Abstract
Escherichia coli requires nickel under anaerobic growth conditions for the synthesis of catalytically active NiFe hydrogenases. Transcription of the NikABCDE nickel transporter, which is required for NiFe hydrogenase synthesis, was previously shown to be upregulated by FNR (fumarate-nit rate regulator) in the absence of oxygen and repressed by the NikR repressor in the presence of high extracellular nickel levels. We present here a detailed analysis of nikABCDE transcriptional regulation and show that it closely correlates with hydrogenase expression levels. We identify a nitrate-dependent mechanism for nikABCDE repression that is linked to the NarLX two-component system. NikR is functional under all nickel conditions tested, but its activity is modulated by the total nickel concentration present as well as by one or more components of the hydrogenase assembly pathway. Unexpectedly, NikR function is independent of NikABCDE function, suggesting that NikABCDE is a hydrogenase-specific nickel transporter, consistent with its original identification as a hydrogenase (hyd) mutant. Further, the results suggest that the hydrogenase assembly pathway is sequestered within the cell. A second nickel import pathway in E. coli is implicated in NikR function.
Several energetically difficult reactions, such as nitrogen or carbon fixation, are catalyzed by enzymes with complex metal cofactors (26). A striking feature of the synthesis of these enzymes is the requirement of intricate assembly pathways that utilize several protein cofactors to ensure the fidelity of catalytic-site assembly (18). Metalloenzyme expression levels can be tightly regulated in response to changes in environmental conditions; for example, the nitrogenase operon is induced by nitrogen availability but repressed in the presence of oxygen (14). This shifting metabolism, combined with the biosynthetic cost of making these enzymes, means that the transcriptional regulation of these pathways is both necessary and complex. Cells are unlikely to synthesize large quantities of apoenzyme in the absence of the required cofactor(s), just as they are unlikely to expend the energy necessary to synthesize the transporter and accessory proteins necessary for cofactor assembly when the apoenzyme is not being expressed.
Escherichia coli exhibits a complex transcriptional response to growth conditions at low oxygen tensions (38). Respiration still occurs, but at lower energetic yield, and it requires the presence of an alternative electron acceptor, such as nitrate, dimethyl sulfoxide (DMSO), trimethylamine oxide (TMAO), or fumarate, and a corresponding terminal reductase (2, 16, 32, 42). E. coli can also ferment carbon sources in the absence of a suitable electron acceptor. E. coli expresses NiFe hydrogenases under anaerobic growth conditions (1, 5, 27, 29, 38) when energetic yields are low, for example, during fermentation or with low-energy-yield electron acceptors such as fumarate. Hydrogenases 1 and 2 (expressed by hya and hyb, respectively) oxidize H2 in the presence of fumarate to generate ATP. Hydrogenase 3 (hyc) is part of a complex with formate dehydrogenase that converts formate to CO2 and H2. The expression of a fourth hydrogenase (hyf) has been observed only under synthetic conditions (30). These hydrogenases require nickel, iron, and organic ligands for catalytic activity (39), and several accessory proteins control the ordered delivery of these cofactors to the active site (3). Nickel is the last cofactor to be inserted into the active site.
The NikABCDE transporter is synthesized under anaerobic conditions to meet the increased demand for nickel resulting from hydrogenase synthesis (24, 43-45). Regulation of nikABCDE expression is positively controlled by FNR (44) and negatively controlled by NikR (9), in both cases by direct protein binding to the nikABCDE promoter (PnikABCDE or Pnik). This arrangement provides two distinct inputs that control nickel uptake. A decrease in oxygen tension results in activation of FNR and upregulation of nikABCDE expression, while the presence of excess nickel activates NikR, which overrides the action of FNR and results in repression of nikABCDE transcription. NikR forms two distinct DNA complexes in vitro in response to different nickel concentrations (4, 8, 9), suggesting that two modes of NikR-dependent repression of Pnik expression might be observed in vivo. The pattern of Pnik regulation determined thus far has indicated that transcription of nikABCDE is simple and is not tightly linked to the regulation of hydrogenase expression, raising the possibility that NikABCDE levels could be unnecessarily high under conditions of known low hydrogenase expression (i.e., growth in nitrate) and/or low extracellular nickel concentrations (inactive NikR). Further, NikR exhibits picomolar affinity for nickel ions (4, 8), raising the question of whether hydrogenase assembly must compete for available nickel ions with such a rapacious intracellular competitor.
Here, we show that NikABCDE levels are under complex transcriptional control, which results in an expression pattern that is closely linked to hydrogenase expression levels. Both NikR-DNA complexes act to repress expression from Pnik, although the activity of the first NikR-DNA complex is observed only under conditions of low hydrogenase expression. Additionally, nitrate represses Pnik expression via the NarLX two-component system. Surprisingly, NikR function does not depend on nickel transport by NikABCDE, suggesting the presence of another nickel transporter in E. coli. A model is presented for hydrogenase assembly, showing its isolation from the rest of the intracellular milieu to the extent that it can be considered to constitute a discrete module or circuit within the cell.
MATERIALS AND METHODS
Strains and plasmids.
E. coli strains used in these experiments are listed in Table 1. Gene deletions for nikR (bases 10 to 399), nikABCDER (bases 16 to 1,560), and corA (bases 61 to 891) were constructed using the method of Datsenko and Wanner (12). Numbering refers to base positions relative to ATG.
TABLE 1.
E. coli strains used in these experiments
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| RZ4500a | MG1655 ΔlacZ | 10 | |
| BW25113b | lacIqrrnBT14 lacZWJ16 hsdR514 araBADAH33 rhaBADLD78 | 12 | |
| BW27873b | BW25113 | narP1312 | 46 |
| BW30008b | BW25113 | narQ1314 | 46 |
| BW27864b | BW25113 | narLX1316 | 46 |
| PC269 | RZ4500 | nikR | This work |
| PC379 | RZ4500 | nikABCDE nikR | This work |
| PC350 | BW25113 | nikR | This work |
| PC420 | BW27864 | nikR narLX1316 | This work |
| PC477 | PC269 | corA | This work |
Source: Patricia J. Kiley, University of Wisconsin—Madison.
Source: E. coli Genetic Stock Center at Yale University (http://cgsc.biology.yale.edu).
A translational Pnik-lacZ fusion was constructed in three steps. A Pnik fragment (∼400 bp) was amplified by PCR from E. coli genomic DNA using oligonucleotides PC118 (5′-CTATGGCCGGCCGGGCAAACCTGCATTTGCGCCGG-3′) and PC638-GC (5′- CCAGTGAATCCGTAATCATGGTCATGGTAACCCCAATGGATTAAAATAGA-3′). lacZ was amplified by PC638 (5′-TCTATTTTAATCCATTGGGGTTACCATGACCATGATTACGGATTCACTGG-3′) and PC639 (5′- AGCTCATTCTAGATTATTTTTGACACCAGACCAACTGG-3′). The underlined bases correspond to EagI and XbaI restriction sites, respectively. The two resulting fragments were purified and combined in a second PCR mixture that contained PC118 and PC639. The resulting 3,633-bp fragment was digested with EagI and XbaI and ligated into pACYC184, cut with the same enzymes to create pPC181. This plasmid is a precise fusion between PnikA and the ATG codon of lacZ. It differs from the previously described pPC163 (9), which contains five codons of the 5′ end of the nikA gene fused to the 5′ end of lacZ. pPC181 has at-least-fourfold-greater expression than pPC163 under identical conditions (data not shown), suggesting that the nikA codons negatively affect the production of LacZ.
Growth media.
M63 salts (5×) were treated overnight with (2 g/liter) Chelex-100 resin (Sigma) to remove trace metals, including nickel. Trace metal nutrients, excluding nickel, were then added back at the following concentrations: 1 mM MgCl2, 100 nM MnCl2, 2 μM FeCl2, 1 μM ZnCl2, 100 nM (NH4)2MoO4, and 100 nM NaSeO3. Nutrient concentrations were individually optimized for maximal growth by measuring optical density at 600 nm (OD600) values after overnight growth (14 to 16 h) at 37°C under anaerobic conditions in capped microcentrifuge tubes with no headspace. Glucose (0.25%) was used as a carbon source and potassium nitrate (KNO3), sodium formate, sodium fumarate, DMSO, or TMAO was added when required, each at a final concentration of 10 mM, except when noted otherwise. Nickel was added by making serial 10-fold dilutions into minimal growth media from a 1 mM stock. Concentrations higher than 10 μM NiCl2 were toxic, as judged by a 20 to 30% decrease in the OD600 after 14 to 16 h of growth at 37°C.
β-Galactosidase assays.
Strains containing pPC181, the Pnik-lacZ fusion, and pNIK103 (9), which provides a low level of NikR expression in the absence of any inducer, were inoculated in the defined media to a starting OD600 of 0.0001 and grown 14 to 16 h at 37°C in capped microcentrifuge tubes with no headspace. For experiments examining the nickel-dependent effects of a nikR deletion, strains lacking chromosomal nikR were transformed with pNIK103 Cys95Ala (7), which produces a stable variant of NikR with a mutation in the high-affinity nickel binding site. For every experiment, two separate aliquots (100 μl) of cells were extracted to measure OD600 and LacZ activity. OD600 values ranged from 0.3 to 0.8, and LacZ activity was constant over this range for a given growth condition. The LacZ activities of cultures grown in microcentrifuge tubes with no headspace were similar to the activities measured in 2-ml cultures grown in 15-ml polyethylene tubes in an anaerobic chamber (data not shown). Data were collected in duplicate or triplicate from separate overnight cultures started from the same inoculum. Error bars indicate standard errors between these measurements. In all experiments, the relative LacZ activity was normalized to the level measured for E. coli RZ4500 grown in media containing glucose alone (3,500 to 4,000 Miller units). For example, a relative LacZ activity of 0.25 is equivalent to ∼1,000 Miller units. Nickel titration data for each growth condition and/or strain were fit to the equation y = {(a − b)/[1 + (KNi/x)n]} where a is the fraction of LacZ activity at a low nickel concentration (i.e., upper baseline), b is the fraction of LacZ activity at a high nickel concentration (i.e., lower baseline), KNi is the nickel concentration required for half-maximum LacZ activity, and n is a cooperativity term required to fit the data set from growth in media containing formate (Fig. 1B).
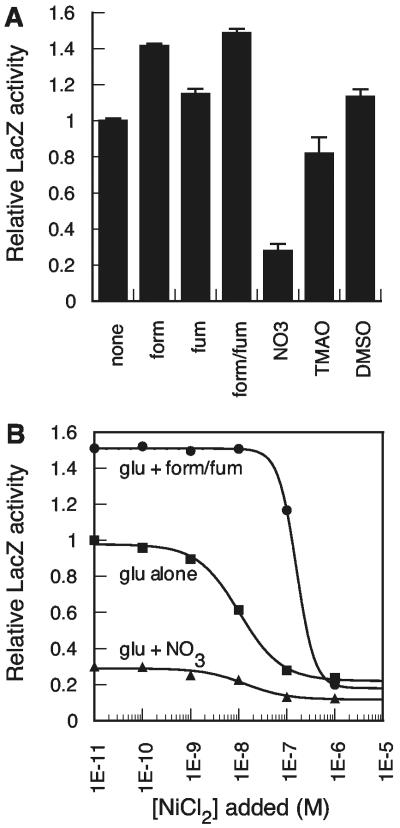
FIG. 1.
Anaerobic Pnik-lacZ expression under varied medium conditions. (A) E. coli RZ4500 containing pPC181 and pNIK103 grown in 0.25% glucose with different electron acceptors. (B) Nickel-dependent repression of Pnik-lacZ expression in E. coli RZ4500. Squares, glucose alone; circles, glucose plus 10 mM formate-10 mM fumarate; triangles, glucose plus 10 mM nitrate. KNi values were 10.3 nM for glucose alone, 13.6 nM for nitrate, and 158.5 nM for formate (n = 2.3). Form, formate; fum, fumarate; form/fum, formate and fumarate; glu, glucose.
RESULTS
Hydrogenase expression in E. coli is regulated by the particular electron acceptors present in the growth medium under anaerobic growth conditions, as well as by formate, which is a product of pyruvate-formate lyase. Hydrogenase activity is low in the presence of nitrate and high in the presence of fumarate and/or formate (1, 5, 29). Changes in hydrogenase expression levels should correlate with a changing requirement for nickel and a corresponding change in NikABCDE levels. E. coli RZ4500 organisms containing a low-copy-number Pnik-lacZ fusion were grown anaerobically in M63 minimal medium to which glucose, an electron acceptor (nitrate, DMSO, TMAO, or fumarate), and/or formate was added. Changes in LacZ activity were compared to the LacZ activity of cells grown in medium containing glucose alone (Fig. 1A). Nitrate repressed Pnik expression by 70%. TMAO resulted in slight repression of PnikABCDE expression, while fumarate and DMSO resulted in slight enhancement of expression. Formate enhanced Pnik expression by 40%. Thus, NikABCDE expression levels correlate with previously observed changes in hydrogenase activity in different media.
The nickel dependence of Pnik expression has previously been examined in LB medium in the absence or presence of 250 μM or higher nickel ion concentrations (9, 43), but a nickel titration has not been carried out under any growth condition. Additionally, the relationship between Pnik expression levels (Fig. 1A) and nickel-dependent repression of expression has not been examined. Pnik-lacZ expression was measured over a range of nickel concentrations in a subset of the growth conditions from Fig. 1A. In all cases, a monophasic decrease in LacZ activity was observed with increasing nickel concentration (Fig. 1B). Interestingly, the KNi value increased substantially in the presence of formate, from 10 nM to 158 nM, and nickel-dependent repression in the presence of formate was more cooperative (n = 2.3) than growth conditions with lower hydrogenase expression (n = 1). These results suggest that the nickel-dependent regulation of Pnik is strongly correlated with the synthesis of active hydrogenase at limiting nickel concentrations and raise the question of whether NikR activity is altered under these conditions as a result of competition for available nickel ions. The absence of a biphasic repression curve suggested that in vivo roles for the distinct Ni-NikR DNA complexes observed in vitro as a function of nickel (4, 8, 9) are not observable under these conditions.
Levels of LacZ expression in LB medium were qualitatively similar (data not shown). However, the maximum LacZ activity was at least twofold lower and the repression curve was shifted to ∼103-fold higher (KNi ≥ 10 μM). Components of rich medium likely influence hydrogenase expression in anaerobically growing E. coli as well as restrict nickel availability (44). The addition of 0.1% peptone to M63 minimal medium resulted in a 2.5-fold decrease in Pnik-lacZ expression but did not affect KNi compared to that after growth in M63 (data not shown). We also observed a twofold pH-dependent difference in Pnik-lacZ expression when M63 medium was buffered to below pH 6.2 (normal pH is 7.5). Similarly, buffered LB medium (100 mM morpholinepropanesulfonic acid, pH 7.2) had 1.6-fold-higher Pnik-lacZ expression than unbuffered LB medium, but this level was still lower than the expression in M63 medium. E. coli organisms acidify LB medium under anaerobic conditions as a function of increasing growth, which influences hydrogenase expression (17) and therefore the nickel requirement of the cell.
Nitrate-dependent regulation of PnikABCDE expression.
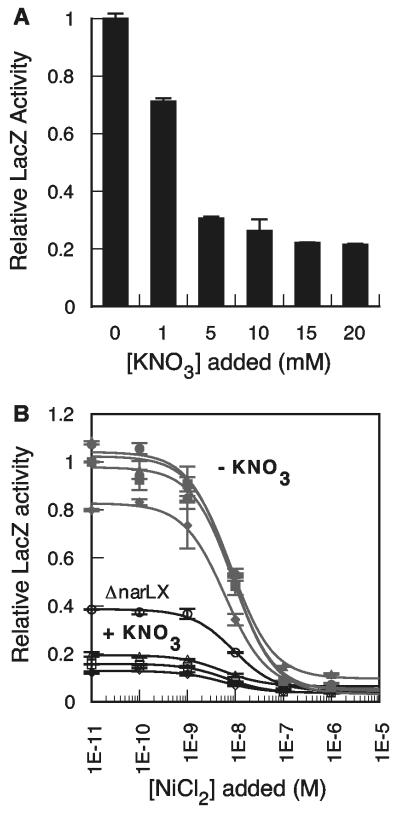
The significant decrease in the basal levels of Pnik-lacZ expression in the presence of nitrate (Fig. 1) suggested a previously unidentified mechanism for the transcriptional regulation of NikABCDE. E. coli responds to a range of extracellular nitrate concentrations via the NarLX and NarPQ two-component systems (33). Nitrate-dependent repression of Pnik-lacZ expression was observed at nitrate concentrations ≥1 mM (Fig. 2A), which is inversely correlated with the NarLX-dependent positive regulation of nitrate reductase (narG) expression (40). To identify whether Pnik regulation was NarLX dependent, Pnik-lacZ expression was assayed in the absence or presence of 15 mM nitrate in mutant strains deleted of the narLX, narP, or narQ gene. Only narLX-deficient E. coli showed a partial loss of repression of Pnik-lacZ, compared to the expression of the narP-deficient, narQ- and narP-deficient, narQ- and narQ-deficient, or parent strain at low nickel concentrations when strains were grown in nitrate-containing media (Fig. 2B). Repression of Pnik was unaffected at high nickel concentrations. Little effect on LacZ activity was seen for the nar mutant strains in the absence of nitrate (Fig. 2B). Thus, the NarLX system, which negatively regulates hydrogenase expression (27, 34), has the same effect on Pnik expression.
FIG. 2.
Anaerobic nitrate-dependent regulation of Pnik-lacZ expression. (A) Pnik-lacZ expression as a function of KNO3 concentration. Nitrate concentrations above 20 mM resulted in decreased OD600 readings. (B) Pnik-lacZ expression in wild-type and nar-deficient strains in glucose (filled gray symbols) or in glucose-15 mM KNO3 (open symbols). Squares, wild type (RZ4500 or BW25113); circles, ΔnarLX (BW27864); triangles, ΔnarP (BW27873); diamonds, ΔnarQ (BW30008). Each strain contained pPC181 and pNIK103. KNi values were between 5.4 and 9.2 nM.
Nitrate (NarLX) and nickel (NikR) independently regulate PnikABCDE expression.
Deletion of the narLX genes did not result in complete restoration of Pnik expression in nitrate-containing growth media. Additionally, the nickel-dependent repression at high nickel concentrations was NarLX independent, raising the question of whether NikR- and NarLX-dependent repression rely on the same operator site in Pnik. NarL is a response regulator that, when phosphorylated, has increased affinity for DNA (22), suggesting a direct mechanism for nitrate-dependent regulation of Pnik. To determine whether NarL- and NikR-dependent regulation of Pnik expression were independent, LacZ levels were measured in nikR-deficient and nikR narLX-deficient strains containing wild-type NikR or a mutant protein, the Cys95Ala protein, which lacks high-affinity nickel-binding activity and shows no DNA binding in vitro with up to 1 μM NiCl2 (7). Cells containing Cys95Ala NikR showed increased LacZ activity with all nickel concentrations (Fig. 3). Cells lacking both NarLX and functional NikR showed constant high levels of Pnik-lacZ, suggesting that NarL and NikR are sufficient to account for the Pnik repression observed under the conditions tested here (Fig. 3). The difference in Pnik-lacZ repression at low nickel concentrations in the narLX-deficient strain indicates that NikR likely binds at a site distinct from that required for NarL-dependent repression. Additionally, mutations in the NikR operator that diminish NikR binding (9) retained nitrate-dependent repression of the Pnik promoter (data not shown). Recent bioinformatics approaches to identify transcription factor binding sites in E. coli have not predicted a NarL-binding site in the region of the nik promoter (6, 19, 23); however, these studies have also not predicted the FNR-binding site (TTGAT-N4-AACAG versus consensus TTGAC-N4-ATCAA) in the nik promoter (24, 44).
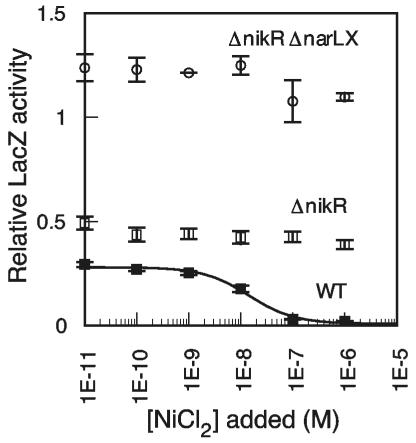
FIG. 3.
NikR and NarLX act independently to repress Pnik-lacZ. Filled squares, wild type (BW25113 plus pNIK103); open squares, ΔnikR (PC269 plus pNIK103 C95A NikR); and open circles, ΔnikR ΔnarLX (PC420 plus pNIK103 C95A NikR). All strains contained pPC181. Strains were grown in minimal medium with 15 mM KNO3. The KNi for the wild-type sample was 14.3 nM.
These data reveal a role for the high-affinity nickel-binding site in NikR function at low total nickel concentrations. They also suggest that NikR is active when nitrate is present in the growth media but that its function is somehow inhibited under conditions that favor the expression of hydrogenase isozymes and their corresponding assembly proteins (Fig. 2B), such as the presence of formate in the growth medium.
NikABCDE is not required for NikR function.
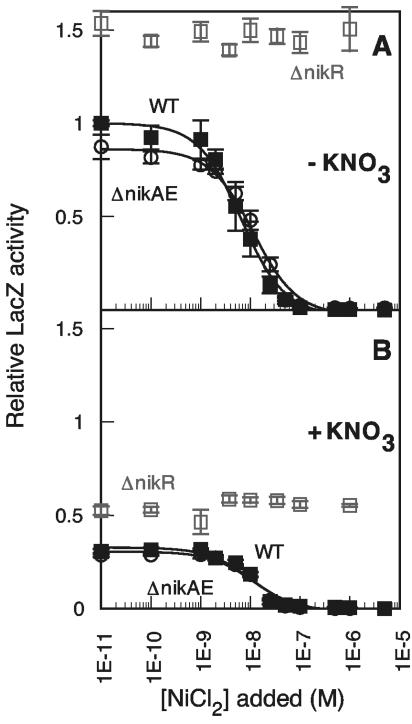
An obvious mechanism for inhibition of NikR function is the exclusion of nickel from inside the cell. Deletion of the nikABCDE operon should dramatically restrict the amount of intracellular nickel available for nickel-binding proteins, including NikR. The effect of the nikABCDE deletion should mimic the effect of the Cys95Ala high-affinity nickel-binding mutant of NikR on Pnik-lacZ activity by reducing the amount of functional NikR in the cell. Deletion of nikABCDE (Fig. 4) or nikA alone (data not shown) had no effect on the nickel-dependent repression of Pnik-lacZ expression, leading to the surprising conclusion that NikR repression is independent of nickel transport by NikABCDE. The nickel- and NikR-dependent repression pattern was not inhibited by the high concentrations of magnesium (4 mM) known to block nickel import by CorA (31), and deletion of corA had no effect on the nickel-dependent repression curves observed here (data not shown). These data suggest the presence of another nickel import pathway in E. coli.
FIG. 4.
NikABCDE-independent regulation of PnikABCDE expression by NikR. (A) Filled squares, Pnik-lacZ expression in glucose for the wild type (WT) (RZ4500 plus pNIK103); open squares, ΔnikR (PC269 plus pNIK103 C95A NikR); and open circles, ΔnikABCDE (PC379 plus pNIK103). (B) Filled squares, Pnik-lacZ expression in glucose plus 15 mM KNO3 for the wild type (RZ4500 plus pNIK103); open squares, ΔnikR (PC269 plus pNIK103 C95A NikR); and open circles, ΔnikABCDE (PC379 plus pNIK103). KNi values were 6.6 nM for the wild type (without KNO3), 11.3 nM for the ΔnikABCDE strain (without KNO3), 10.3 nM for the wild type (with KNO3), and 11.7 nM for the ΔnikABCDE strain (with KNO3).
DISCUSSION
A complex and hierarchical set of inputs controls gene expression in anaerobically growing E. coli cells (38). This intricate regulation results in the synthesis of an enzyme complement that produces the highest energy yield under a given growth condition. E. coli NiFe hydrogenases are upregulated under fermentative growth conditions or in the presence of a low-energy-yield electron acceptor, such as fumarate. Here, we have shown that transcription of the NikABCDE nickel transporter, which is essential for hydrogenase activity (24, 44, 45), is regulated by several distinct mechanisms in order to match the hydrogenase expression level of the cell.
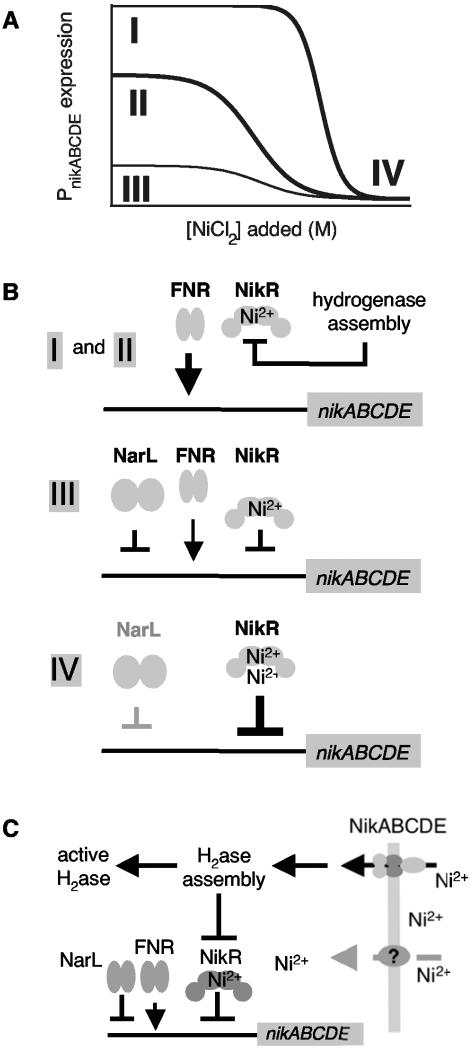
The regulation of NikABCDE synthesis under different growth conditions is summarized in Fig. 5A and B with curves and diagrams labeled I to IV. FNR upregulates NikABCDE (I and II) in the absence of oxygen. In the presence of nitrate (II), NikABCDE synthesis is repressed by both the NarLX two-component system and NikR. Hydrogenase expression is repressed in the presence of nitrate in favor of the synthesis of nitrate reductase (38), which catalyzes the reduction of nitrate as the terminal electron transfer step in the absence of oxygen. NikR further represses NikABCDE expression at higher nickel concentrations (IV), providing nearly complete repression under these conditions. NarLX-dependent repression is absent when nitrate is not present in the growth medium, while NikR-dependent repression depends on hydrogenase expression levels (I and II). The loss of NikR-dependent repression does not appear to be due to a loss of nickel-containing NikR. Rather, NikR function seems to be inhibited by components of the hydrogenase assembly pathway. In particular, the levels of these components are increased in the presence of formate, which induces expression of the hyd-3 operon (28).
FIG. 5.
Model for nikABCDE transcriptional regulation. (A and B) Transcriptional regulation of PnikABCDE expression as a function of nickel concentration under different anaerobic growth conditions. There are three general states of promoter occupancy. At low nickel concentrations in the absence of nitrate (curves and diagrams I and II), FNR upregulates transcription from PnikABCDE and some component of the hydrogenase assembly pathway blocks NikR function. The degree of NikR inhibition depends upon the level of hydrogenase expression. In the presence of nitrate at low nickel concentrations (III), NarL and NikR strongly repress transcription. At high nickel concentrations (IV), NikR is the dominant regulator of PnikABCDE expression regardless of the presence of nitrate (i.e., NarL). (C) Model of the coordination of hydrogenase assembly and NikABCDE-dependent nickel transport in anaerobically growing E. coli based on the data presented here. H2ase, hydrogenase.
There are at least four proteins that either positively or negatively control NikABCDE expression. The activities of these proteins are controlled in some manner by a small molecule: FNR (positive) is inactivated by O2, NarL (negative) is phosphorylated by NarX in the presence of NO3−, NikR (negative) is activated by Ni2+, and NikR function is inhibited by one or more formate-inducible hydrogenase assembly components. This multilayered regulation provides a way for NikABCDE-dependent nickel uptake to be tuned to the hydrogenase requirements of the cell as well as to the external nickel concentration (Fig. 5).
Surprisingly, NikR function does not depend on nickel import by NikABCDE at any nickel concentration or under any growth condition tested here. The high affinity of NikR for metal ions (8, 41) suggests different possibilities for the activation of NikR in the absence of added nickel. Either nickel ions enter the cell by some previously unidentified pathway or NikR is activated in vivo by a different transition metal. The growth medium used in these experiments was treated to remove nickel, but the addition of other metal supplements after this treatment would have resulted in some very low level of nickel being added back to the medium. It is not possible, based on the data presented here, to differentiate between the two NikR activation mechanisms described above. However, in the complete absence of any added nickel, Pnik-lacZ expression was 10 to 15% higher than with 10 pM nickel, suggesting that E. coli cells can sense even very low extracellular nickel concentrations. At higher nickel concentrations, NikR repression exhibits a consistent dependence on the added nickel concentration. This observation provides strong evidence for a second pathway for nickel import into E. coli that is independent of the previously identified NikABCDE and CorA routes for nickel import.
The tight nickel-binding affinity exhibited by NikR in vitro poses a paradox with regard to intracellular metal trafficking. Because NikR can bind nickel at a concentration (<5 pM) well below that corresponding to a single nickel ion inside the cell (∼1 nM), there might be competition for nickel ions between functional and regulatory pathways. The data presented here suggest that there is no such competition. Instead, the hydrogenase assembly pathway sequesters nickel ions within the cell beginning with the NikABCDE transporter, and another nickel transporter is required for NikR function and likely establishes a second “pool” of nickel ions within the cell. Previously, it was shown that the presence of a functional NikABCDE system is required for the correct insertion of nickel into hydrogenase 3 in the face of competition from high extracellular concentrations of Zn2+ (21), suggesting a tight link between nickel transport and hydrogenase assembly. The original identification and designation of nikABCDE as hydC (43) may have presaged its apparently specific function. The model presented here suggests a nickel-independent mechanism for “competition” between functional and regulatory pathways, in which hydrogenase assembly components block NikR function when hydrogenase synthesis is high and nickel concentrations are limiting (Fig. 5). The hydrogenase assembly pathway is complex (3), and it is likely that more than one component affects NikR function (J. L. Rowe and P. T. Chivers, unpublished results).
E. coli likely requires a second nickel uptake pathway not linked to hydrogenase expression. Glyoxalase I (GlxI), has maximal activity in vitro in the presence of nickel ions (11, 13, 15, 35). This enzyme, which detoxifies methylglyoxal produced from dihydroxyacetone phosphate, is expressed under aerobic growth conditions (20) and thus requires an independent source of nickel for its activity. This source of nickel may also be important for NikR function.
A number of microbial genomes carry a nikR ortholog. However, the nickel requirements of these microbes will be different based on the differing complements of nickel enzymes encoded by their genomes. For example, Helicobacter pylori has a large requirement for nickel to support both urease and hydrogenase activity, both of which are essential for the colonization of the stomach (25, 37). Methanogens have an absolute requirement for nickel in three enzymes involved in their central carbon metabolism (36). This diversity of nickel-related physiology raises interesting questions about the biochemical properties of different species of NikR, their corresponding biological roles, and differences in the regulatory inputs controlling intracellular nickel homeostasis in these different microbes. The nitrate-dependent regulation of nickel uptake observed in E. coli will not be a common feature of all microbes, because many NikR-encoding microbes lack both the ability to respire nitrate and a nitrate-responsive two-component system. Further, in microbes that have more than one high-abundance nickel enzyme or nickel enzyme assembly pathway, such as H. pylori or methanogenic archaea, nickel must be trafficked to multiple sites before NikR regulation of uptake can be allowed to occur. Differences in the numbers of nickel transporters and their structures may also influence how each microbe responds to nickel. Thus, the results from E. coli provide a conceptual model of how NikR might function in a microbial cell but illustrate that its function is not solely governed by its in vitro ligand-binding properties.
Acknowledgments
We thank members of the Chivers lab for helpful discussions and comments on the manuscript.
We acknowledge the Washington University School of Medicine for financial support.
REFERENCES
- 1.Ballantine, S. P., and D. H. Boxer. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 163:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco, F., C. Iobbi, G. Giordano, M. Chippaux, and V. Bonnefoy. 1989. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol. Gen. Genet. 218:249-256. [DOI] [PubMed] [Google Scholar]
- 3.Blokesch, M., A. Paschos, E. Theodoratou, A. Bauer, M. Hube, S. Huth, and A. Bock. 2002. Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30:674-680. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, S. L., and D. B. Zamble. 2004. Metal-selective DNA-binding response of Escherichia coli NikR. Biochemistry 43:10029-10038. [DOI] [PubMed] [Google Scholar]
- 5.Brøndsted, L., and T. Atlung. 1994. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J. Bacteriol. 176:5423-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulyk, M. L., A. M. McGuire, N. Masuda, and G. M. Church. 2004. A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli. Genome Res. 14:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington, P. E., P. T. Chivers, F. Al-Mjeni, R. T. Sauer, and M. J. Maroney. 2003. Nickel coordination is regulated by the DNA-bound state of NikR. Nat. Struct. Biol. 10:126-130. [DOI] [PubMed] [Google Scholar]
- 8.Chivers, P. T., and R. T. Sauer. 2002. NikR repressor: high-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chem. Biol. 9:1141-1148. [DOI] [PubMed] [Google Scholar]
- 9.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 10.Choe, M., and W. S. Reznikoff. 1991. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J. Bacteriol. 173:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clugston, S. L., J. F. Barnard, R. Kinach, D. Miedema, R. Ruman, E. Daub, and J. F. Honek. 1998. Overproduction and characterization of a dimeric nonzinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions. Biochemistry 37:8754-8763. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K., and B. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, G., S. L. Clugston, J. F. Honek, and M. J. Maroney. 2000. XAS investigation of the nickel active site structure in Escherichia coli glyoxalase I. Inorg. Chem. 39:2962-2963. [DOI] [PubMed] [Google Scholar]
- 14.Halbleib, C. M., and P. W. Ludden. 2000. Regulation of biological nitrogen fixation. J. Nutr. 130:1081-1084. [DOI] [PubMed] [Google Scholar]
- 15.He, M. M., S. L. Clugston, J. F. Honek, and B. W. Matthews. 2000. Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry 39:8719-8727. [DOI] [PubMed] [Google Scholar]
- 16.Iobbi-Nivol, C., J. Pommier, J. Simala-Grant, V. Mejean, and G. Giordano. 1996. High substrate specificity and induction characteristics of trimethylamine-N-oxide reductase of Escherichia coli. Biochim. Biophys. Acta 1294:77-82. [DOI] [PubMed] [Google Scholar]
- 17.King, P. W., and A. E. Przybyla. 1999. Response of hya expression to external pH in Escherichia coli. J. Bacteriol. 181:5250-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchar, J., and R. P. Hausinger. 2004. Biosynthesis of metal sites. Chem. Rev. 104:509-525. [DOI] [PubMed] [Google Scholar]
- 19.Li, H., V. Rhodius, C. Gross, and E. D. Siggia. 2002. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc. Natl. Acad. Sci. USA 99:11772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean, M. J., L. S. Ness, G. P. Ferguson, and I. R. Booth. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563-571. [DOI] [PubMed] [Google Scholar]
- 21.Magalon, A., M. Blokesch, E. Zehelein, and A. Bock. 2001. Fidelity of metal insertion into hydrogenases. FEBS Lett. 499:73-76. [DOI] [PubMed] [Google Scholar]
- 22.Maris, A. E., M. R. Sawaya, M. Kaczor-Grzeskowiak, M. R. Jarvis, S. M. Bearson, M. L. Kopka, I. Schroder, R. P. Gunsalus, and R. E. Dickerson. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771-778. [DOI] [PubMed] [Google Scholar]
- 23.McGuire, A. M., J. D. Hughes, and G. M. Church. 2000. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10:744-757. [DOI] [PubMed] [Google Scholar]
- 24.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 25.Olson, J. W., and R. J. Maier. 2002. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298:1788-1790. [DOI] [PubMed] [Google Scholar]
- 26.Rees, D. C. 2002. Great metalloclusters in enzymology. Annu. Rev. Biochem. 71:221-246. [DOI] [PubMed] [Google Scholar]
- 27.Richard, D. J., G. Sawers, F. Sargent, L. McWalter, and D. H. Boxer. 1999. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145:2903-2912. [DOI] [PubMed] [Google Scholar]
- 28.Rossmann, R., G. Sawers, and A. Bock. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807-2814. [DOI] [PubMed] [Google Scholar]
- 29.Sawers, R. G., S. P. Ballantine, and D. H. Boxer. 1985. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J. Bacteriol. 164:1324-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Self, W. T., A. Hasona, and K. T. Shanmugam. 2004. Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J. Bacteriol. 186:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snavely, M. D., S. A. Gravina, T. T. Cheung, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266:824-829. [PubMed] [Google Scholar]
- 32.Spencer, M. E., and J. R. Guest. 1974. Proteins of the inner membrane of Escherichia coli: changes in composition associated with anaerobic growth and fumarate reductase amber mutation. J. Bacteriol. 117:954-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart, V. 2003. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, V., and B. L. Berg. 1988. Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase gene expression in Escherichia coli K-12. J. Bacteriol. 170:4437-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukdeo, N., S. L. Clugston, E. Daub, and J. F. Honek. 2004. Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes. Biochem. J. 384:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 37.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 39.Volbeda, A., and J. C. Fontecilla-Camps. 2003. The active site and catalytic mechanism of NiFe hydrogenases. Dalton Trans. 2003:4030-4038. [Google Scholar]
- 40.Wang, H., C.-P. Tseng, and R. P. Gunsalus. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181:5303-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, S. C., A. V. Dias, S. L. Bloom, and D. B. Zamble. 2004. Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry 43:10018-10028. [DOI] [PubMed] [Google Scholar]
- 42.Weiner, J. H., D. P. MacIsaac, R. E. Bishop, and P. T. Bilous. 1988. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J. Bacteriol. 170:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, L. F., and M. A. Mandrand-Berthelot. 1986. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie 68:167-179. [DOI] [PubMed] [Google Scholar]
- 44.Wu, L. F., M. A. Mandrand-Berthelot, R. Waugh, C. J. Edmonds, S. E. Holt, and D. H. Boxer. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709-1718. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L. F., C. Navarro, and M. A. Mandrand-Berthelot. 1991. The hydC region contains a multi-cistronic operon (nik) involved in nickel transport in Escherichia coli. Gene 107:37-42. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, L., X.-H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]