Abstract
Pseudomonas syringae pv. phaseolicola, a gram-negative bacterial plant pathogen, is the causal agent of halo blight of bean. In this study, we report on the genome sequence of P. syringae pv. phaseolicola isolate 1448A, which encodes 5,353 open reading frames (ORFs) on one circular chromosome (5,928,787 bp) and two plasmids (131,950 bp and 51,711 bp). Comparative analyses with a phylogenetically divergent pathovar, P. syringae pv. tomato DC3000, revealed a strong degree of conservation at the gene and genome levels. In total, 4,133 ORFs were identified as putative orthologs in these two pathovars using a reciprocal best-hit method, with 3,941 ORFs present in conserved, syntenic blocks. Although these two pathovars are highly similar at the physiological level, they have distinct host ranges; 1448A causes disease in beans, and DC3000 is pathogenic on tomato and Arabidopsis. Examination of the complement of ORFs encoding virulence, fitness, and survival factors revealed a substantial, but not complete, overlap between these two pathovars. Another distinguishing feature between the two pathovars is their distinctive sets of transposable elements. With access to a fifth complete pseudomonad genome sequence, we were able to identify 3,567 ORFs that likely comprise the core Pseudomonas genome and 365 ORFs that are P. syringae specific.
The gram-negative plant-pathogenic species Pseudomonas syringae is comprised of at least 50 pathovars that can be distinguished by their host ranges (30). Many P. syringae pathovars also contain several races characterized by their avirulence on different host cultivars. Genetic control of host specificity at the race-cultivar level, and possibly the pathovar-host species level, is conditioned by “gene-for-gene” interactions between avirulence genes in the pathogen and the corresponding resistance genes in the plant (35). In the last 2 decades, a number of pathogen avirulence genes, as well as the corresponding host resistance genes, have been cloned and identified (9, 13). Resistance gene products, regardless of whether they encode resistance to viral, bacterial, fungal, nematode, or insect pathogens, share similar structures and with few exceptions contain a leucine-rich repeat region (reviewed in reference 37), suggesting a conserved mechanism(s) for pathogen recognition and signal transduction events. In contrast, avirulence gene products share little sequence similarity, although it is well known that bacterial avirulence gene products, along with other virulence factors collectively termed effectors, are injected into the host cell via the specialized type III secretion system (TTSS) that is conserved among plant and animal pathogens (15). The P. syringae effectors are designated Avr (avirulence) or Hop (Hrp outer protein) according to a recently adopted nomenclature system (38). P. syringae effectors collectively are important to virulence, and increasing evidence suggests that these proteins are involved in suppression of host defense responses in compatible interactions with host plants (2).
P. syringae pv. phaseolicola is the seed-borne causative agent of halo blight disease in the common bean (Phaseolus vulgaris), which is a devastating disease in several developing countries. Typical symptoms on bean leaves in a compatible interaction are water-soaked lesions that are surrounded by a yellow halo produced by the release of phaseolotoxin, a chlorosis-inducing phytotoxin that affects hosts and nonhosts (7). The combinations of functional effector and resistance genes in races of P. syringae pv. phaseolicola and cultivars of Phaseolus, respectively, determine the outcome of the plant-pathogen interaction (66). Classification of strains of P. syringae pv. phaseolicola into races is based on their differential abilities to cause disease in diagnostic cultivars of Phaseolus. P. syringae pv. phaseolicola strain 1448A belongs to the race 6 pathotype, members of which are virulent on all P. vulgaris varieties examined (64). As a consequence, race 6 has been selected as the recipient strain of choice in the identification of effector genes from other races based on their avirulence phenotypes (66).
P. syringae pv. tomato DC3000 is the causal agent of bacterial speck of tomato and has been developed into a robust model organism for the study of plant-pathogen interactions. Several features of the tomato bacterial speck system have contributed to its utility as a premier model system for studying plant-pathogen interactions. For example, (i) the first plant disease resistance gene (Pto) was cloned from tomato and confers resistance to P. syringae pv. tomato isolates containing avrPto (40), (ii) P. syringae pv. tomato DC3000 can infect the model plant species Arabidopsis thaliana (69), for which a near-complete genome is available (4), and (iii) the complete genome sequence has been determined for the DC3000 isolate (10), analysis of which has revealed ∼300 open reading frames (ORFs) implicated in virulence, 71 of which are components of the TTSS or are effectors with the capacity to be translocated into the host cell. To date, DC3000 has by far the largest number of putative and confirmed effector molecules known in any bacterial pathogen of animals or plants. An important question now is what combination of effectors and other virulence factors controls the host specificities of P. syringae pv. phaseolicola and P. syringae pv. tomato?
Comparative genomics of closely related isolates or species of pathogenic bacteria represents a powerful tool for rapid identification of genes involved in host specificity and virulence (8, 16, 46). Likewise, comparative genomics between two P. syringae pathovars has the potential to provide new insights into both shared and pathovar-specific genes involved in host-pathogen interactions. Phylogenetic analyses reveal that the P. syringae pathovars fall into three major clusters and that P. syringae pv. tomato and P. syringae pv. phaseolicola represent two of these clusters (53, 54), strengthening the value of comparing the genomes of DC3000 and 1448A.
In this study, we describe the sequence and annotation of P. syringae pv. phaseolicola 1448A and compare its genome with those of P. syringae pv. tomato DC3000 and other Pseudomonas spp. Our comparisons have revealed not only conserved components of the P. syringae core genome, but also those components unique to each pathogen. These data provide a foundation for detailed functional analyses of host specificity and virulence mechanisms among the P. syringae pathovars.
MATERIALS AND METHODS
Sequencing.
Strain 1448A was isolated in 1985 from P. vulgaris in Ethiopia (65). Prior to high-throughput sequencing, the pathogenicity of 1448A on the common bean was confirmed (data not shown). Sequencing and annotation were performed essentially as described previously (10). In brief, two shotgun libraries (insert sizes of 1.5 to 3.5 and 8 to 14 kbp) were constructed in modified pBR322 plasmids using total DNA isolated from 1448A. DNA was prepared from the shotgun clones using a modified alkaline lysis method, and templates were sequenced from both ends using standard high-throughput sequencing methods on ABI 3730xl sequencers (ABI, Foster City, California). In total, 51,402 small-insert (average edited read length, 829 bp) and 24,779 large-insert (average edited read length, 813 bp) sequences were generated. The sequences were trimmed to remove vector and low-quality regions. The sequences were assembled using the Celera Assembler (42). Gaps were closed using a combination of resequencing, alternative chemistries, PCR, PCR-directed resequencing, and transposon-mediated sequencing.
Bioinformatics.
The genome was annotated using a standard set of processes as described previously (10). In brief, ORFs were identified using the Glimmer algorithm (17), and predicted proteins were searched against a nonredundant amino acid database. Domains were identified using HMMer with the Pfam (6) and TIGRfam (28) databases. Initially, transitive annotation was used to annotate ORFs with a high degree of sequence identity to P. syringae pv. tomato DC3000, a genome that has been manually curated by annotators at the structural and functional levels previously (10). Using alignment with MUMmer (http://www.tigr.org/software/mummer/; default 20-nucleotide [nt] mum cutoff), we identified 3,793 ORFs that aligned with the DC3000 genome. Using a stringent set of filtering criteria, such as role category and HMM matches, we transitively annotated 1,494 of the 1448A ORFs using the DC3000 annotation. A subset of these ORFs, as well as other ORFs, was manually curated using the available evidence and output from Glimmer (http://www.tigr.org/software/glimmer/). In total, 2,383 ORFs were manually curated. Functional-role categories (48) were assigned using available evidence. To estimate expansion of gene families within the predicted proteome, paralogous families were constructed (44). Insertion sequences (IS elements) were classified according to transposase gene similarity using BLAST analysis with the ISFinder database (http://www-is.biotoul.fr/). The presence of putative homologs of 1448A ORFs in the DC3000 genome (GenBank accession numbers AE016853, AE016854, and AE016855) was determined by BLASTP (3) using the following cutoff criterion: E < 10−15. Orthology was inferred by the presence of reciprocal best BLASTP hits in the two pathovars. Conservation of gene order in the chromosomes of 1448A and DC3000 was established using the syntenic-block algorithm from the Sybil software package (http://sybil.sourceforge.net), with a minimum syntenic-block size of four ORFs and a maximum gap size (the maximum number of adjacent nonmatching genes that may appear in a block) of two ORFs.
Nucleotide sequence accession numbers.
The sequence and annotation of the 1448A genome has been submitted to GenBank under accession numbers CP000058 to -60.
RESULTS AND DISCUSSION
General features of the 1448A genome.
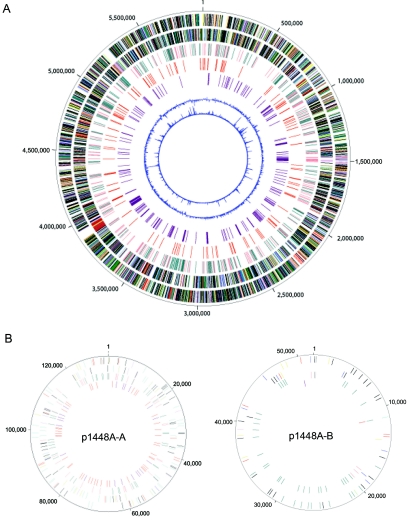
The 1448A genome consists of one circular chromosome (5,928,785 bp) and two plasmids, p1448A-A (131,950 bp) and p1448A-B (51,711 bp) (Table 1 and Fig. 1). In total, 5,353 ORFs were identified in the 1448A genome. A putative function was assigned to 3,626 (68%) ORFs, with the remaining ORFs (1,727 total) designated as hypothetical proteins (224; 4%), conserved hypothetical proteins (822; 15%), or proteins of unknown function (681; 13%). The classification of 1448A ORFs into biological-role categories is summarized in Fig. S1 in the supplemental material. Transport and binding proteins constitute the largest role category found in 1448A (728 ORFs; 14%), followed by genes involved in cellular processes (708 ORFs; 13%). Approximately 10% of the total ORFs have a regulatory role (535 ORFs) and presumably facilitate adaptation to the different environments faced by 1448A as a pathogen, an epiphyte, and a seed-borne bacterium during different phases of its life cycle. Approximately 5% (256) of the total number of ORFs fall within the mobile genetic element category, with 201 ORFs involved in transposition constituting the majority of this role category. The 1,348 paralogous families identified in 1448A, encompassing 3,177 ORFs (59%), indicate the extent of gene duplication within the genome. Apart from ORFs encoding proteins, the 1448A genome also contains 64 tRNAs, five rRNA operons, and one structural RNA.
TABLE 1.
General features of the P. syringae pv. phaseolicola 1448A genome and comparison with the P. syringae pv. tomato DC3000 genome
| Feature | P. syringae pv. phaseolicola 1448A | P. syringae pv. tomato DC3000 | ||||
|---|---|---|---|---|---|---|
| Molecule | Chromosome | p1448A-A | p1448A-B | Chromosome | pDC3000A | pDC3000B |
| Size (bp) | 5,928,787 | 131,950 | 51,711 | 6,397,126 | 73,661 | 67,473 |
| G+C% | 58.0 | 54.1 | 56.0 | 58.4 | 55.1 | 56.1 |
| % Coding regions | 87.0 | 78.0 | 84.9 | 86.8 | 81.9 | 84.7 |
| No. of ORFs | 5,144 | 149 | 60 | 5,615 | 71 | 77 |
| Avg ORF length (bp) | 1,007 | 660 | 719 | 988 | 849 | 742 |
| No. of ORFs with annotated function (%) | 3,492 (68) | 97 (65) | 37 (62) | 3,402 (61) | 52 (73) | 46 (60) |
FIG. 1.
Features of the P. syringae pv. phaseolicola 1448A genome. (A) The chromosome. The ORFs on the positive and negative strands are depicted on the outermost and second circle of the figure, respectively. The ORFs are color coded based on the major grouping of role categories as follows: amino acid biosynthesis (salmon); biosynthesis of cofactors, prosthetic groups, and carriers (light blue); cell envelope (light green); cellular processes (red); central intermediary metabolism (brown); DNA metabolism (yellow); energy metabolism (green); fatty acid and phospholipid metabolism (purple); mobile genetic elements (cyan); protein fate/synthesis (pink); purines, pyrimidines, nucleosides, and nucleotides (orange); regulatory functions (navy blue); transcription (gray); transport and binding proteins (teal); and unknown function, conserved hypothetical, and hypothetical (black). The third circle represents the ORFs specific to the P. syringae pathovars (salmon) and the ORFs specific to 1448A (teal) in comparison to the predicted proteomes of four complete Pseudomonas genomes (BLASTP E value < 10−5). The fourth circle represents IS elements. The fifth circle represents virulence factors (purple). The sixth circle is the GC skew, and the seventh circle represents atypical nucleotides. (B) Plasmids within 1448A. The three outermost circles in the figure are the same as in panel A. The fourth circle depicts IS elements (red) and ORFs contributing to 1448A virulence (purple).
Comparative genomics with P. syringae pv. tomato DC3000.
Access to complete genomes of pathovars from two of the three P. syringae phylogenetic clusters (53, 54) provides the opportunity to expand comparison to the whole-genome level. With respect to size and other genomic features, 1448A is generally similar to the tomato pathogen DC3000 (Table 1 and Fig. 1; see Table S1 in the supplemental material), with 83% of the ORFs shared between DC3000 and 1448A (BLASTP cutoff, E < 10−15). Furthermore, classification of the ORFs from DC3000 and 1448A into functional-role categories reveals that the genomes of these two pathovars are highly conserved with respect to the ability to encode proteins required for basic physiological and metabolic functions (see Fig. S1 in the supplemental material). Indeed, an analysis of 154 biological processes included in the Genome Properties System (29; http://www.tigr.org/Genome_Properties), a computational method to identify the presence/absence of metabolic pathways and systems in prokaryotic genomes, detected only two differences between the strains. One difference was the presence of an intein in DC3000 (PSPTO3229) where none is found in 1448A. The second difference was an authenticated point mutation in 1448A in the gene for N-formylglutamate amidohydrolase (PSPPH4868), resulting in a truncation and raising the possibility that 1448A may be unable to degrade histidine to glutamate by this pathway (see Table S2 in the supplemental material). In the sections below, we report on features of the 1448A genome important to its pathogenic lifestyle and, when relevant, compare these to the related but distinct pathovar DC3000.
Virulence-implicated ORFs in 1448A and DC3000.
We previously identified 298 ORFs implicated in virulence in the DC3000 genome (10). Within the virulence category, we include effectors (both Avr and Hop) and other proteins of potential benefit to pathogenesis, including those involved in epiphytic fitness. Searches of the 1448A genome using the DC3000 virulence ORFs revealed that 240 (81%) of the DC3000 virulence ORFs are present in 1448A, including genes for many Hop effectors; secretion pathways I, II, and III; cell wall-degrading enzymes; adhesins; and nonribosomal peptide synthases (NRPSs) (see Table S3 in the supplemental material). Examples of virulence factors present in DC3000 but absent from 1448A include genes involved in the synthesis and regulation of the phytotoxin coronatine. On the other hand, a vestigial virulence-related gene, inaZ, which encodes the ice nucleation protein that is responsible for frost damage in plants (30), is present in 1448A but not in DC3000. However, the inaZ gene in 1448A (PSPPH1596 and PSPPH1598) is unlikely to encode a functional gene product, given that the reading frame is interrupted by a transposase (PSPPH1597; ISPsy18 transposase; Mutator family).
Although many virulence factors, such as the hrp and hrc genes encoding the TTSS, are highly conserved in both sequence and location between the two pathovars, the sizes and locations of others vary substantially. For example, the effector candidate HopAS1 (PSPPH4736; 1,361 amino acids) is truncated in DC3000 due to a point mutation but is present in a longer version in 1448A. HopX1 (PSPPH1296; formerly AvrPphE) is present and functional in DC3000 but appears to be nonfunctional as an avirulence determinant in 1448A, as previously reported for race 6 strains of P. syringae pv. phaseolicola (62). The effector gene inventories of DC3000 and 1448A are large and characteristically contain a mix of functional and nonfunctional genes that differ among strains and pathovars (10, 14, 26, 55, 57; M. Vencato et al., unpublished data). As experimentally documented and discussed elsewhere, 1448A harbors at least 22 genes (see Table S3 in the supplemental material) encoding proteins that can be delivered by the TTSS into plant cells (14; Vencato et al., unpublished data).
Comparison of virulence genes that are plasmid borne in 1448A with homologs in DC3000.
With regard to differences in location, genes encoding putative homologs of most of the virulence factors that are located on plasmids p1448A-A and p1448A-B are chromosomally located in DC3000. For example, genes encoding 1448A TTSS effectors HopD1 and HopQ1 are located on plasmid p1448A-A (PSPPHA0010 and -A0012), whereas their orthologs in DC3000 (PSPTO0876 and -0877) are clustered with other virulence factor genes on a chromosomal genomic island (34). Similarly, of the four shared virulence factors that are plasmid borne in DC3000, all but one, a copy of levansucrase (PSPPHA0027 on p1448A-A), are found on the 1448A chromosome.
In general, the plasmids present in the two pathovars exhibit significant differences in both size and content. Plasmids p1448A-A and p1448A-B belong to the pPT23A family found in DC3000 and other P. syringae pathovars (56, 71). The small plasmid (p1448A-B) carries the rulAB genes (PSPPHB0003 and -B0004) for UV resistance but lacks type III effector genes. By contrast, in DC3000, the rulAB genes are located on the plasmid that is enriched in virulence factors (pDC3000A) and rulB has a disrupted reading frame. Plasmid p1448A-B does carry an ORF with a potential role in virulence, PSPPHB0059, a homolog of an ExeA-like protein (PMA4326B11) present on plasmid pPMA4326B in P. syringae pv. maculicola (61). ExeA, along with ExeB, is required for type II secretion of the toxin aerolysin in the pathogen Aeromonas hydrophila (33). No homologs of ExeB were found in 1448A, but a fragment of an ExeA-like ORF (PSPPHA0124) is present in the pathogenicity island (PAI) on p1448A-A.
Other genes of potential significance to pathogenicity found on plasmid p1448A-A but not in DC3000 include homologs of the atypical fimbrial genes found in Salmonella species, safBCD (PSPPHA0063 to -65) and sinR (PSPPHA0062). A fourth gene, safA, frequently but not exclusively associated with this locus in Salmonella, is absent from 1448A. The safA and safD genes encode fimbrial structural proteins, whereas the safBC genes encode usher and chaperone proteins and sinR encodes a LysR transcriptional regulator. All were identified on a 47-kb genomic island in Salmonella enterica serovar Typhimurium and were adjacent to a transposase gene, upstream of safA. The synteny in p1448A-A is inverted compared to Salmonella, with sinR located upstream of safA rather than downstream of safD. Transposases are located on either side of the gene cluster (PSPPHA0061 and PSPPHA0068). It has been recognized that the saf and sinR genes are horizontally transferred (20, 27), but a role in virulence has yet to be determined. The horizontal transfer of the saf genes has been linked to the evolution of Salmonella serovars and may be associated with host adaptation. Homologs of saf genes have not been reported previously in a plant-pathogenic pseudomonad.
Comparison of virulence genes in the plasmid-borne PAI between P. syringae pv. phaseolicola strains 1448A (race 6) and 1449B (race 7).
Knowledge of the 1448A genome sequence also provides an opportunity to compare gene content and organization with a race 7 strain of P. syringae pv. phaseolicola, 1449B. This strain contains a well-characterized PAI on its 154-kb plasmid (32) that includes the known effector genes hopAB1, avrB2, hopF1, avrD, and ORF4, whose product has cysteine protease motifs (31, 58). Also located on the plasmid, but physically separated from the PAI, is hopD1. The main difference between the 1449B PAI and the homologous region in 1448A is the absence from 1448A of a 9,471-nt sequence that carries hopF1. The deletion was flanked on its left and right borders by a chimeric ORF with homology to IS1492b (Pseudomonas putida) and IS1090 (Ralstonia eutropha) and a region containing several transposon gene fragments, including IS801, respectively (49). Loss of hopF1 helps to explain the virulence of 1448A to the bean cultivar Red Mexican, which carries the matching R1 gene (66). In 1448A, an insertion of 10 nt in the coding sequence of the homolog of ORF3 in 1449B (32) results in a deduced 218-amino-acid protein (PSPPHA0122; type III effector HopAW1) that includes the catalytic triad CHD motif indicative of the cysteine protease activity found in several effectors. The H and D components of the triad are absent from the peptide encoded by the truncated ORF3 in 1449B (accession number AAD47205).
Attachment factors potentially contributing to virulence.
Bacterial attachment and colonization represent essential stages in the pathogenic processes of many bacteria and depend upon the presence of multiple factors. Attachment factors previously identified in DC3000 include type IV pili, alginate, nonalginate capsular polysaccharide, exopolysaccharides, and filamentous hemagglutinin, many of which are also found in 1448A. For example, all genes required for alginate biosynthesis are present in both pathovars, as well as in Pseudomonas aeruginosa, although regulation of its production in P. syringae probably differs from that in P. aeruginosa (45). Similarly, a full complement of genes required for synthesis of type IV pili appears to be present, and pilus biosynthesis has been experimentally confirmed in both DC3000 and another strain of P. syringae pv. phaseolicola (50). Although hemagglutinins have not been shown to play a significant role in pseudomonad virulence, three genes encoding filamentous hemagglutinin-like proteins are present in 1448A, as are three genes implicated in biosynthesis of capsular polysaccharides and two involved in exopolysaccharide biosynthesis.
Secretion systems associated with virulence.
An unusual feature of the 1448A chromosome is the presence of a cluster of 12 genes (PSPPH2519 to -2538) encoding TTSS components whose closest homologs are variously in Rhizobium, Photorhabdus, and Aeromonas spp. or in P. aeruginosa. Mutations in numerous hrp and hrc genes encoding the Hrp TTSS abolish the ability of P. syringae pv. phaseolicola to cause disease in beans or to elicit the hypersensitive response in nonhost or resistant plants (39), which suggests that this second, putative TTSS does not deliver effectors into plant cells. No sequences associated with HrpL-responsive promoters are associated with the second set of TTSS genes, and the expression and functions of these genes remain to be tested.
In addition to the TTSS, three other major secretion pathways (I, II, and IV) are found in many pathogenic bacteria, including other pseudomonads (18). Like DC3000, 1448A encodes multiple type I secretion pathway components, although protein substrates implicated in virulence have yet to be identified. The 1448A genome also contains genes for two distinct type II secretion pathways, an occurrence previously observed in P. aeruginosa (5). Candidate substrates for the type II pathways include two cellulases, two pectate lyases, a pectin lyase, and a polygalacturonase. By contrast, DC3000 contains genes only for a single type II pathway, and the cell wall-degrading enzymes representing the most likely secretion substrates are limited to two cellulases and a pectin lyase.
Type IV secretion systems are employed by various bacteria to secrete diverse substrates and are closely related to systems involved in conjugal transfer of DNA (11, 12). In contrast to DC3000, but similar to P. syringae pv. maculicola (61), 1448A has a large number of genes with high similarity to the type IV secretion genes of Agrobacterium tumefaciens (the virB operon and virD4) (71). These genes are found on both plasmids in 1448A, with the large plasmid containing a subset of the virB operon and the small plasmid containing the entire operon, along with the homolog of virD4. However, the fact that these genes are on plasmids, the presence of a virB5 homolog found only in systems transferring DNA (11), and the presence of a small protein between virB5 and virB6 found only in conjugal systems (11) strongly suggest that the Type IV homologs in 1448A are involved in the process of conjugal transfer of DNA rather than virulence-related protein translocation.
Virulence-implicated metabolic, biosynthetic, and regulatory capabilities.
The known sources of carbon potentially available to P. syringae while growing in the host apoplast (intercellular space) are sucrose and gamma-aminobutyric acid (GABA) (25, 59), with the latter also serving as a nitrogen source. The genes required to transport and metabolize these substrates have been reported in DC3000 (10, 34) and are also present in 1448A. As seen in DC3000, the genes encoding the sucrose porin precursor and sucrose-6-phosphate hydrolase (PSPPH5187 and -5192) are clustered, along with a putative regulator (PSPPH5193) and other sugar transporters, on the 1448A chromosome. As in DC3000, 1448A has one GABA permease (PSPPH4937) and multiple copies of GABA transaminase (PSPPH0095, -3457 [authentic frameshift], and -5040) and succinate-semialdehyde dehydrogenase (PSPPH0096, -2572, and -5038 [authentic frameshift]).
The ethylene-forming enzyme (EFE) 1-aminocyclopropane-1-carboxylate (ACC) oxidase (EC 1.14.17.4) has been reported in some strains of P. syringae pv. phaseolicola, such as PK2 (22), and other P. syringae pathovars (68). EFE, which catalyzes the conversion of the precursor 1-aminocyclopropane-1-carboxylate to ethylene in the final step in the synthesis of the phytohormone, is not present in 1448A or DC3000. However, the enzyme ACC deaminase (EC 3.5.99.7), which also uses ACC as a substrate, is present in 1448A (PSPPH1761) and DC3000 (PSPTO3675). Transgenic tomato plants expressing ACC deaminase from Pseudomonas sp. strain 6G5 have demonstrated delayed ripening of tomato fruit due to reduced ethylene synthesis (36).
The genes encoding the enzymes IaaM and IaaH, required for the production of the plant hormone indole-3-acetic acid (IAA; auxin), are present in both pathovars; however, the enzyme indoleacetate-lysine ligase (IaaL), which converts IAA to IAA-lysine, is present in DC3000 (10) but not in 1448A. In DC3000, iaaL is part of the Hrp regulon (21), is located adjacent to the transposable element ISPsy7, and appears to have been inserted into a portion of the genome that otherwise shares a high degree of gene synteny with 1448A.
The wss operon (wssA-J) in P. fluorescens SBW25, a plant growth-promoting pseudomonad, encodes the genes required for the synthesis of an acetylated cellulose-like polymer that is associated with fitness of the microbe in the rhizosphere and phyllosphere (23, 60). The homologs of the genes comprising the wss operon, except wssJ, are present in DC3000 (PSPTO1026 to -1034) but not in 1448A.
Global regulatory proteins associated with P. syringae virulence or epiphytic fitness include the alternative sigma factors HrpL and RpoN, the two-component regulators GacS and GacA, quorum-sensing components AhlI and AhlR, and the TetR family activator of the quorum-sensing system AefR (47, 70). Genes encoding all of these proteins are found in 1448A (see Table S1 in the supplemental material). HrpL is unique to P. syringae and other phytopathogens with group I Hrp systems, such as Erwinia amylovora (1), and genes activated by HrpL are addressed elsewhere (14; Vencato et al., unpublished data).
Nonribosomal peptide and polyketide synthases.
Several P. syringae pv. phaseolicola strains are known to produce the phytotoxin phaseolotoxin, which acts by inhibiting the activity of ornithine carbamoylphosphate transferase (OCTase), an enzyme in the pathway for synthesis of arginine (19). Toxigenic strains produce a phaseolotoxin-insensitive OCTase encoded by argK (41), which is part of the argK-tox gene cluster (54) that is involved in the biosynthesis of phaseolotoxin. Gonzalez et al. (24) demonstrated via PCR that 1448A contains the argK-tox cluster. However, although they reported that the strain does not produce phaseolotoxin, recent assays indicate that 1448A does produce phaseolotoxin (R. Jackson, unpublished data). As expected, 1448A contains two OCTases, one of which is encoded by argK (PSPPH4319) clustered with other loci associated with toxin production. The presence of a transposase immediately downstream of argK supports the acquisition of the cluster by horizontal transfer (54). DC3000, which does not synthesize the toxin, contains only a single OCTase (PSPTO4164) that is an ortholog of the phaseolotoxin-sensitive enzyme (PSPPH3895) in 1448A.
The 1448A genome has eight regions that contain genes with predicted NRPS and polyketide synthase domains (see Table S4 in the supplemental material). Three of these, including the argK-tox region, appear to be unique to 1448A compared to other sequenced genomes. Syntenic homologs of the remaining five regions are found in the genomes of either DC3000, P. syringae pv. syringae B728a, or both. The homologs include the gene cluster for the biosynthesis of pyoverdin, which is found in all three strains, and for yersiniabactin, which is found in 1448A and DC3000. A cluster that is common to 1448A and B728a (>90% similarity; PSPPH1749 to -1752) contains two NRPS genes, one hybrid NRPS-polyketide synthase, and a possible dioxygenase. Of particular interest is a region shared between 1448A and B728a (>90% similarity and perfect gene synteny; PSPPH2703-2720), which contains a series of genes, each of which is 40 to 80% similar to the genes of the phaseolotoxin-producing argK-tox region, which encodes homologs of the dCTP deaminases, fatty acid desaturases, ornithine aminotransferase, a phosphatase-like protein, a PEP synthase-like protein, and eight proteins of unknown function. Several of the argK-tox genes are missing from this cluster, including argK itself and the gene for Arg-Lys amidinotransferase, and the gene order appears to have undergone at least two rearrangement events. This “pseudophaseolotoxin” region itself sits within a larger region (PSPPH2736 to -2860) that appears to be a lateral-transfer “hotspot,” showing evidence of at least seven different transfer events with distinguishable origins, and is quite close to the yersiniabactin gene cluster (PSPPH2892 to -2904).
Mobile genetic elements.
The percentage of ORFs in 1448A (5%) included in the category of mobile genetic elements is less than that seen in DC3000 (7%). However, as in DC3000, IS elements comprise the majority of ORFs in this role category. Most of the transposases found in 1448A are different from those identified in DC3000, which indicates that they were acquired after the divergence of the lineages leading to the present pathovars. ISPsy18 transposases, which are not present in DC3000, belong to the Mutator family and comprise the most abundant family of IS elements (48 intact copies; 60 total) in the 1448A genome. These transposases belong to the IS256 family and are most similar to ISRSO7 transposase found in Ralstonia solanacearum (52). Many of the ISPsy18 transposases are either inserted into ORFs or linked to genes with disrupted reading frames (see Table S5 in the supplemental material).
The transposase for insertion sequence element IS801 is present in a number of pseudomonads and has been characterized in P. syringae pv. phaseolicola strain LR781 (51). IS801, a member of the IS91 family, is not present in DC3000, although the IS91 family is represented by the transposases ISPsy3 and ISPsy4. A total of 19 intact and disrupted versions of the IS801 transposase are present in 1448A. The full-length transposase is 410 amino acids, similar to IS801, first isolated from plasmid pMMC7105 of P. syringae pv. phaseolicola strain LR781 (51). Of the four copies of the full-length IS801, one is present on p1448A-A (PSPPHA0083). One of the three chromosomal copies is located close to the origin of replication, upstream of dnaA, the gene encoding the chromosomal replication protein. The 15 copies of IS801 with disrupted reading frames include 10 identical copies of a shorter version of IS801 (IS801s; stop codon after the first 156 amino acids) which are distributed on the chromosome and p1448A-A. Interestingly, if the stop codon in IS801s (TAA in all cases) is replaced by TTC (the codon for phenylalanine), the reading frame then encodes the full-length IS801. Since IS801s retains the left and right ends of IS801 and is present in multiple copies on plasmid p1448A-A and the chromosome, it is likely that the truncated copies of IS801 are capable of being transposed by the intact versions of IS801. The small plasmid, p1448A-B, does not contain IS801.
Some regions of the genomes in the two pathovars appear to be preferred sites for the insertion of mobile genetic elements (see Table S6 in the supplemental material). For example, unrelated transposon proteins are located adjacent to PSPPH5203 (in 1448A) and PSPTO5595 (encoding glucosamine-fructose-6-phosphate aminotransferase in DC3000), which define one end of a syntenic block near the origins of replication in the two genomes (see below). Similarly, a GTP-binding protein, YchF (PSPTO1101 and PSPPH0988), is adjacent to a site-specific recombinase in DC3000 and a transposase in 1448A. Through close analysis of the plasmids, we have identified a genetic signature sequence of 133 nt that is linked to several effector genes in 1448A and other P. syringae pathovars. The 133-nt sequence was first identified in the 1448A PAI, close to avrPphC and next to the right junction of the avrPphF deletion (see Fig. S2 in the supplemental material). BLAST analysis for short, nearly exact matches showed that this sequence was mosaic in that part of it that forms the right flank of IS801, while the remainder is found within or next to recombinase integrases. Copies of the sequence were found next to hopH1, hopC1, hopAM1-1 (P. syringae pv. tomato DC3000), avrPpiA1 (P. syringae pv. pisi), and avrRpm1 (P. syringae pv. maculicola), and in each case, the sequence was either part of or next to a phage integrase/site-specific recombinase gene. The sequence may therefore form a common target for integration events, allowing acquisition and deletion of effectors.
The large plasmid (p1448A-A) contains a 4.1-kb region that has been duplicated (coordinates 2,726 to 6,858 and 59,496 to 63,628; PSPPHA0004 to -A0009 and PSPPHA0070 to -A0075; 100% identity at the nucleotide level). Genes present in this region include two polygalacturonases (PSPPHA0006 and -A0072) and truncated versions of the type III effector HopW1 (PSPPHA0009 and -A0075). Full-length copies of HopW1 genes were not identified in 1448A or DC3000. Both duplicated regions are associated with mobile genetic elements, suggesting that they may have been acquired by independent horizontal-transfer events. The duplicated region close to the origin of replication is bordered by an IS801 transposase (PSPPHA0003) on one side and type III effectors (PSPPHA0010 and -A0012; HopD1 and HopQ1 and -2, respectively) and transposases (PSPPHA0013 to -A0015) on the other. The second duplicated region is flanked by a truncated resolvase (PSPPHA0076) on one side and a transposase and a stability/partitioning determinant (PSPPHA0068 and -A0069) on the other.
Identification of the core Pseudomonas genome and ORFs unique to P. syringae.
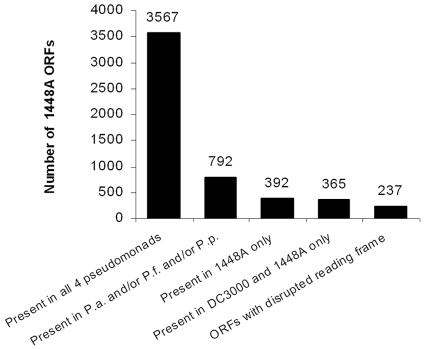
To identify the core Pseudomonas genome and characterize the P. syringae-specific portion of 1448A, the peptide complement of 1448A was compared to the predicted proteomes of the four complete pseudomonad genomes (10, 43, 44a, 63). The results of this comparative analysis are presented in Table S7 in the supplemental material. Using a BLASTP E value cutoff of 10−5, a total of 3,567 ORFs (67%) were found to be present in all four reference species, 365 (7%) were specific to the two P. syringae pathovars and were found in both, and 392 ORFs (7%) were specific to 1448A (Fig. 2).
FIG. 2.
Comparison of the P. syringae pv. phaseolicola 1448A genome with four other finished Pseudomonas genomes (P.a., P. aeruginosa PAO1; P.p., P. putida KT2440; P.f., P. fluorescens Pf-5; DC3000, P. syringae pv. tomato DC3000).
The ORFs that were specific and common to the two P. syringae pathovars included those encoding several predicted TTSS effectors, pectic enzymes, insecticidal toxins, regulatory proteins, and lipoproteins. Several of these genes encode virulence factors, discussed above, that are likely to be of particular importance to plant pathogens. The shared TTSS effectors represent a subset that appears to be broadly conserved among P. syringae pathovars (Vencato et al., unpublished data). The pectic enzymes have the potential to play multiple roles in modifying plant cell walls during pathogenesis, although extensive wall disruption has not been reported. The function of insecticidal toxins, previously identified in DC3000 (10), is unknown, but insects may benefit P. syringae as vectors or compete with the bacteria as herbivores (30). The regulatory proteins that are either specific to P. syringae or unique to 1448A will be particularly interesting to investigate because of their potential roles in regulating virulence-related functions and host specificity. The P. syringae-specific lipoproteins are also interesting, given the prevalence of studies predicting bacterial envelope functions among genes reported to be conserved in plant-associated bacteria but lacking in reference genomes (67).
Because a BLASTP E value cutoff of 10−5 was used in this analysis, the ORFs that are unique to one or both of the strains are unlikely to have domains in common with ORFs in the other pseudomonads, which makes these unique ORFs of particular interest in exploring pathogenesis. However, two caveats must be noted. First, 1448A ORFs that are absent from DC3000 but present in one or more of the other pseudomonads may be potentially involved in fitness functions not directly related to virulence. Thus, some of the ORFs that are unique to 1448A or DC3000 could also be involved in various fitness functions. Second, some of the ORFs that are likely to be important in P. syringae pathogenicity may be members of ubiquitous gene families, such as ABC transporters with predicted specificities for plant-derived sugars and nonribosomal peptide synthases, and therefore not recognized as being unique to one or both strains in this analysis.
Comparative analysis of 1448A and DC3000 based on orthology and synteny.
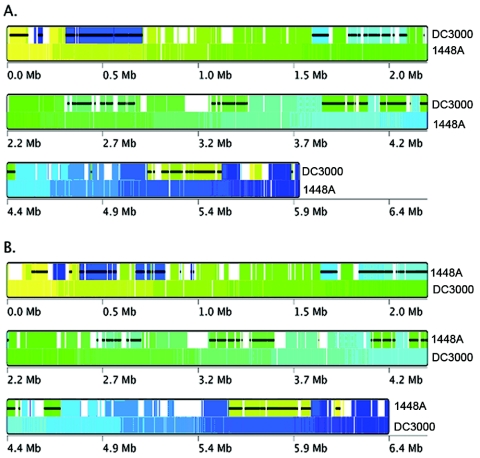
Relatedness between the two pathovars was further analyzed using reciprocal best hits and a more stringent BLASTP cutoff to identify putative orthologs. At the whole-genome level, reciprocal BLASTP analysis (E value < 10−15) of the predicted functional ORFs in the 1448A and DC3000 genomes revealed that 4,133 ORFs (81% of predicted 1448A ORFs and 74% of predicted functional DC3000 ORFs) were orthologous. An additional 290 ORFs in 1448A have putative paralogs in DC3000, whereas 693 ORFs are unique to 1448A. Conversely, 1,018 DC3000 ORFs are absent from 1448A. The genes unique to 1448A predominantly encode proteins of unknown function (381; 55% of unique ORFs), including 198 hypothetical proteins and 122 conserved hypothetical proteins, as well as transposable elements (93; 13%). Complete lists of ORFs conserved between the two pathovars, unique to 1448A, and unique to DC3000 can be found in Tables S1 and S8 in the supplemental material. BLASTP analyses provide a one-dimensional perspective on gene content similarity. However, conservation of gene order and location are also important indications of genome conservation. Using the Sybil syntenic-block algorithm, 3,941 of 5,144 chromosomal ORFs (77%) were found to be syntenic in location between these two pathovars (Fig. 3). Thus, although the 1448A and DC3000 genomes represent divergent clades of the P. syringae group, their genomes are highly conserved.
FIG. 3.
Syntenic relationships between P. syringae pathovars. Regions of collinearity in the chromosomes of the two pathovars were identified using the Sybil syntenic-block algorithm. The syntenic blocks were color coded by their positions in the reference chromosome (bottom track). The reference chromosome is P. syringae pv. phaseolicola 1448A in panel A and P. syringae pv. tomato DC3000 in panel B. The black horizontal lines within the syntenic blocks in the target chromosome (top track) indicate which blocks are inverted with respect to the reference genome. The white blocks indicate regions not shared by the reference chromosome.
Identification of recombination hotspots.
In a previous study, the DC3000 genome was compared with the genomes of the saprophyte P. putida and the animal pathogen P. aeruginosa for the purpose of identifying lineage-specific regions (LSRs) enriched for genes unique to DC3000 (34). The availability of a genome sequence for a second pathovar now provides the opportunity to determine if the DC3000 LSRs point to regions more widely conserved among the P. syringae pathovars yet lacking in other pseudomonads and therefore represent a distinguishing fingerprint for the P. syringae species. Not only would mapping of shared regions aid in the localization of genes responsible for P. syringae-specific phenotypes, but the genetic makeup of these regions could also shed light on the timing of gene acquisition relative to speciation and pathovar differentiation.
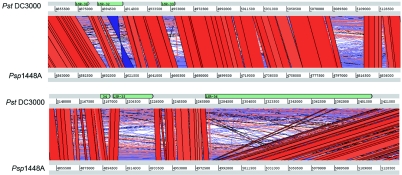
As previously mentioned, the DC3000 and 1448A genomes exhibit extensive regions of conserved sequence and gene order. However, a plot of the predicted DC3000 LSRs relative to the genome alignment reveals that the DC3000 LSR locations do not generally correspond to regions of syntenic conservation between DC3000 and 1448A. In fact, 29/44 DC3000 LSRs exhibit <10% syntenic conservation with the corresponding regions in 1448A. Of the 10/44 LSRs with >70% sequence similarity, only 4 were syntenically conserved between the two pathovars. Representative examples of LSRs with low syntenic conservation are illustrated in Fig. 4.
FIG. 4.
Alignment of the P. syringae pv. tomato DC3000 (Pst DC3000) and P. syringae pv. phaseolicola 1448A (Psp 1448A) genomes in the region encompassing the P. syringae pv. tomato DC3000 LSRs 31 to 36. The red bars indicate colinear regions of similarity, and the blue bars indicate regions of similarity that are inverted in their relative orientations. Locations of LSRs are indicated in green.
The DC3000 LSRs were initially characterized with the goal of identifying novel genes involved in DC3000-specific and/or P. syringae-specific virulence, and indeed, the locations of known virulence genes were found to correlate with their locations. However, with the exception of the main Hrp island (LSR13), the lack of LSR conservation between DC3000 and 1448A suggests that relative position within the genome may be of limited use as a guide to shared, novel virulence factors. This is supported by the observation that locations of previously identified virulence genes common to the two pathovars are not strongly conserved. For example, of the 17 Hop virulence proteins present in both pathovars, only 8 are conserved in their relative positions within the genome, and 1 of these, HopAF1, though conserved in relation to neighboring genes, is inverted in its orientation.
Although previously hypothesized to represent conserved syntenic P. syringae-specific cassettes of genes, the LSRs may, in fact, represent recombination hotspots within the genus Pseudomonas, accounting not only for the differences between DC3000 and species such as P. putida and P. aeruginosa, but also for differences among the P. syringae pathovars. Comparison with the genome sequences of additional Pseudomonas species, including the soon-to-be-completed P. syringae pv. syringae B728a genome, should provide further insight into this hypothesis.
Conclusions.
Analysis of the genome of the bean pathogen P. syringae pv. phaseolicola 1448A revealed a large degree of conservation with other pseudomonads, especially P. syringae pv. tomato DC3000. However, divergence in ORF content was observed and could be the basis for differential host ranges between the pathovars. Numerous differences in the suite of genes involved in virulence, fitness, survival, and synthesis of natural products were found. Coupled with the differential ORFs with unknown functions, these genes provide a resource for future investigations into pathogenicity and host range specificity.
ADDENDUM IN PROOF
An article by Feil et al. (H. Feil et al., Proc. Natl. Acad. Sci. USA 102:11064-11069, 2005) describing the complete genome sequence of P. syringae pv. syringae B728a was recently published.
Supplementary Material
Acknowledgments
This work was supported by funds from the National Science Foundation Plant Genome Research Program DBI-0077622 to A.C., C.R.B., S.C., and A.K.C. J.M. and R.W.J. received support from the United Kingdom Biotechnology and Biological Sciences Research Council.
We are grateful for the assistance of personnel in the J. Craig Venter Science Foundation Joint Technology Center, the TIGR Informatics Department, and the TIGR IT Support Group.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815. [DOI] [PubMed] [Google Scholar]
- 5.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, C. L., F. Alarcon-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya, A., S. Stilwagen, N. Ivanova, M. D'Souza, A. Bernal, A. Lykidis, V. Kapatral, I. Anderson, N. Larsen, T. Los, G. Reznik, E. Selkov, Jr., T. L. Walunas, H. Feil, W. S. Feil, A. Purcell, J. L. Lassez, T. L. Hawkins, R. Haselkorn, R. Overbeek, P. F. Predki, and N. C. Kyrpides. 2002. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. USA 99:12403-12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonas, U., and T. Lahaye. 2002. Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr. Opin. Microbiol. 5:44-50. [DOI] [PubMed] [Google Scholar]
- 10.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Vanaken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete sequence of the tomato and Arabidopsis pathogen, Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, D. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 12.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, J. H., A. K. Goel, S. R. Grant, and J. L. Dangl. 2004. Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr. Opin. Microbiol. 7:11-18. [DOI] [PubMed] [Google Scholar]
- 14.Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu, S. Gombar, S. R. Grant, F. M. Ausubel, and J. L. Dangl. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 16.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 17.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duong, F., E. Bonnet, V. Geli, A. Lazdunski, M. Murgier, and A. Filloux. 2001. The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene 262:147-153. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson, A. R., and J. S. Johnston. 1980. Phaseolotoxin: chlorosis, ornithine accumulation and inhibition of ornithine carbamoyltransferase in different plants. Physiol. Plant Pathol. 16:269-275. [Google Scholar]
- 20.Folkesson, A., A. Advani, S. Sukupolvi, J. D. Pfeifer, S. Normark, and S. Lofdahl. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol. Microbiol. 33:612-622. [DOI] [PubMed] [Google Scholar]
- 21.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N. C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda, H., T. Ogawa, K. Ishihara, T. Fujii, K. Nagahama, T. Omata, Y. Inoue, S. Tanase, and Y. Morino. 1992. Molecular cloning in Escherichia coli, expression, and nucleotide sequence of the gene for the ethylene-forming enzyme of Pseudomonas syringae pv. phaseolicola PK2. Biochem. Biophys. Res. Commun. 188:826-832. [DOI] [PubMed] [Google Scholar]
- 23.Gal, M., G. M. Preston, R. C. Massey, A. J. Spiers, and P. B. Rainey. 2003. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 12:3109-3121. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez, A. I., M. Perez de la Vega, M. L. Ruiz, and C. Polanco. 2003. Analysis of the argK-tox gene cluster in nontoxigenic strains of Pseudomonas syringae pv. phaseolicola. Appl. Environ. Microbiol. 69:4979-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottwald, J. R., P. J. Krysan, J. C. Young, R. F. Evert, and M. R. Sussman. 2000. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97:13979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenberg, J. T., and B. A. Vinatzer. 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6:20-28. [DOI] [PubMed] [Google Scholar]
- 27.Groisman, E. A., M. A. Sturmoski, F. R. Solomon, R. Lin, and H. Ochman. 1993. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc. Natl. Acad. Sci. USA 90:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haft, D. H., J. D. Selengut, and O. White. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haft, D. H., J. D. Selengut, L. M. Brinkac, N. Zafar, and O. White. 2005. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 21:293-306. [DOI] [PubMed] [Google Scholar]
- 30.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotson, A., and M. B. Mudgett. 2004. Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. 7:384-390. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma, D. L. Arnold, M. J. Gibbon, J. Murillo, J. D. Taylor, and A. Vivian. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. USA 96:10875-10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahagirdar, R., and S. P. Howard. 1994. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J. Bacteriol. 176:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joardar, V., M. Lindeberg, D. J. Schneider, A. Collmer, and C. R. Buell. 2005. Lineage specific regions in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 6:53-64. [DOI] [PubMed] [Google Scholar]
- 35.Keen, N. T. 1990. Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24:447-463. [DOI] [PubMed] [Google Scholar]
- 36.Klee, H. J., M. B. Hayford, K. A. Kretzmer, G. F. Barry, and G. M. Kishore. 1991. Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell 3:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann, P. 2002. Structure and evolution of plant disease resistance genes. J. Appl. Genet. 43:403-414. [PubMed] [Google Scholar]
- 38.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 39.Lindgren, P. B., R. C. Peet, and N. J. Panopoulos. 1986. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, G. B., S. H. Brommonschenkel, J. Chunwongse, A. Frary, M. W. Ganal, R. Spivey, T. Wu, E. D. Earle, and S. D. Tanksley. 1993. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432-1436. [DOI] [PubMed] [Google Scholar]
- 41.Mosqueda, G., G. Van den Broeck, O. Saucedo, A. M. Bailey, A. Alvarez-Morales, and L. Herrera-Estrella. 1990. Isolation and characterization of the gene from Pseudomonas syringae pv. phaseolicola encoding the phaseolotoxin-insensitive ornithine carbamoyltransferase. Mol. Gen. Genet. 222:461-466. [DOI] [PubMed] [Google Scholar]
- 42.Myers, E. W., G. G. Sutton, A. L. Delcher, I. M. Dew, D. P. Fasulo, M. J. Flanigan, S. A. Kravitz, C. M. Mobarry, K. H. Reinert, K. A. Remington, E. L. Anson, R. A. Bolanos, H. H. Chou, C. M. Jordan, A. L. Halpern, S. Lonardi, E. M. Beasley, R. C. Brandon, L. Chen, P. J. Dunn, Z. Lai, Y. Liang, D. R. Nusskern, M. Zhan, Q. Zhang, X. Zheng, G. M. Rubin, M. D. Adams, and J. C. Venter. 2000. A whole-genome assembly of Drosophila. Science 287:2196-2204. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 44.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Paulsen, I. T., et al. 2005. Complete genome sequence of the plant commensual Pseudomonas fluorescens pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penaloza-Vazquez, A., S. P. Kidambi, A. M. Chakrabarty, and C. L. Bender. 1997. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179:4464-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 47.Quinones, B., C. J. Pujol, and S. E. Lindow. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant-Microbe Interact. 17:521-531. [DOI] [PubMed] [Google Scholar]
- 48.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivas, L. A., J. Mansfield, G. Tsiamis, R. W. Jackson, and J. Murillo. 2005. Changes in race-specific virulence in Pseudomonas syringae pv. phaseolicola are associated with a chimeric transposable element and rare deletion events in a plasmid-borne pathogenicity island. Appl. Environ. Microbiol. 71:3778-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roine, E., D. N. Nunn, L. Paulin, and M. Romantschuk. 1996. Characterization of genes required for pilus expression in Pseudomonas syringae pathovar phaseolicola. J. Bacteriol. 178:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romantschuk, M., G. Y. Richter, P. Mukhopadhyay, and D. Mills. 1991. IS801, an insertion sequence element isolated from Pseudomonas syringae pathovar phaseolicola. Mol. Microbiol. 5:617-622. [DOI] [PubMed] [Google Scholar]
- 52.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 53.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawada, H., F. Suzuki, I. Matsuda, and N. Saitou. 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49:627-644. [DOI] [PubMed] [Google Scholar]
- 55.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sesma, A., G. W. Sundin, and J. Murillo. 2000. Phylogeny of the replication regions of pPT23A-like plasmids from Pseudomonas syringae. Microbiology 146:2375-2384. [DOI] [PubMed] [Google Scholar]
- 57.Shan, L., H. S. Oh, J. Chen, M. Guo, J. Zhou, J. R. Alfano, A. Collmer, X. Jia, and X. Tang. 2004. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant-Microbe Interact. 17:447-455. [DOI] [PubMed] [Google Scholar]
- 58.Shao, F., P. M. Merritt, Z. Bao, R. W. Innes, and J. E. Dixon. 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109:575-588. [DOI] [PubMed] [Google Scholar]
- 59.Solomon, P. S., and R. P. Oliver. 2001. The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum. Planta 213:241-249. [DOI] [PubMed] [Google Scholar]
- 60.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 61.Stavrinides, J., and D. S. Guttman. 2004. Nucleotide sequence and evolution of the five-plasmid complement of the phytopathogen Pseudomonas syringae pv. maculicola ES4326. J. Bacteriol. 186:5101-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens, C., M. A. Bennett, E. Athanassopoulos, G. Tsiamis, J. D. Taylor, and J. W. Mansfield. 1998. Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol. Microbiol. 29:165-177. [DOI] [PubMed] [Google Scholar]
- 63.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 64.Taylor, J. D., D. M. Teverson, D. J. Allen, and M. A. Pastor-Corrales. 1996. Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathol. 45:469-478. [Google Scholar]
- 65.Teverson, D. M. 1991. Genetics of pathogenicity and resistance in the halo-blight disease of beans in Africa. Ph.D. thesis. University of Birmingham, Birmingham, United Kingdom.
- 66.Tsiamis, G., J. W. Mansfield, R. Hockenhull, R. W. Jackson, A. Sesma, E. Athanassopoulos, M. A. Bennett, C. Stevens, A. Vivian, J. D. Taylor, and J. Murillo. 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19:3204-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Sluys, M. A., C. B. Monteiro-Vitorello, L. E. Camargo, C. F. Menck, A. C. Da Silva, J. A. Ferro, M. C. Oliveira, J. C. Setubal, J. P. Kitajima, and A. J. Simpson. 2002. Comparative genomic analysis of plant-associated bacteria. Annu. Rev. Phytopathol. 40:169-189. [DOI] [PubMed] [Google Scholar]
- 68.Weingart, H., B. Volksch, and M. S. Ullrich. 1999. Comparison of ethylene production by Pseudomonas syringae and Ralstonia solanacearum. Phytopathology 89:360-365. [DOI] [PubMed] [Google Scholar]
- 69.Whalen, M. C., R. W. Innes, A. F. Bent, and B. J. Staskawicz. 1991. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willis, D. K., and T. G. Kinscherf. 2004. Global regulation in Pseudomonas syringae, p. 223-238. In J. L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic, New York, N.Y. [Google Scholar]
- 71.Zhao, Y., Z. Ma, and G. W. Sundin. 2005. Comparative genomic analysis of the pPT23A plasmid family of Pseudomonas syringae. J. Bacteriol. 187:2113-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.