Abstract
We demonstrate that the nuclear topological arrangement of chromosome territories (CTs) has been conserved during primate evolution over a period of about 30 million years. Recent evidence shows that the positioning of chromatin in human lymphocyte nuclei is correlated with gene density. For example, human chromosome 19 territories, which contain mainly gene-dense and early replicating chromatin, are located toward the nuclear center, whereas chromosome 18 territories, which consist mainly of gene-poor and later replicating chromatin, is located close to the nuclear border. In this study, we subjected seven different primate species to comparative analysis of the radial distribution pattern of human chromosome 18- and 19-homologous chromatin by three-dimensional fluorescence in situ hybridization. Our data demonstrate that gene-density-correlated radial chromatin arrangements were conserved during higher-primate genome evolution, irrespective of the major karyotypic rearrangements that occurred in different phylogenetic lineages. The evolutionarily conserved positioning of homologous chromosomes or chromosome segments in related species supports evidence for a functionally relevant higher-order chromatin arrangement that is correlated with gene-density.
The chromatin of individual chromosomes is organized in chromosome territories (CTs) that are essential components of the higher-order chromatin architecture of the vertebrate cell nucleus (reviewed in refs. 1–5). Recently, the extent to which evolutionarily conserved, cell type, cell cycle, and species-specific motifs of chromatin arrangements may exist, has become the focus of intense studies (6–9). In mammals, two principal components of mitotic chromosomes can be distinguished: G-light bands (also called R-bands) replicate early during S-phase and contain most of the housekeeping but relatively few tissue-specific genes. G-dark bands replicate later, are gene poor, and contain tissue-specific genes (10). Recently it has been demonstrated that these chromosome bands are maintained in interphase nuclei as focal chromatin aggregations (11) built up by a number of chromatin domains in the order of ≈1 Mb. These domains apparently persist through all interphase stages, show distinct nuclear localization patterns, and may provide an important component of the higher-order nuclear architecture (refs. 11–14, for review see ref. 2). The nuclear location of human (Homo sapiens, HSA) chromosomes 18 and 19 CTs (further referred to as HSA18 and HSA19) has become of special interest in this respect. These chromosomes are of similar DNA content (86 Mb and 72 Mb, respectively; ref. 15) but differ strongly in their gene content and replication timing: most of HSA19 chromatin belongs to G-light bands, is gene-dense [20.5 genes per megabase pair (Mbp)], whereas most of HSA18 chromatin represents G-dark bands and consists mainly of gene-poor chromatin (4.3 genes per Mbp; ref. 10 and http://www.ensembl.org/Homo_sapiens/). In human lymphocyte nuclei, which exhibit an almost spherical shape, HSA19 CTs are consistently localized toward the nuclear center without any detectable attachment to the nuclear envelope, whereas the HSA18 CTs are positioned close to the nuclear border (7, 8). In this study we demonstrate the evolutionary conservation of radial nuclear arrangements for chromosomes or chromosome segments homologous to HSA18 and 19 in seven higher-primate species. The last common ancestor of these species dates back approximately 30–40 million years ago. This evolutionary conservation argues for a still unknown functional significance of distinct radial higher-order chromatin arrangements.
Materials and Methods
Cell Lines and Slide Preparation.
Epstein–Barr virus-transformed lymphoblastoid cell lines were obtained from human (H. sapiens, HSA; LB-3), chimpanzee (Pan troglodytes, PTR; EB176 (JC), ECACC No.89072704), gorilla [Gorilla gorilla, GGO; EB (JC), ECACC No.89072703], orangutan (Pongo pygmaeus, PPY; EB185 (JC), ECACC No.89072705),white-handed gibbon (Hylobates lar, HLA), cotton-top tamarin (Saguinus oedipus, SOE; B95–8, ECACC No.85011419), common marmoset (Callithrix jacchus, CJA) (kindly provided by M. Rocchi), and squirrel monkey (Saimiri sciureus, SSC). All cell lines were karyotypically normal, except for cell line B95–8, derived from Saguinus oedipus, which exhibited five marker chromosomes (16, 17) of which none involved chromosomes relevant for this study. For S-phase detection, cells were pulse-labeled with 5-bromodeoxyuridine (BrdUrd) for 1 h before fixation. For preparing three-dimensionally-preserved cell nuclei, cells were seeded onto polylysine-coated slides and fixed in 4% paraformaldehyde in 0.3× PBS. Permeabilization steps included treatment of cells in 0.5% Triton X-100 in PBS, 20% glycerol in PBS, repeated freezing/thawing in liquid nitrogen, and incubation in 0.1 M HCl as described in detail elsewhere (18).
Probe Preparation, Three-Dimensional Fluorescence In Situ Hybridization (3D-FISH) and Detection.
For each target primate species, painting probes for the delineation of chromosomes or chromosome segments homologous to HSA18, HSA19, and HSA1q32→qter were generated by degenerate oligonucleotide primed PCR (19) from flow-sorted chromosomes. Because human paint probes were not found appropriate to achieve intensely painted CTs useful for light optical sectioning and 3D reconstruction in distantly related primate species, for any given target primate species we used HSA18- and HSA19-homologous probes that were established from a closely related primate species (Table 1, Figs. 1 and 2 a–c).
Table 1.
DNA probes used for the respective target species
| Target species | Paint probes used for the delineation of homologous chromosomal regions to
|
||
|---|---|---|---|
| HSA18 | HSA19 | HSA1q32→qter | |
| Human (HSA) | GGO16 | PPY20 | — |
| Chimpanzee (PTR) | GGO16 | PPY20 | — |
| Gorilla (GGO) | HSA18 | PPY20 | — |
| Orangutan (PPY) | GGO16 | HSA19 | — |
| Gibbon (HLA) | GGO16 | PPY20 | — |
| Marmoset (CJA) | LLA4* | SOE5* | LLA25* |
| Tamarin (SOE) | LLA4* | LLA13* | LLA25* |
| Squirrel monkey (SSC) | LLA4* | SOE5* | LLA25* |
For the delineation of the indicated target chromatin in the respective species, chromosome-specific painting probes derived from human (HSA), gorilla (GGO), orangutan (PPY), and the New World monkeys tamarin (SOE) and wooly monkey (Lagothrix lagothricha, LLA) were used.
LLA4 is homologous to HSA18 and HSA8p, LLA13/SOE 5 is homologous to HSA19, and LLA25 is homologous to HSA1q32→qter.
Figure 1.
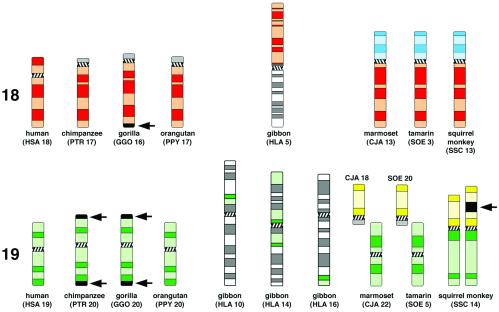
Idiogramatic illustration of G-banded primate chromosomes or chromosomal subregions homolgous to human chromosome 18 (red; Upper) or 19 (green; Lower). Arrows indicate heterochromatin blocks not present in the human and orangutan chromosomes. P-arms of CJA13, SOE3, and SSC13 (highlighted light-blue) are homologous to HSA8p. SSC14p, CJA18, and SOE20 (highlighted yellow) are homologous to HSA1q32→qter. The investigated squirrel monkey individual showed an interstitial repeat heteromorphism in SSC14p (arrow). Chromosome nomenclature followed refs. 16, 20, 21, and 24.
Figure 2.
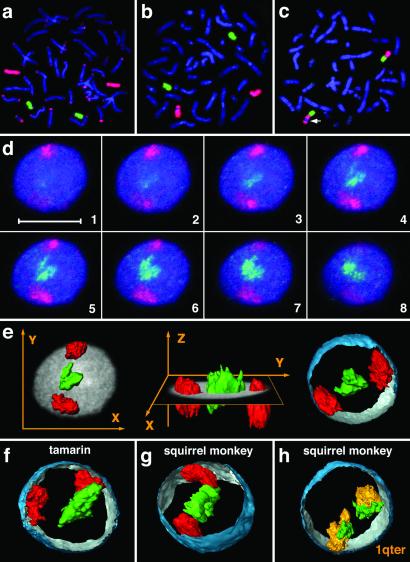
Visualization of chromosomes and chromosome territories. (a and b) Hybridization of GGO16 (red) and PPY20 (green) to gorilla and human metaphase preparations. (a) In the gorilla, extensive cross hybridization of subtelomeric heterochromatin was observed when employing a gorilla paint (red), whereas the orangutan paint (green) yielded no cross hybridization. (b) In human, both probes produced highly specific hybridization signals. These experiments demonstrate the necessity to use probes derived from evolutionarily closely related, but different species than the target species for the unequivocal and exclusive delineation of homologous chromatin in subsequent 3D-FISH experiments (see Materials and Methods for details). (c) In the squirrel monkey, SOE5 (green) and LLA25 (red) visualize syntenic association of HSA19 (green) and HSA1q32→qter (red) homologous material. The polymorphic heterochromatin block is marked by an arrow (see also Fig. 1 and Discussion for details). (d) Gallery of 200-nm serial light optical sections (every third section shown) through a marmoset lymphoblastoid cell nucleus after painting of HSA18 (red) and HSA19 (green) homologous chromosome material, DNA counterstain shown in blue. (Scale bar = 10 μm.) (e) Three-dimensional reconstructions of painted CTs presented in d. In the X, Y view (Left) and X, Z view (Center), the mid-plane section of the counterstained nucleus is added as a gray shade. (Right) CTs together with a partial 3D reconstruction of the DNA-counterstained nuclear border (outside, blue; inside, silver-gray). (f and g) Three-dimensional reconstructions of chromosome material homologous to HSA18 and HSA19 in a tamarin and squirrel monkey nuclei. (h) Three-dimensional reconstruction of the two SSC14 territories in a squirrel monkey nucleus after two-color FISH of SSC14q (green; homologous to HSA19) and SSC14p (orange; homologous to HSA1q32→qter; compare Fig. 1). The origin of paint probes used for each species is described in Table 1.
Further, it was previously reported for human chromosomes that painting probes derived from closely related nonhuman primates yield equal signal intensities compared with human probes, but superior specificity even in the absence of suppression with Cot1-DNA, because repeat sequences that cause cross hybridization show a higher rate of evolutionary divergence compared with euchromatic DNA (17). We therefore avoided the hybridization of paint probes to target nuclei from the same species (see Table 1). Employing this approach, an excellent specificity and signal-to-noise ratio was obtained in all cross-species 3D-FISH experiments. All primate paint probes were previously characterized by reverse painting to human metaphases (16, 17, 20, 21). Three-dimensional FISH, detection of labeled probes and of incorporated BrdUrd, and DNA counterstaining was performed according to the protocols described elsewhere (7, 18). Only BrdUrd-positive nuclei indicating their S-phase stage were selected for further evaluation.
Confocal Microscopy.
Nuclei were scanned with an axial distance of 200 nm using a three-channel laser scanning confocal microscope (Zeiss LSM 410). For each optical section, images were collected sequentially for all three fluorochromes. Stacks of 8-bit gray-scale two-dimensional images were obtained with a pixel size of 66 nm. Displayed overlays of confocal images were processed with Adobe Photoshop 5.5. Three-dimensional reconstructions of CT image stacks were performed by using AMIRA 2.2 TGS (http://www.amiravis.com/). The nuclear periphery was reconstructed from thresholded images of the DNA counterstain (Figs. 2 and 3).
Figure 3.
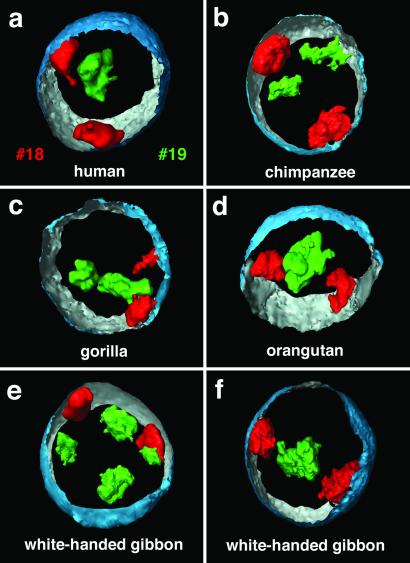
Three-dimensionally reconstructed CTs in great apes and white-handed gibbon nuclei. (a) Three-dimensional positioning of HSA18 (red) and HSA19 (green) CTs in a human lymphoblastoid cell nucleus with the partially reconstructed nuclear border (outside, blue; inside, silver-gray). (b–d) Three-dimensional positioning of HSA18 and HSA19-homologous CTs in nuclei of great apes. In correspondence with the situation in human lymphoblastoid cell nuclei, the HSA18-homologous CTs were always positioned close to the nuclear border. The relative locations of the CTs, however, varied from a close neighborhood to opposite positions (for example, compare b and c). (e and f) Two gibbon nuclei exemplify the variability but still consistently interior location of reshuffled HSA19-homologous chromosome segments in this species of lesser apes in contrast to the peripherally located HSA18-homologous segments. The origin of paint probes used for each species is described in Table 1.
Quantitative Evaluation of the 3D Positioning of Painted Territories.
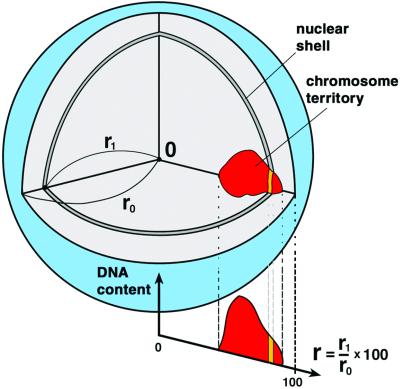
A detailed description of the quantitative 3D evaluation of light optical serial sections was published elsewhere (7). Briefly, a voxel (volume element)-based algorithm was applied (Fig. 4). As a first step, the center and the border of the nucleus were determined by using the 3D data set of the DNA-counterstain fluorescence in the following way; first, the fluorescence intensity gravity center of the counterstain voxels after automatic thresholding was calculated. For the interactive segmentation of the nuclear border, a straight line was drawn from the gravity center toward each voxel considered, and the nuclear center was then determined as the geometrical center of the segmented voxels. In the second step, segmentation of CTs was performed in each 3D stack representing the color channels for the respective painted CTs. The segmented nuclear space was divided into 25 equidistant shells with a thickness of Δr = 1/25 r0. For comparison of nuclei with different shape and size, the distance between the nuclear center and any point on the segmented nuclear border along the straight lines mentioned above, was given as r0 = 100. For each voxel located in the nuclear interior, the relative distance r from the nuclear center was calculated as a fraction of r0. A shell at a given r contains all nuclear voxels with a distance between r − Δr/2 and r + Δr/2. For each shell all voxels assigned to a given CT were identified and the fluorescence intensities derived from the respective emission spectrum were summed up. This procedure yielded the individual DNA-shell contents for painted CTs as well as the overall DNA content reflected by the DNA counterstain. For better comparison of different nuclei, the sum of the voxel intensities measured in each nucleus was set to 100% for each fluorochrome. When this normalization is used, the average relative DNA content in nuclear shells as a function of the relative distance r from the 3D center represents the average radial distribution of the DNA representing the painted CTs or of the overall DNA in the entire set of evaluated nuclei. As an estimate for the variation obtained for each relative distance r, the standard deviation of the mean DNA shell content was calculated. The data collected for each species were plotted as graphs (Figs. 5 and 6).
Figure 4.
Scheme of 3D evaluation of radial chromosome territory arrangements in spherical cell nuclei. (for details see Materials and Methods).
Figure 5.
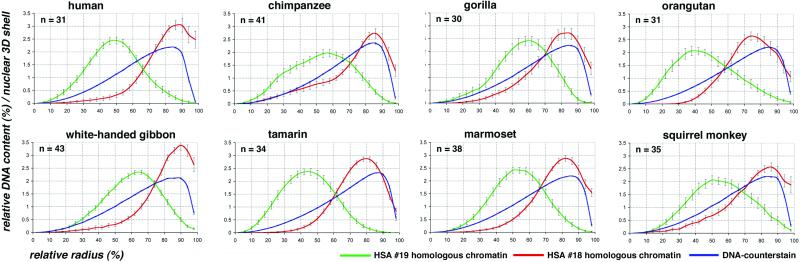
Quantitative 3D evaluation of radial chromatin arrangements in primate cell nuclei with HSA18 and HSA19 homologues. Radial chromatin arrangements observed after painting with HSA18- and HSA19-homologous probes evaluated in 25 radial concentric nuclear shells (compare Fig. 4). The abscissa denotes the relative radius r of the nuclear shells, the ordinate denotes the normalized sum of the intensities in the voxels for a respective fluorochrome belonging to a given shell. For normalization, the area underlying the curve for each color (total relative DNA content) was set to 100. n = number of 3D evaluated nuclei. In all species, a highly significant difference (P < 0.001) was noted between the radial positioning of HSA18 (red) and HSA19 (green) homologous chromatin. HSA18-homologous chromosome material is consistently distributed closer to the nuclear border, whereas HSA19-homologous material is distributed toward the nuclear interior. Bars indicate standard deviations of the mean for each shell. Blue curves represent counterstained DNA.
Figure 6.
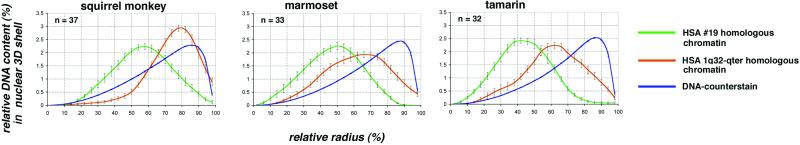
Quantitative 3D evaluation in New World monkey species nuclei with HSA19 and HSA1q32→qter homologues. (Left) Quantitative 3D evaluation of SSC14 positioning in squirrel monkey cell nuclei after painting of SSC14p (red) with a HSA1q32→qter-homologous probe and of SSC14q (green) with a HSA19-homologous probe (compare Fig. 1). (Center and Right) Quantitative 3D evaluation after painting of separate chromosomes homologous to HSA1q32→qter (red) and HSA19 (green) homologous segments in marmoset and tamarin cell nuclei (for the description of the abscissa and the ordinate, see Fig. 5).
Results
Delineation of Human Chromosome 18 and 19 Homologous Territories in Primates.
We chose lymphoblastoid cell lines from human and from seven other higher-primate species that exhibit distinct differences in their pattern of karyotype evolution (22): three great apes (chimpanzee, gorilla, and orangutan), the white-handed gibbon, and three New World monkeys (cotton-top tamarin, common marmoset, and squirrel monkey). In comparison to the human karyotype, great ape chromosomes are strongly conserved and differ from human mainly by intrachromosomal rearrangements (23), whereas gibbon karyotypes show a high degree of chromosome reshuffling (24). Compared with gibbons, New World monkeys show a moderate rate of chromosomal changes, mainly Robertsonian type or tandem rearrangements (21). Fig. 1 shows individual higher-primate chromosomes and chromosomal segments homologous to HSA18 and HSA19, respectively, as well as chromosomal material homologous to HSA1q32→qter and HSA8p (see below).
Employing two-color FISH, we delineated CTs and chromosomal segments, respectively, homologous to HSA18 or HSA19 in 3D preserved lymphoblastoid cell nuclei from all of the primate species mentioned above. As the extent to which radial chromosome territory arrangements in these nuclei may change during the cell cycle is not clear at present, all analyses described below were restricted to cells at a clearly identifiable stage of the cell cycle, namely S-phase. Three-dimensional preserved cell nuclei were then hybridized with differentially labeled painting probes specific for HSA18 or HSA19 or their primate homologues (Table 1). Four different fluorochromes were used for the visualization of painted CTs, BrdUrd-labeled DNA, and overall nuclear DNA. Light optical serial sections were recorded from BrdUrd-positive nuclei with a laser scanning confocal microscope. As an example, Fig. 2d shows an image gallery from a marmoset lymphoblastoid cell nucleus demonstrating the interior location of HSA19-homologous CTs and the peripheral location of the HSA18-homologous chromosome segments. Fig. 2e shows representative 3D reconstructions of this nucleus. Figs. 2 f–g and 3 indicate the same radial arrangement of HSA18- and HSA19-homologous chromatin in lymphoblastoid cell nuclei in all species studied.
Quantitative 3D Evaluation of Primate Homologous CTs.
For each species, this positioning was quantitatively evaluated in a set of nuclei (31 to 43 nuclei per species) after defining the geometrical center and border of each nucleus and dividing its space into 25 radial concentric shells (Fig. 4). The relative contribution of each painted CT to each of these shells was calculated as described in Materials and Methods. For comparison, the relative contribution of nuclear DNA was also recorded in each shell. The resulting data are graphically presented in Figs. 5 and 6.
In cell nuclei from human and great apes, we observed homologous HSA18 chromatin at the nuclear periphery with a maximum DNA content between 75% and 88% of the relative radius, whereas homologous HSA19 chromatin was observed in the nuclear interior with a maximum DNA content between 38% and 58% (Table 2, Fig. 5). The findings obtained for human and great apes lymphoblastoid cell nuclei are fully consistent with previous studies of human lymphocytes and show that transformed lymphoblastoid cell lines retain the distinctly different radial arrangement of HSA18 and HSA19 CTs observed in nuclei of both noncycling B and T cells from peripheral blood and phytohemagglutinin-stimulated, cycling normal T-lymphocytes (7, 8).
Table 2.
Radial distribution of human chromosome 18 and 19 homologous CTs
| Species | HSA18 homologues, %* | HSA19 homologues, %* | HSA1q32→1qter homologues, %* | Nuclei with conserved radial arrangement† |
|---|---|---|---|---|
| HSA | 88 | 48 | — | 31/31 (100%) |
| PTR | 86 | 58 | — | 38/41 (92.7%) |
| GGO | 85 | 60 | — | 30/30 (100%) |
| PPY | 75 | 38 | — | 31/31 (100%) |
| HLA | 90 | 64 | — | 43/43 (100%) |
| SOE | 79‡ | 42§ | 62 | 34/34 (100%) |
| CJA | 82‡ | 51§ | 65 | 38/38 (100%) |
| SSC | 86‡ | 54§ | 79 | 33/35 (94.3%) |
Peaks of maximum DNA content at the relative distance from the nuclear center.
Individual nuclei where CTs 18 gravity centers show a more peripheral location than CTs 19.
These painted CTs contain also HSA8p-homologous chromosome segments.
In lesser apes (gibbons), the HSA18-homologue is conserved but translocated, whereas the HSA19 homologue is fragmented (24). In the white-handed gibbon a HSA18-homologous segment is associated with a HSA1p32→q22 homologous segment to form the metacentric gibbon chromosome 5 (Fig. 1). Gibbon chromosomes 10 and 16 each carry one segment of HSA19-homologous chromatin, whereas gibbon chromosome 14 contains two HSA19-homologous segments, one of them including the centromeric heterochromatin (Fig. 1). According to these complex rearrangements, most nuclei revealed four to eight painted regions homologous to HSA19 (Fig. 3e). These regions were located toward the nuclear interior with a content maximum at 64% of the relative radius (Fig. 5, Table 2). Some nuclei even showed one large cluster of a centrally located painted region (Fig. 3f). In contrast, HSA18-homologous chromatin was always located in proximity to the nuclear envelope with a DNA content maximum at 90% (Table 2, Figs. 3 e and f and 5).
In New World monkeys, the HSA18 homologues are entirely conserved but translocated to the HSA8p (7.3 genes per Mbp; http://www.ensembl.org/Homo_sapiens/) (Fig. 1). This chromosome form represents the ancestral condition of this primate infraorder (22). The probes used for these species painted these chromosomes along their entire length, i.e., both HSA18- and HSA8p-homologous materials. In the three New World monkey species included in this study the HSA18- and HSA8p-homologous chromosome segments revealed maximum DNA contents at relative radii between 79% and 86%, and were thus within the same range as found for the great and lesser apes. HSA19-homologous chromosomes or chromosome segments showed maximum DNA contents at relative radii between 42% and 54% (Table 2, Fig. 5). In the squirrel monkey, HSA19-homologous chromatin is represented by SSC14q, and SSC14p is represented by a HSA1q32→qter-homologous segment. Two-color painting of squirrel monkey chromosomes 14p and 14q revealed a specific orientation of these CTs: HSA1q32→qter-homologous chromosome segments with a gene density of 9.5 genes per Mbp (http://www.ensembl.org/Homo_sapiens/) were typically located closer toward the nuclear periphery with a DNA content maximum at around 79% of the relative radius, whereas the HSA19-homologous segments with a gene density of 20.5 genes per Mbp were located toward the nuclear centre with a maximum DNA content at 54% of the relative radius (Figs. 2h and 6). Similar radial orientations were observed in tamarin and marmoset cell nuclei (Fig. 6), where HSA19 and HSA1q32→qter-homologous chromatin represent separate chromosome entities (Fig. 1).
Discussion
Our results demonstrate that the distinctly different radial distribution patterns that have been found for CTs 18 and 19 in human lymphocyte and lymphoblastoid cell nuclei have been conserved for HSA18- and HSA19-homologous chromatin during higher-primate evolution. In all species analyzed, HSA18-homologous chromatin was found at the nuclear periphery and HSA19-homologous chromatin was found toward the nuclear interior. This radial distribution pattern was thus maintained over a period of at least 30 million of years, irrespective of the extensive chromosomal rearrangements that occurred during the evolution of higher primates. Our results fit the hypothesis that radial chromatin arrangements reflect differences in gene density (6). Additional evidence supporting this hypothesis is provided by our observation of a specific orientation of the relatively gene-poor HSA1q32→qter and the gene-dense HSA19-homologous chromatin segment, which form chromosome 14 in the squirrel monkey. This hypothesis also holds true for somatic translocation events. For example, a somatic t(18;19) translocated chromosome also maintained the original nuclear orientation of the translocation partners with a peripheral location of HSA18 and an internal location of the HSA19 region (8).
It is a well established fact that the positioning of genes close to heterochromatin blocks can strongly affect their transcription (25), and it has also been argued that heterochromatin blocks may play a role with regard to the evolving nuclear architecture (26). Chromosome translocations that join heterochromatic segments with gene-dense chromatin segments may therefore lead to radial chromatin shifts depending on the size and composition of the respective segments. For example, gene-poor chromatin and heterochromatin has often been noted at the nuclear periphery (5). Accordingly, the joining of a heterochromatin block with a gene-dense chromosome segment, previously located in a gene-dense interior nuclear compartment, may result in a positional shift of the latter toward the peripheral nuclear compartment, possibly affecting its transcriptional activity. Our present study provides two possible examples for such a shift. First, the squirrel monkey chromosome 14p (homologous to HSA1q32→qter) is heteromorphic in the cell line used in this study (Figs. 1 and 2c). One 14 p-arm shows a large additional heterochromatic band, which is neither present in the other homologous squirrel monkey chromosome 14 nor in the corresponding counterparts of marmoset chromosome 18 and tamarin chromosome 20 (Fig. 1). This additional band may possibly explain why squirrel monkey 14p chromatin was distributed on average more toward the nuclear periphery (maximum DNA content at 79%) than marmoset 18 and tamarin 20 CTs showing their maximum DNA content at 65% and 62%, respectively (Table 2, Fig. 6). Second, we observed a more exterior position of CTs of HSA19 homologues in chimpanzee and gorilla (at relative radius of 58% and 60%, respectively) as compared with human and orangutan (at 48% and 38%, respectively; Table 2, Fig. 5). Chimpanzee and gorilla homologues contain large regions of terminal heterochromatin (Figs. 1, arrows, and 2a), whereas these blocks are absent in the respective human and orangutan chromosomes (Fig. 1). It is intriguing to look for other examples of chromosome evolution as well as chromosomally rearranged tumor cells, where the chromatin context adjacent to a given gene-poor or gene-rich chromosome segment changes, and to test the consequences for radial positioning and gene function.
Finally, the finding of specific radial CT arrangements is not limited to primate cell nuclei, but was also reported for chicken cell nuclei (9). In Gallus gallus domesticus, microchromosomes are early replicating and considerably more gene-dense than the gene-poor and later replicating macrochromosomes. We noted the location of microchromosome territories preferentially in the nuclear interior surrounded by the more peripherally located macrochromosome territories. This gene-density-correlated radial higher-order chromatin arrangement in chicken cell nuclei shows that the evolutionary conservation of nonrandom radial arrangements is compatible with drastic changes in karyotype evolution that have occurred before the separation of the evolutionary branches that led to present days mammals and birds. In this context, it is interesting to note that syntenic regions of HSA19 have been assigned to chicken microchromosomes, whereas syntenic regions of HSA18 have been assigned to the chicken macrochromosomes 2 and Z (27, 28).
The evidence for an evolutionary conservation of gene-density-correlated radial chromatin arrangements argues for a functional significance. Possible underlying molecular mechanisms responsible for the establishment and maintenance of these higher-order chromatin arrangements remain to be elucidated. In this context, it is interesting to test whether the density of expressed genes rather than of all genes plays a major role. The observation of different positions of the active and inactive X chromosome in female cell nuclei argues for such a possibility. In addition, the different CG content of gene-dense and gene-poor chromosome segments should be considered (29).
Acknowledgments
This study was supported by a stipend of the Japanese government, Science and Technology Agency (to H.T.), Deutsche Forschungsgemeinschaft Grant Cr 59/20-1 (to T.C.), European Community Grant FIGH-CT 1999-00011 (to C.C.), and by a grant in part, Health Sciences Research Grants, Ministry of Health, Labor and Welfare, Japan.
Abbreviations
- CTs
chromosome territories, 3D-FISH
- three-dimensional fluorescence in situ hybridization.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chevret E, Volpi E V, Sheer D. Cytogenet Cell Genet. 2000;90:13–21. doi: 10.1159/000015654. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer C. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 3.Cremer T, Kreth G, Koester H, Fink R H A, Heintzmann R, Cremer M, Solovei I V, Zink D, Cremer C. Crit Rev Eukaryotic Gene Expression. 2000;12:179–212. [PubMed] [Google Scholar]
- 4.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 5.Leitch A R. Microbiol Mol Biol Rev. 2000;64:138–152. doi: 10.1128/mmbr.64.1.138-152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle S, Gilchrist S, Bridger J M, Mahy N L, Ellis J A, Bickmore W A. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 7.Cremer M, v. Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- 8.Croft J A, Bridger J M, Boyle S, Perry P, Teague P, Bickmore W A. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habermann F, Cremer M, Walter J, Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I. Chromosome Res. 2001;9:569–584. doi: 10.1023/a:1012447318535. [DOI] [PubMed] [Google Scholar]
- 10.Craig J M, Bickmore W A. Nat Genet. 1994;7:376–382. doi: 10.1038/ng0794-376. [DOI] [PubMed] [Google Scholar]
- 11.Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner B M, Zink D. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson D A. BioEssays. 1995;17:587–591. doi: 10.1002/bies.950170704. [DOI] [PubMed] [Google Scholar]
- 13.Nakayasu H, Berezney R. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Keefe R T, Henderson S C, Spector D L. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Müller S, Neusser M, O'Brien P C M, Wienberg J. Chromosome Res. 2001;9:689–693. doi: 10.1023/a:1012960525326. [DOI] [PubMed] [Google Scholar]
- 17.Müller S, O'Brien P C, Ferguson-Smith M A, Wienberg J. Hum Genet. 1997;101:149–153. doi: 10.1007/s004390050604. [DOI] [PubMed] [Google Scholar]
- 18.Solovei I, Walter J, Cremer M, Habermann F, Schermelleh L, Cremer T. In: FISH: A Practical Approach. Squire J, Beatty B, Mai S, editors. Oxford: Oxford Univ. Press; 2002. , in press. [Google Scholar]
- 19.Telenius H, Carter N P, Bebb C E, Nordenskjold M, Ponder P A I, Tunnacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 20.Stanyon R, Consigliere S, Müller S, Morescalchi A, Neusser M, Wienberg J. Am J Primatol. 2000;50:95–107. doi: 10.1002/(SICI)1098-2345(200002)50:2<95::AID-AJP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Stanyon R, Consigliere S, Bigoni F, Ferguson-Smith M, O'Brien P C, Wienberg J. Chromosome Res. 2001;9:97–106. doi: 10.1023/a:1009274802086. [DOI] [PubMed] [Google Scholar]
- 22.Wienberg J, Stanyon R. ILAR J. 1998;39:77–91. doi: 10.1093/ilar.39.2-3.77. [DOI] [PubMed] [Google Scholar]
- 23.Yunis J J, Prakash O. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 24.Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Proc Natl Acad Sci USA. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown K E, Baxter J, Graf D, Merkenschlager M, Fisher A G. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 26.Manuelidis L. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- 27.Burt D W, Bruley C, Dunn I C, Jones C T, Ramage A, Law A S, Morrice D R, Paton I R, Smith J, Windsor D, et al. Nature (London) 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- 28.Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, Schartl M, Haaf T, Weigend S, Fries R, Buerstedde J M, et al. Cytogenet Cell Genet. 2000;90:169–218. doi: 10.1159/000056772. [DOI] [PubMed] [Google Scholar]
- 29.Bernardi G. Gene. 2001;276:3–13. doi: 10.1016/s0378-1119(01)00644-8. [DOI] [PubMed] [Google Scholar]