the NsiI site. A second fragment was made by PCR using λJL163 as the template with primers prmdope-35 and croleft as described previously (32). Second, these fragments were cloned into pJWL334 cut with NsiI and BglII in one of two ways; either they were cut with BsmAI and ligated and the product was amplified with cIright and croleft, followed by digestion with NsiI and BglII, or fragments were cut with NsiI and BsmAI (left fragment) or BsmAI and BglII (right fragment) and the resulting fragments were cloned into pJWL334 in a three-piece ligation. Third, pools were introduced into JL468 by CaCl2 transformation, followed by growth overnight in LB-ampicillin, yielding a culture containing a mixture of plasmids with variant PRM alleles. These pools contained only about 8,000 to 12,000 members, so that not all of the 65,356 (48) possible PRM sequences were represented.
Isolation of Δ(PRM) plasmid and phage.
A phage containing a deletion of PRM and the first several codons of cI was made in two steps. First, a plasmid, pJWL702, was made with two inserts, both of which were PCR products using λJL163 as the template. One insert was made with primers croleft and delprmright, followed by cutting with BglII and ClaI, the other with primers cIright and delprmleft, followed by cutting with NsiI and ClaI. These two fragments were cloned into pJWL334 cut with NsiI and BglII. The resulting deletion, termed Δ(PRM cI-K4) or Δ(PRM) for brevity, removed λ sequence starting just to the left of OR2 and extending leftward through codon 4 of the cI gene (codons in cI are numbered by convention with the AUG start codon as 0, since the N-terminal Met is removed after translation [38]). Second, a phage-by-plasmid cross was done between pJWL702 and λJL163; clear plaques were purified, and deletion of PRM was verified by DNA sequencing. The resulting phage was λJL801.
Phages with the same structure but carrying the cI D38N or D38R allele were made in the same way, except that PCR products were made using primers delprmleft and PstIcI80right, which puts a PstI site at the position of cI codon 80 (giving a longer region of homology distal to D38 for crossing over), with pJWL706 and λJL800 (see below), respectively, as the templates, followed by cutting with ClaI and NsiI and cloning into pJWL702 cut with the same enzymes, yielding pJWL707 and pJWL704, respectively. These were crossed with λJL163; clear plaques were purified, and deletion of PRM and the presence of the cI allele were verified by sequencing, yielding λJL804 and λJL802, respectively.
Isolation of λ cI D38N and D38R.
The D38N and D38R mutations were made with silent changes in Ala37, changing the codons for Ala37 and Asp38 from GCAGAC (the codon for D38 is underlined) to GCCAAC and GCGCGC, creating sites for BstXI and BssHII, respectively. For each, two primers were made including these changes. PCR product 1 was made with D38Nright or D38Rright and croleft; PCR product 2 was made with D38Nleft or D38Rleft and PstIcI80right. Product 1 was cut with BglII and BstXI or BssHII, product 2 was cut with PstI and BstXI or BssHII, and the resulting fragments were cloned into pJWL334 cut with BglII and PstI. The resulting plasmids were pJWL701 (D38R) and pJWL706 (D38N).
To isolate λ cI D38R, λJL163 was crossed with pJWL701; clear plaques were isolated from the progeny, and the presence of the D38R mutation was verified by sequencing of the cI-cro interval, yielding λJL800. To isolate λcI D38N, λJL804 was crossed with pJWL706; turbid plaques were picked and sequenced, yielding λJL815.
Isolation of phages with PRM variants.
Several phage-by-plasmid crosses were carried out between λJL801, λJL802, or λJL804 and the doped PRM pools carrying the corresponding cI allele. In such crosses, we have found that recombinants generally are about 0.05% of the total; hence, if we assume that all 8,000 PRM variants are represented about equally in the plasmid pool, a given PRM sequence would be carried on roughly 5 × 10−3/8,000, or roughly 10−6, of the total phages in the cross progeny.
Enrichment procedure.
The enrichment procedure was designed to enrich for PRM variants that could lysogenize and undergo prophage induction at UV doses similar to those of the wild type. hfl mutant strain JL6039 was grown in LBMM to 2 × 108 cells/ml, concentrated by centrifugation, and resuspended in 1/10 volume of TMG. Cells (109) were infected with ∼108 cross progeny. After 20 min at room temperature, cells were diluted 10-fold in LBGM and shaken for 30 min at 37°C; kanamycin was then added to 30 μg/ml, an aliquot was plated on kanamycin plates to verify lysogenization, and the culture was shaken overnight at 37°C.
An aliquot was grown to mid-exponential phase in LBGM and then irradiated with UV light (5 J/m2 for pools with cI+ or cI D38R, 10 J/m2 for those with cI D38N) as previously described (32), followed by centrifugation and resuspension in an equal volume of LBGM. The culture was diluted 10-fold and shaken overnight at 37°C. Cells were diluted, grown to mid-exponential phase, and UV irradiated at 20 J/m2. After 2 h, cultures were treated with chloroform and plated on overnight plating lawns. Individual plaques were then purified and characterized as described in Results.
Backcrosses.
PCR of phages with various PRM alleles was done using cIright and croleft. The product was cut with NsiI and BglII and cloned into pJWL334, the plasmid was crossed with a Δ(PRM) phage carrying the desired cI allele, turbid plaques were isolated, and the presence of the cI and PRM alleles was verified by sequencing.
Screening for set point of prophage induction.
Exponentially growing cells were concentrated twofold in TMG, and a small drop was applied to a kanamycin plate, which was tipped to allow the liquid to run across the plate. Streaks were exposed to graded amounts of UV irradiation by shielding with an opaque card, followed by incubation for 16 h at 37°C. Induction was assayed by the gradual thinning of the streak, and the pattern shown by each mutant was compared with a wild-type strain on the same plate.
UV induction.
Single lysogens in JL2497 were identified and UV induction was carried out as previously described (32).
Reporter constructs and β-galactosidase assays.
Reporter constructs were made, single lysogens were prepared, and enzyme assays were done as previously described (32), except that the indicator strains (JL6994 to JL6997) were derived from JL6142 (Table 1).
RESULTS
Approach.
Our goal was to characterize derivatives of λ in which CI was no longer able to stimulate its own expression. As a first step, we made a phage bearing the cI D38N mutation, which was previously shown (13) to abolish positive control. We found that λ cI D38N could form stable lysogens, but that these were extremely easy to induce at low levels of DNA damage (see below). Accordingly, we sought to isolate suppressors that would restore a more normal set point for prophage induction. Rather than carrying out an enrichment for such phage, we chose to use site-directed mutagenesis, with the following rationale.
The most direct result of positive control is to elevate CI levels above the level produced by unstimulated PRM. Our previous study with wild-type CI suggested that increasing the strength of PRM would increase the set point for prophage induction (32). In that study, we isolated variants with altered PRM promoters that yielded relatively normal behavior of the λ regulatory circuitry, and stronger promoters generally led to higher set points. We reasoned that with λ cI D38N we could likewise isolate PRM variants that would restore prophage induction with a set point more like that of wild-type λ. We chose the approach of doping PRM (Fig. 2). This approach was applied using phages and plasmids bearing wild-type cI and with the cI D38N and cI D38R alleles. We will first describe the approach and then the results obtained with D38N.
FIG. 2.
Isolation of pools with mutated PRM. (A) Sequence of the PRM promoter, inverted from the usual order of the λ map. Locations of OR3 and the end of OR2 are shown in bold; the −35 and −10 regions of PRM are underlined; the N′s represent positions that were randomized. Above the sequence is that of a consensus promoter; positions in which PRM differs are underlined. (B) Genetic crosses. λ Δ(PRM) was crossed with a pool of cells containing PRM variants made as previously described (see Materials and Methods and Results). Maps of the OR region in plasmids and phages are to scale. The dashed line between the two parents shows the double crossover yielding the desired recombinant, shown at the bottom. Ns and Bg, NsiI and BglII, respectively. (C) Selection and enrichment for cross progeny forming stable lysogens with a set point near the wild type (see text for details) m.o.i., multiplicity of infection.
In this approach, pools of variant PRM alleles were made on plasmids, yielding a pool of cells with the variants (denoted PRM* in Fig. 2). As in our previous study (32), only positions in PRM lying outside OR2 and OR3 were randomized, to avoid effects on the binding of CI or Cro to these sites. Then a λ mutant carrying a deletion of PRM was crossed with the pool of cells containing mutant plasmids by infection of the mixed culture (Fig. 2). In each cross, both plasmids and phages carried the same cI allele. A pool of those cross progeny able to lysogenize was made by infecting an hfl mutant strain (in which lysogens can arise after single infection) at a low multiplicity and selecting for kanamycin resistance. This pool of lysogens was treated with a low dose of UV light to induce lysogens that are induced much more easily than the wild type; surviving lysogens were grown overnight and treated with a higher dose of UV light to induce those with a set point near that of the wild type. The resulting pool of phages was plated, and turbid plaques were purified and characterized. In each case, the starting pools did not contain all possible PRM sequences, so that we almost certainly did not identify all of the possible PRM variants that could pass this enrichment.
We used the following criteria for qualitatively normal behavior. First, the phage should be able to grow lytically, not a demanding criterion since CI has no role in lytic growth. Second was the ability to establish and maintain a stable lysogenic state. Expression of CI from PRM is required for maintenance of this state. Third was the ability to undergo prophage induction after UV irradiation. A final criterion was that the set point for this process (the UV dose at which the yield of phage was about half its maximal value) should resemble that of the wild type.
PRM variants containing D38N.
Thirty different isolates carrying D38N were purified, and the DNA sequence of the OR region was determined for each. Twelve different PRM alleles were identified (Table 2). Preliminary analysis indicated that isolates with three of these alleles were induced with a much lower set point than the wild type. For six of the remaining nine mutants, we first carried out a backcross with the Δ(PRM) parent to ensure that no other mutations were present in the isolates (see Materials and Methods). We refer to isolates by giving the PRM allele and the cI allele they carry, such as λ PRM-NP2 cI D38N; for the sake of brevity, they are indicated in the figures in a shorthand form, such as NP2 D38N.
TABLE 2.
Sequences and properties of PRM variants isolated with cI D38Na
| PRM allele | −35 region | −10 region | cI allele or no. of isolates | Set point | Maximum level of β-galactosidase with cI D38N (except as noted)
|
|
|---|---|---|---|---|---|---|
| −OL | +OL | |||||
| WT | TAGA | GATT | cI+ | 12 | 266, 280 (cI+) | 190, 168 (cI+) |
| WT | TAGA | GATT | cI D38N | ∼0.8 | 54 (cI D38N) | 34 (cI D38N) |
| Mutants containing a PRM variant and cI D38N | ||||||
| NP1b | GGAA | AGAT | 4 | 15 | NA | NA |
| NP2 | GCTG | TATT | 1 | 7 | 140 | 85 |
| NP3 | CATT | GAAT | 2 | 4 | ND | ND |
| NP4 | CCTT | CCAT | 1 | 3.5 | 102 | 73 |
| NP5 | CTAA | GAAT | 2 | 8 | 171 | 114 |
| NP6 | CCCA | TGAT | 1 | 5 | ND | 120 (not shown) |
| NP7 | TACC | TACT | 1 | 4 | ND | ND |
| NP8 | GTGT | GTAT | 1 | 15 | ND | 217 (not shown) |
| NP9 | GCAC | GGAT | 14 | 8 | 179 | 117 |
| NP10 | CCAA | GAAT | 1 | 14 | 246 | 174 |
| NP11 | CTCA | TGCT | 1 | 3 | ND | ND |
| NP12 | CTGA | TAAT | 1 | 8 | ND | 78 (not shown) |
Only positions 3 to 6 for each of the −35 and −10 regions are shown. Positions 1 and 2 are TA in both cases. The set point is the dose of UV giving 50% of the maximal phage yield. ND, not done; NA, not applicable.
The mutant carrying PRM-NP1 also has a mutation of OR2 (CAACGCGCACGGTGT; the underlined base is A in the wild type).
Isolates were further characterized in two ways. First, we determined UV dose-response curves for lysogens containing a single prophage to test whether their behavior resembled that of wild-type lysogens; these tests examined the system behavior of the phage. Second, we analyzed the various PRM alleles in an uncoupled reporter gene assay to determine both the relative strengths of the mutant promoters and their responses to wild-type and mutant CI proteins.
For lysogens of each mutant, prophage induction was carried out with graded doses of UV light. In this experiment, the wild-type lysogen exhibits threshold behavior with a particular set point. Mutants have been isolated (27, 32; unpublished data) that affect either the set point, the yield of phage after induction, or both; nearly all mutants examined to date exhibit threshold behavior.
The phage bearing cI D38N and various PRM alleles also showed threshold behavior. The λ PRM+ cI D38N phage, as noted above, was induced at very low UV doses, with threshold behavior and a set point of ∼0.8 J/m2. Lysogens of phage with cI D38N and variant PRMs were characterized by a range of phage yields and set points (Fig. 3; Table 2). From these findings, we conclude first that it is possible to isolate λ variants that lack a contact normally required for positive control and yet show relatively normal behavior and second that these can exhibit threshold behavior with a range of set points.
FIG. 3.
UV induction of selected mutants with cI D38N. UV induction curves were determined for single lysogens as described in Materials and Methods. Results of typical experiments are shown; data were obtained in a single experiment. Here and in Fig. 4 to 6, mutants are referred to by giving the PRM allele followed by the cI allele; for example, NP2 D38N is shorthand for PRM-NP2 cI D38N. WT, wild type.
Although several PRM alleles could suppress the very low set point of λ cI D38N PRM+, it remained possible in principle that we had selected a special class of promoters for which the mutant CI could support positive control. To test this idea, we compared the strength and regulation of these variant PRM alleles with those of wild-type PRM in an uncoupled system using PRM::lacZ protein fusions (32). We also installed a variant of the OL region distal to the lacZ gene to allow looping between OR and OL. Looping leads to partial repression of PRM due to cooperative binding of CI to OR3, facilitated by other CI dimers in the looped structure (cf. Fig. 1). To test the response of the mutant promoters to graded levels of CI, we supplied wild-type or D38N mutant CI from a lacP::cI fusion on a plasmid and provided graded amounts of isopropyl-β-d-thiogalactopyranoside (IPTG) to induce CI expression. In addition, we tested the response both to wild-type CI and to D38N mutant protein.
These studies revealed several properties of the variant PRMs. First, they were stimulated only slightly or not at all by the presence of CI D38N (Fig. 4A), as observed here and previously (4, 13) with the wild-type PRM. This finding indicates that we have not enriched for a special class of PRM alleles that suppress the activation defect of D38N. Second, all were stimulated by the presence of wild-type CI (Fig. 4B). Third, all were stronger than wild-type PRM, as would be expected if they were to provide sufficient levels of CI to maintain a lysogenic state with stability similar to that of the wild type. As found previously (32), the promoter strength was correlated with the set point (Table 2).
FIG. 4.
Activities of selected promoters and responses to various levels of CI D38N or wild-type (WT) CI. Single lysogens of phages bearing PRM::lacZ protein fusions were prepared. For each, three host strains were used. The first, JL6994, contained no CI; the second and third contained pJWL709 or pA3B2, which are regulated by the Lac repressor and make low levels of CI D38N or wild-type CI, respectively. A small amount of CI is made in the absence of IPTG. Cells were grown in the presence of the indicated levels of IPTG, and β-galactosidase levels were measured as described in Materials and Methods. Results of a typical experiment are shown. (A) Activity in the presence of an OL variant located distal to the lacZ reporter with CI D38N. (B) Activity in the presence of OL with wild-type CI. (C) Activity in the absence of OL with cI D38N.
To examine the properties of these PRM variants in the absence of looping, we also assayed LacZ expression in versions of the reporter constructs lacking OL. In these assays, there was a modest stimulation of wild-type and mutant derivatives of PRM by D38N CI (Fig. 4C), implying that this protein retains a small amount of stimulatory activity. Again, promoter strength was correlated with the set point. Wild-type CI stimulated all the variant PRMs to much greater extents (data not shown), as also seen in the presence of OL (Fig. 4B).
We conclude that these mutant promoters are not markedly activated by D38N mutant CI. Therefore, positive control is not necessary for qualitatively normal behavior of the λ circuitry. In particular, positive control is apparently not necessary for threshold behavior in prophage induction.
Lysogenizing derivatives of λcI D38R.
We next analyzed a mutant CI that we expected to have a more drastic effect on positive control. Since D38 in CI makes an electrostatic interaction with R596 in σ70, we expected that a CI derivative with a positive charge at residue 38 might repel R596. Indeed, we found that λ cI D38R made clear plaques and was unable to lysogenize. We then made a pool of recombinant phage with variant PRM alleles as described above and isolated lysogenizing variants. Following enrichment as before, turbid plaques were isolated, their OR regions were sequenced, and their ability to form stable lysogens was assessed by streaking plaques onto kanamycin plates. Among 57 lysogenizing variants sequenced, only four different sequences were identified (PRM-RP1 to PRM-RP4; Table 3) F that allowed the formation of stable lysogens, as assessed by colony size; lysogens of several other isolates (not described here) formed small colonies, a phenotype we attribute (32; unpublished data) to lysogens being unstable. These four variants were backcrossed as before, and recombinants were further characterized.
TABLE 3.
Sequences and properties of PRM variants isolated with cI D38Ra
| PRM allele | −35 region | −10 region | No. of isolates | Set point with:
|
Maximum level of β-galactosidase with cI D38N (+OL) | |
|---|---|---|---|---|---|---|
| cI D38R | cI D38N | |||||
| RP1 | CCCT | TAAT | 9 | 3 | 18 | 327 |
| RP2 | TGAC | TAAT | 34 | 4 | 20 | 246 |
| RP3 | CACG | TACT | 10 | 2.5 | 15 | 169 |
| RP4 | TGAC | CAAT | 4 | 2.5 | 14 | 149 |
See Table 2, footnote a.
In UV induction experiments, all were easier to induce than a wild-type λ lysogen (Fig. 5A). When each PRM variant was combined with cI D38N, the mutants were somewhat harder to induce than wild-type λ (Fig. 5B); in combination with wild-type cI, they were still more difficult to induce, giving small bursts only at high UV doses (data not shown).
FIG. 5.
UV induction and promoter activities of RP mutants. (A) UV induction of single lysogens. All phages carried cI D38R and the indicated promoter allele, except for the wild type (WT). (B) UV induction of single lysogens. Except for the wild type, all phages carried cI D38N and the indicated promoter allele. (C) Promoter activity of D38R suppressors in the presence of CI D38R. Single lysogens of phages bearing PRM::lacZ protein fusions were prepared and assayed as described in legend to Fig. 4, except that the second strain carried pJWL710, which expresses CI D38R. In the wild-type control, wild-type CI was provided from plasmid pA3B2. All phages carried a variant of OL located distal to the lacZ reporter. For each curve, the PRM allele in the reporter fusion is given, followed by the allele of cI carried on the plasmid. (D and E) Promoter activities of D38R suppressors in the presence of the wild type (D) or CI D38N (E). The experiment was as in panel C, except that cells with the CI-bearing plasmid carried pJWL709 or pA3B2, which expresses CI D38N or wild-type CI, respectively. Curves are labeled as for panel C.
In reporter gene assays, we found first that the four variant PRM alleles were relatively strong promoters in the absence of CI (Fig. 5C and D). Second, CI D38R acted as a repressor of wild-type PRM, as might be expected if there is charge repulsion between CI D38R and Arg596 in σ70 (Fig. 5C). A slight repressive effect of this same mutation was previously seen in a similar assay system (4). Third, CI D38R also repressed the four PRM variants with which it was isolated. Fourth, in contrast, wild-type CI was able to stimulate these same variants (Fig. 5D) and CI D38N had little effect on their expression (Fig. 5E).
We conclude first that these variant PRM alleles do not counteract the repressive effects of CI D38R bound to OR2; that is, they do not greatly alter the interaction between CI and σ70. Second, these variants probably achieve a high enough level of CI to maintain stable lysogeny by virtue of being strong promoters. Third, it is likely that these variants are engaged in a sort of “balancing act” in which the levels of CI are determined by two opposing forces (see Discussion). Again, the strength of these promoters was roughly correlated with the set point for prophage induction, in that the stronger promoters were found in those phage with higher set points (Tables 2 and 3).
PRM variants with little positive control in the presence of wild-type CI.
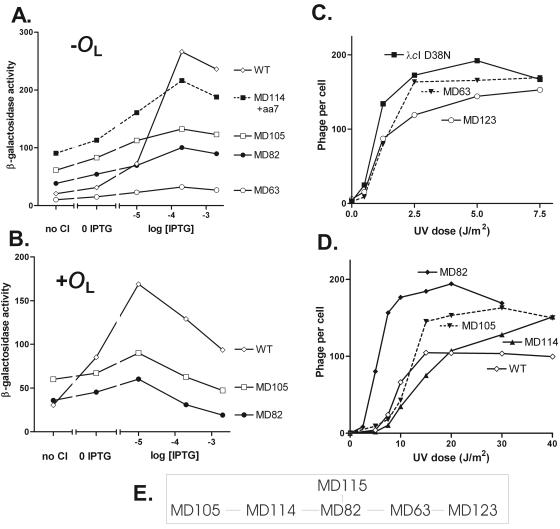
In this section, we describe another approach to eliminating positive autoregulation by CI. Using methodology different from that employed here, we previously isolated variant phages with many different PRM sequences altered in the same positions as in this work (32). Several of these phage variants had biological properties similar to those of wild-type λ. However, in uncoupled assays of promoter function, they were stimulated by CI to various extents, ranging from 3- to 4-fold to about 12-fold. In order to test whether certain PRM variants might show a wider range of responses to stimulation by CI than the previous set, we used the same approach as described above to isolate many more variants with altered PRM sequences in a phage with wild-type CI. We sequenced the OR region of 98 variants and identified phage with 60 different PRM alleles. These alleles were termed PRM-MD63 to PRM-MD122. In addition, stable lysogens of several other phage were isolated from the initial lysogenization procedure; these phage carried alleles termed PRM-MD123 to PRM-MD128. Their properties and sequences are given in Table S2 of the supplemental material; a subset is described in Table 4.
TABLE 4.
PRM variants selected as stably lysogenizing and enriched for set point near wild typea
| PRM allele | −35 region | −10 region | No. of differences from:
|
|
|---|---|---|---|---|
| W | C | |||
| Wild type | GATA | GATT | 0 | 3 |
| MD63 | ACCA | GGAT | 5 | 4 |
| MD82 | ACCA | GAAT | 4 | 3 |
| MD105 | ACCC | TAAT | 6 | 3 |
| MD114 | ACCA | TAAT | 5 | 2 |
| MD115b | ACGA | GAAT | 4 | 4 |
| MD123c | ACCC | GGAT | 6 | 5 |
Only positions 3 to 6 for each of the −35 and −10 regions are shown. Positions 1 and 2 are TA in both cases. The number of differences in these eight positions from wild-type PRM and from the consensus are given by the letters W and C, respectively.
The mutant carrying PRM-MD115 had an additional mutation in OR3 (TATCCCTTGTGGTGATA; the underlined base is C in the wild type).
The mutant carrying PRM-MD123 was selected as one that gave stable lysogens but did not undergo the enrichment procedure shown in Fig. 2B.
As expected from the way these isolates were obtained, all of them could form stable lysogens, as judged by the formation of large colonies on plates containing kanamycin. In addition, all were able to undergo prophage induction, as judged by a rapid plate screen (see Materials and Methods). We carried out UV induction experiments for single lysogens of most of the mutants with set points resembling that of the wild type in the plate test (data not shown; Table S2). These showed a relatively wide range of set points as judged by this quantitative assay. From these data, as from the previous set (32), we conclude that the set point can vary markedly and that changes in PRM can contribute to its value. Our findings reinforce previous conclusions that a very wide range of PRM sequences is compatible with relatively normal λ behavior.
We determined the strengths of variant PRMs in a reporter gene assay. In preliminary assays, PRM-MD105 showed a very weak response to wild-type CI, so we assayed several other variants with PRM sequences related (see Fig. 6E) to that of PRM-MD105. Data for PRM-MD105 and three other variants in the absence of a distal OL site are shown in Fig. 6A. Strikingly, these variants showed only a slight response to CI in this assay. For PRM-MD105 and PRM-MD82, we repeated this assay on a template with an OL site distal to lacZ in order to assess promoter strength in the presence of looping-mediated repression (Fig. 6B). In this assay, these two promoters were stimulated only slightly by CI. We conclude first that the sequence of PRM is capable of modulating the stimulatory effect of CI and second that, as above, positive control is not required for proper operation of the λ gene regulatory circuitry.
FIG. 6.
Promoter activities of PRM variants with weak responses to CI, and UV induction of prophages bearing these promoters. Experiments were carried out as described in the legends to Fig. 3 and 4, except that CI was wild type (WT) in all cases. (A and B) Promoter activity in the absence (A) or presence (B) of a distal OL site. The PRM-MD114 fusion carried a silent mutation in Leu7 of cI and hence cannot be compared directly to the data in panel D, since it is possible that this change affects the apparent promoter strength (for a similar mutation, see reference 32). Data are presented to illustrate the slight response to CI for this promoter. (C and D) UV induction curves for derivatives that are induced at low (C) or higher (D) set points. All data were obtained in a single experiment and are separated for clarity. (E) Evolutionary pathways among PRM variants. Alleles differing by a single base change are connected by single lines.
Finally, we measured the UV dose response of lysogens of most of the phages with alleles related by single changes to PRM-MD82 and PRM-MD105 (Fig. 6C and D). These showed a range of set points, and as with all the mutants described here, these gave threshold behavior. Again, the set points were roughly correlated with the strength of PRM in the cases we examined.
DISCUSSION
We present several lines of evidence that positive control is not essential for proper operation of the λ regulatory circuitry. Our findings have implications for the mechanism of positive control, for its role in λ gene regulatory circuitry, and for systems behavior. They also suggest mechanisms by which complex gene regulatory circuits might have evolved and gained in complexity.
Mechanism of positive control.
Structural evidence (16) shows a small contact region between CI and σ70 bound to the −35 region. This region includes the contact we altered and lies close to the DNA. The authors suggest that favorable interaction between the two proteins requires precise alignment and suggest that this alignment can only be achieved in the wild-type case at a step after formation of the closed complex, since CI stimulates the isomerization step (kf) and not the initial binding step (KB) (7, 11, 40).
We found that several related changes in the promoter sequence allowed only a small amount of positive control. As discussed previously for other PRM variants with less drastic effects on positive control (32), this could occur by various mechanisms. The most straightforward is that the two proteins are misaligned and cannot interact (15), either because the specific contacts between σ70 and the bases in the −35 region are altered, thereby repositioning σ70, or by altering the detailed DNA structure near the contact. Alternatively, CI apparently stimulates an early step in the isomerization of the closed complex to the open complex (kf); perhaps a promoter mutation could cause a later step in open-complex formation to be rate limiting (15). Finally, PRM has an extended −10 region (37); conceivably, these particular mutations have largely abrogated the requirement for a −35 region, at least in cases, like some of these, in which the −10 region is a consensus or nearly so and the −35 region lies far from the consensus.
Systems behavior and the role of positive control in λ gene regulatory circuitry.
We have described several mutants in which positive control is largely absent and in which the set point for prophage induction is similar to that of the wild type (Fig. 3 and 5B), with a range of values spanning that of the wild type. Although some of the mutants (such as those carrying PRM-NP5 and PRM-NP10; Fig. 3) have reduced burst sizes relative to the wild type, others, including that with PRM-NP7 (not shown), have burst sizes like those of the wild type. We conclude that positive control is not an essential feature of the λ circuitry by the criteria we have used here and that mutants lacking it have qualitatively normal behavior.
If positive control is not essential, why is it present? Positive control is a form of positive feedback, and it may be expected to play some role in stabilizing a bistable circuit by helping to drive the system toward either of the two stable states (3, 10). For circuits to be bistable, they need to involve some form of feedback (10)—either positive feedback or double-negative feedback (see below). In addition, they need to involve some form of nonlinearity, that is, a response that is greater than proportional to the input.
Both feedback and nonlinearity are present in the λ circuitry in several forms. The first is positive autoregulation of cI, as studied here. Second is relatively weak dimerization of the CI repressor, which results in a nonlinear response of CI dimer levels to the total concentration of CI. Cro dimerizes even more weakly (5, 21). Third is cooperative binding of CI to adjacent operators and higher-order cooperative interactions between CI bound at OR and at OL. These confer a sigmoid (nonlinear) binding curve for binding to operators. Finally, the circuitry involves double-negative feedback, since CI represses the synthesis of its own repressor, Cro, and vice versa. In the present studies, we have removed only one of these features, so our data do not provide a critical test of the importance of positive feedback or nonlinearity for bistability.
In any case, it is reasonable to conclude that several mechanisms act together to provide positive feedback and nonlinearity to stabilize states of the λ circuitry. Accordingly, these may be partially redundant, and removal of any one feature might confer only a slight defect on the system. Importantly, this type of redundancy differs from the more usual form of redundancy, in which different gene products execute biochemically related functions. We propose to term it “circuit- level redundancy.”
In this view, positive control may offer a selective advantage that is too slight to be detected in our experiments; even small advantages are decisive over the course of evolution. This is particularly difficult to evaluate experimentally, since we do not study λ or its host in the natural environment. Nonetheless, we now discuss current ideas about the role of positive control and suggest other plausible roles acting either in a different part of the λ life cycle than those we examined or under different conditions.
One plausible role for positive control in the λ circuitry is that it may play a role in the lysis-lysogeny decision. It permits PRM to be weak and thereby largely prevents synthesis of CI from unstimulated PRM. With a constitutive PRM of the same strength as stimulated wild-type PRM, enough CI might be made in a small fraction of cells to lead to the lysogenic response. Hence, the frequency of lysogenization might change. In the context of the modern λ circuitry, this might in addition lead to an unproductive infection, for the following reason. It is believed that λ assesses the physiological conditions of the infected cell, by mechanisms that are poorly understood (8), and that this assessment leads to a particular level of the CII activator. High levels of CII lead to expression of cI from the PRE promoter, leading to the lysogenic regulatory state. If CI were made from a stronger PRM promoter, this might lead to a lysogenic response, even in cells that otherwise would favor the lytic response and would contain little or no CII. However, this would not lead to the formation of a stable lysogen, since expression of Int protein (integrase) after infection requires the action of CII at the PI promoter. In the absence of Int, an abortive lysogen would form in which the circularized phage genome would be repressed by CI and unable to replicate but not integrated. This problem would be especially severe following a single infection, in which the wild type lysogenizes at a low frequency.
We tested whether several of the phages isolated in this work were able to follow the lysogenic pathway with elevated frequency. We found a modest increase in lysogenization frequency relative to the wild type after a single infection of exponentially growing cells and a further modest increase when Int was provided from a plasmid (data not shown). It is possible that these small effects reflect a role for positive control that would increase fitness, but this issue needs further study under a range of physiological conditions. Since CII also stimulates the paQ promoter, thereby counteracting the lytic pathway (14, 19), the increase in lysogenization observed might also have been small because this promoter was not active.
A second suggested role for positive control is in prophage induction (18, 39). During this process, CI levels drop due to RecA-mediated cleavage, eventually becoming low enough that OR2 is not fully occupied. At this point, PRM is rather abruptly no longer stimulated, largely preventing replenishment of CI by new synthesis, leading to switch-like behavior that helps make the switch irreversible. Our evidence that lysogens lacking positive control can switch suggests that positive control is not essential, at least after an acute stress such as UV induction (see also the next paragraph). It is more difficult in this case to know what criteria to use to assess fitness. However, it is possible that positive control might make a difference in ways that we could not detect.
We suggest two more possible advantages of positive control that would not have been detected here. First, it is plausible that SOS induction often occurs in the natural environment not by an acute dose of DNA damage, as in our experiments, but rather by chronic low-level exposure of cells to DNA-damaging agents. At an intermediate level of DNA damage, lysogens might be in a “subinduced” state (2, 24), leading to an “undecided” mode in which the levels of CI are reduced by cleavage to low levels but the outcome in a given cell would be determined by stochastic events. Such a system might persist in regulatory limbo for a long time. Positive feedback in such a situation would tend to drive the system in one direction or the other, and positive control could contribute to this.
A more speculative role for positive control is one that is unlikely to be important in the modern λ circuitry. This circuitry can exist in an “anti-immune” state in which Cro acts to repress cI expression. This state, although stable, is less stable than the lysogenic state, switching back to the latter at a detectable frequency (9, 33). The anti-immune state can be observed when early lytic gene expression is blocked by mutations in N and in a DNA replication gene (O or P). We suggest that phages such as those containing D38N and a stronger PRM promoter might have a less stable anti-immune state. When Cro is transiently unbound from both OR3 and OR2, PRM would be expressed at higher rates than in the wild type, leading to increased synthesis of CI and to a higher likelihood of switching. This expectation is supported by computer simulation of the λ circuitry (J. W. Little and A. P. Arkin, unpublished data). Since, as stated, the anti-immune state is probably irrelevant to the normal λ circuitry, we surmise that positive control could have arisen as a result of selective pressure on a bistable circuit operating in a different context, perhaps preceding the use of this circuit in a lambdoid phage. In this view, positive control arose in a context different from its present one and was part of the circuitry when it was co-opted by λ, so that it could not readily be dispensed with initially. Perhaps it later assumed a different set of roles that have maintained it over the course of evolution. In this view, the complexity of the λ system did not arise in that context, as we discuss in the final section, but in a prior context; in such a case, the arguments developed below would apply instead to that prior context.
In addition to removing positive control, we also developed a novel type of negative control loop in the λ circuitry. In several lysogenizing variants of λ carrying cI D38R and relatively strong PRM promoters, CI D38R repressed these promoters (Fig. 5C), strongly suggesting that they are under negative autoregulation when CI D38R is bound to OR2. This presents a regulatory paradox for the following reason. For a lysogen to be stable, CI must be bound to OR1 and/or OR2 to repress PR. However, cooperative binding with a CI dimer at OR1 leads to occupancy of OR2; that is, cooperativity largely prevents CI from being bound only at OR1, except at a low CI level. Hence, at levels of CI low enough to allow OR2 to be free, OR1 is likely to be free a substantial fraction of the time as well, allowing RNA polymerase to bind to PR. This suggests that the pattern of CI binding in these lysogens is constantly in flux and that the CI level is likely to be far lower than that in a normal λ lysogen.
We surmise, but have not tested directly, that the pattern of CI expression after UV induction also differs markedly from that in the wild type as follows. As the CI levels begin to drop due to cleavage, CI falls off OR2, derepressing PRM. Since these promoters are as strong as or stronger than stimulated wild-type PRM, derepression tends to restore CI levels, counterbalancing the effects of cleavage to a far greater extent than in the wild type. Eventually, however, expression of cro from PR occurs, the resulting Cro binds to OR3, and the regulatory state can switch.
Implications for evolution of gene regulatory circuitry.
Complex systems have almost certainly arisen from simpler ones during the course of evolution. Our present findings are compatible with the view that certain features of modern gene regulatory circuits are refinements of a basic ground plan. We have proposed (27) a two-stage pathway for evolution of complex circuits in which a simple circuit arises that offers a selective advantage, followed by refinement and elaboration in a later stage. In this view, an initial circuit is likely to have relatively few components and few interactions and to have a simple structure that can arise by chance assembly from simpler components or modules (see also reference 1). Based in part on the present data, we suggest that the second stage of this pathway can involve at least two related types of changes. First, qualitatively new features, such as positive autoregulation, can be grafted onto a pre-existing circuit, increasing its complexity. This may have the effect of increasing its ability to be bistable or to exhibit other useful system properties such as threshold behavior. Second, the parameters characterizing the system, such as the affinity of DNA-binding proteins for their sites or promoter strengths, can be refined. This type of change is more likely if the system is robust, that is, if its behavior is relatively insensitive to parameter changes, since it would be easier to refine the system's properties without destroying its operation.
In the first stage of this two-stage pathway, a simple circuit would be far more likely to arise by chance during evolution, for at least three reasons. The first is that robustness would allow an initial circuit to take a wide variety of forms (27). Second, if the circuit lacked the refinements seen in typical modern circuits, the circuit would also be far more likely to arise than if these refinements were initially necessary. Finally, we have previously described (32) an example of “sequence tolerance,” a feature complementary to robustness, in which a range of sequences for a cis-acting site (PRM in our case) provide similar parameter values; again, this feature makes an even wider range of initial forms possible.
In the second, refinement, stage, addition of new features and parameter changes can occur, among other means, by changes in the proteins or in their cis-acting sites. In our studies showing that the λ regulatory circuit is robust (27), we found that a phage termed λOR323 (with an OR3 site at the position of OR1) could evolve toward the wild type in response to selective pressure for better lytic growth by mutating the site at OR1. This same phage can also evolve toward the wild type, under selective pressure for a set point in prophage induction closer than that of the wild type, by changing a different position (position 5) in the site at OR1 (our unpublished data). New features or parameter changes could also occur by changing the arrangement or spacing of cis-acting sites, thereby altering the interactions among the proteins binding to the sites. For instance, with the CI repressor of the lambdoid phage HK022, reducing the spacing between binding sites changes the mode of cooperative binding from pairwise to an “extended” mode in which a bound dimer can contact other dimers at two flanking sites rather than being restricted to contacting one dimer (28). Changes in proteins, such as adding an interaction with little energy gain such as cooperativity or positive control, as studied here, are also feasible mechanisms for adding new features.
We suggest a specific pathway with four steps by which a phage lacking positive autoregulation, but with a constitutive PRM of a strength comparable to that of stimulated wild-type PRM, could evolve to a form resembling λ. This pathway takes advantage of the modular organization of lambdoid phages, which should allow a given immunity region to be associated with a different set of modules at different times, allowing that region to be subjected to different selective pressures. The pathway utilizes the finding (29) that certain prophages producing Shiga toxin have a low set point, perhaps allowing them to undergo spontaneous induction readily (29, 32). In the first step of the proposed pathway, the immunity region would become associated with a late-gene module producing Shiga toxin. Second, PRM would mutate to a weaker promoter under selective pressure to reduce the set point. Third, a change in CI (like N38→D) would create a contact supporting positive autoregulation, increasing CI levels and changing the set point for prophage induction to that of wild-type λ. Finally, module recombination would occur to remove the Shiga toxin gene, making this set point advantageous, as we presume it is for λ.
In separate studies, we have also found that two other features of the λ circuitry initially believed to be essential for its proper operation can likewise be viewed as refinements that are not necessary for the proper qualitative operation of the circuitry. The differential occupancy of CI and Cro for the various OR operators can be removed by making the sequences of OR1 and OR3 the same (27). Moreover, recent studies (A. C. Watson and J. W. Little, unpublished data) indicate that a loss of cooperative DNA binding by CI can be suppressed by changes in cis-acting sites in the OR region, resulting in near-normal behavior. This implies that cooperativity is also not an essential feature. We are currently testing whether both cooperativity and positive control can be removed with the proper combination of suppressors, further simplifying the λ circuitry.
Supplementary Material
Acknowledgments
We are grateful to Shota Atsumi, Carol Dieckmann, Kim Giese, Gary Gussin, Andrea Watson, and an anonymous reviewer for comments on the manuscript; to Gary Gussin for helpful discussions; and to Anca Segall for plasmids.
This work was supported by grant GM24178 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atsumi, S., and J. W. Little. 2004. Regulatory circuit design and evolution using phage λ. Genes Dev. 18:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailone, A., A. Levine, and R. Devoret. 1979. Inactivation of prophage λ repressor in vivo. J. Mol. Biol. 131:553-572. [DOI] [PubMed] [Google Scholar]
- 3.Becskei, A., B. Séraphin, and L. Serrano. 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20:2528-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman, F. D., C. Shang, and M. Ptashne. 1989. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell 58:1163-1171. [DOI] [PubMed] [Google Scholar]
- 5.Darling, P. J., J. M. Holt, and G. K. Ackers. 2000. Coupled energetics of lambda cro repressor self-assembly and site-specific DNA operator binding I: analysis of cro dimerization from nanomolar to micromolar concentrations. Biochemistry 39:11500-11507. [DOI] [PubMed] [Google Scholar]
- 6.Dodd, I. B., A. J. Perkins, D. Tsemitsidis, and J. B. Egan. 2001. Octamerization of lambda CI repressor is needed for effective repression of PRM and efficient switching from lysogeny. Genes Dev. 15:3013-3022.11711436 [Google Scholar]
- 7.Dove, S. L., F. W. Huang, and A. Hochschild. 2000. Mechanism for a transcriptional activator that works at the isomerization step. Proc. Natl. Acad. Sci. USA 97:13215-13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echols, H. 1986. Bacteriophage λ development: temporal switches and the choice of lysis or lysogeny. Annu. Rev. Genet. 2:26-30. [Google Scholar]
- 9.Eisen, H., P. Brachet, L. Pereira da Silva, and F. Jacob. 1970. Regulation of repressor expression in λ. Proc. Natl. Acad. Sci. USA 66:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrell, J. E., Jr. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140-148. [DOI] [PubMed] [Google Scholar]
- 11.Hawley, D. K., and W. R. McClure. 1982. Mechanism of activation of transcription initiation from the lambda PRM promoter. J. Mol. Biol. 157:493-525. [DOI] [PubMed] [Google Scholar]
- 12.Herskowitz, I., and D. Hagen. 1980. The lysis-lysogeny decision of phage λ: explicit programming and responsiveness. Annu. Rev. Genet. 14:399-445. [DOI] [PubMed] [Google Scholar]
- 13.Hochschild, A., N. Irwin, and M. Ptashne. 1983. Repressor structure and the mechanism of positive control. Cell 32:319-325. [DOI] [PubMed] [Google Scholar]
- 14.Hoopes, B. C., and W. R. McClure. 1985. A cII-dependent promoter is located within the Q gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 82:3134-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, J. J., S. Brown, and G. N. Gussin. 1988. Characterization of a doubly mutant derivative of the lambda PRM promoter: effects of mutations on activation of PRM. J. Mol. Biol. 200:695-708. [DOI] [PubMed] [Google Scholar]
- 16.Jain, D., B. E. Nickels, A. Hochschild, and S. A. Darst. 2004. Structure of a ternary transcription activation complex. Mol. Cell 13:45-53. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, A. D., B. J. Meyer, and M. Ptashne. 1979. Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc. Natl. Acad. Sci. USA 76:5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, A. D., A. R. Poteete, G. Lauer, R. T. Sauer, G. K. Ackers, and M. Ptashne. 1981. λ repressor and cro—components of an efficient molecular switch. Nature 294:217-223. [DOI] [PubMed] [Google Scholar]
- 19.Kobiler, O., A. Rokney, N. Friedman, D. L. Court, J. Stavans, and A. B. Oppenheim. 2005. Quantitative kinetic analysis of the bacteriophage λ genetic network. Proc. Natl. Acad. Sci. USA 102:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuldell, N., and A. Hochschild. 1994. Amino acid substitutions in the −35 recognition motif of σ70 that result in defects in phage lambda repressor-stimulated transcription. J. Bacteriol. 176:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeFevre, K. R., and M. H. J. Cordes. 2003. Retroevolution of lambda Cro toward a stable monomer. Proc. Natl. Acad. Sci. USA 100:2345-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., H. Moyle, and M. M. Susskind. 1994. Target of the transcriptional activation function of phage lambda cI protein. Science 263:75-77. [DOI] [PubMed] [Google Scholar]
- 23.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 81:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little, J. W. 1983. The SOS regulatory system: control of its state by the level of RecA protease. J. Mol. Biol. 167:791-808. [DOI] [PubMed] [Google Scholar]
- 25.Little, J. W. 1993. LexA cleavage and other self-processing reactions. J. Bacteriol. 175:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 27.Little, J. W., D. P. Shepley, and D. W. Wert. 1999. Robustness of a gene regulatory circuit. EMBO J. 18:4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Z., and J. W. Little. 1998. The spacing between binding sites controls the mode of cooperative DNA-protein interactions: implications for evolution of regulatory circuitry. J. Mol. Biol. 278:331-338. [DOI] [PubMed] [Google Scholar]
- 29.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691-1704. [DOI] [PubMed] [Google Scholar]
- 30.Mao, C., and J. W. Little. 1998. Mutations affecting cooperative DNA binding of phage HK022 CI repressor. J. Mol. Biol. 279:31-48. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, B. J., and M. Ptashne. 1980. Gene regulation at the right operator (OR) of bacteriophage λ III. λ repressor directly activates gene transcription. J. Mol. Biol. 139:195-205. [DOI] [PubMed] [Google Scholar]
- 32.Michalowski, C. B., M. D. Short, and J. W. Little. 2004. Sequence tolerance of the phage λ PRM promoter: implications for evolution of gene regulatory circuitry. J. Bacteriol. 186:7988-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neubauer, Z., and E. Calef. 1970. Immunity phase-shift in defective lysogens: non-mutational hereditary change of early regulation of lambda prophage. J. Mol. Biol. 51:1-13. [DOI] [PubMed] [Google Scholar]
- 34.Ptashne, M. 2004. A genetic switch: phage lambda revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Revet, B., B. Von Wilcken-Bergmann, H. Bessert, A. Barker, and B. Müller-Hill. 1999. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr. Biol. 9:151-154. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, J. W., and R. Devoret. 1983. Lysogenic induction, p. 123-144. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sanderson, A., J. E. Mitchell, S. D. Minchin, and S. J. W. Busby. 2003. Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 544:199-205. [DOI] [PubMed] [Google Scholar]
- 38.Sauer, R. T., and R. Anderegg. 1978. Primary structure of the lambda repressor. Biochemistry 17:1092-1100. [DOI] [PubMed] [Google Scholar]
- 39.Shea, M. A., and G. K. Ackers. 1985. The OR control system of bacteriophage lambda: a physical-chemical model for gene regulation. J. Mol. Biol. 181:211-230. [DOI] [PubMed] [Google Scholar]
- 40.Shih, M. C., and G. N. Gussin. 1983. Mutations affecting two different steps in transcription initiation at the phage lambda PRM promoter. Proc. Natl. Acad. Sci. USA 80:496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whipple, F. W., N. H. Kuldell, L. A. Cheatham, and A. Hochschild. 1994. Specificity determinants for the interaction of λ repressor and P22 repressor dimers. Genes Dev. 8:1212-1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.