Abstract
Phase variation of type 1 fimbriae of Escherichia coli requires the site-specific recombination of a short invertible element. Inversion is catalyzed by FimB (switching in either direction) or FimE (inversion mainly from on to off) and is influenced by auxiliary factors integration host factor (IHF) and leucine-responsive regulatory protein (Lrp). These proteins bind to sites (IHF site II and Lrp sites 1 and 2) within the invertible element to stimulate recombination, presumably by bending the DNA to enhance synapses. Interaction of Lrp with a third site (site 3) cooperatively with sites 1 and 2 (termed complex 1) impedes recombination. Inversion is stimulated by the branched-chain amino acids (particularly leucine) and alanine, and according to a current model, the amino acids promote the selective loss of Lrp from site 3 (complex 2). Here we show that the central portion of the fim invertible element, situated between Lrp site 3 and IHF site II, is dispensable for FimB recombination but that this region is also required for full amino acid stimulation of inversion. Further work reveals that the region is likely to contain multiple regulatory elements. Lrp site 3 is shown to bind the regulatory protein with low affinity, and a mutation that enhances binding to this element is found both to diminish the stimulatory effects of IVLA on FimB recombination and to inhibit recombination in the absence of the amino acids. The results obtained emphasize the importance of Lrp site 3 as a control element but also highlight the complexity of the regulatory system that affects this site.
Phase variation (the reversible on-to-off switching in gene expression that produces a mixed population) is a common mode of control among virulence factors such as adhesins. The regulation of genes by phase variation is apparently a strategy to maximize growth and survival in unpredictable environments (15). However, phase variation should nevertheless be controlled in response to environmental cues that convey information about the likely advantages or disadvantages of producing the factor concerned (8). We have shown previously that the DNA inversion that controls the phase variation of type 1 fimbriation in Escherichia coli (fim) is regulated in response to temperature as well as the presence of the branched-chain amino acids isoleucine (I), valine (V), and particularly leucine (L) as well as alanine (A) (21). More recently, we have shown that off-to-on phase switching of fim is suppressed specifically by sialic acid (N-acetylneuraminic acid [Neu5Ac]) and by N-acetylglucosamine (17, 44).
Phase variation of fim involves the site-specific inversion of a small (around 314-bp) element which contains the promoter for fimA, the gene encoding the major structural subunit of the fimbriae (1, 29). The recombination is catalyzed by FimB (on-to-off and off-to-on inversion) or by FimE (on-to-off inversion mainly), recombinase proteins which share 50% identity with each other and which resemble the lambda integrase family of site-specific recombinases (2, 14, 16, 30, 36). FimB and FimE bind to half-sites on either side of sites of DNA cleavage and strand exchange (inverted repeat right [IRR] and inverted repeat left [IRL]) to catalyze inversion (19). By analogy with other site-specific recombination systems, the juxtaposition of these loci is likely to be required for DNA cleavage, strand exchange, and rejoining (41). Inversion is also influenced by the site-specific DNA binding proteins integration host factor (IHF) (4, 14, 16, 31, 46) and leucine-responsive regulatory protein (Lrp) (5, 48, 50). Whereas Lrp binds cooperatively to three sites within the invertible region situated proximal to IRR in the off orientation, IHF interacts with sites both adjacent to site I and within site II, the invertible region (Fig. 1) (20, 40). Mutations in each of these binding sites, with the exception of Lrp site 3, inhibit fim recombination. IHF and Lrp both introduce sharp bends into DNA, and accordingly we suppose that the combined interaction of these two proteins with the invertible element facilitates recombination by bringing IRR and IRL into a juxtaposition at synapses (4).
FIG. 1.
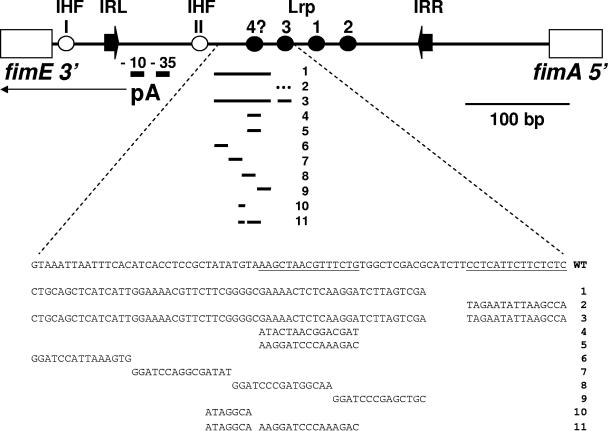
The organization of the fim invertible element in the off orientation. Shown are characterized Lrp binding sites 1, 2, and 3 and potential Lrp site 4 (dark circles); IHF binding sites I and II (light circles); and IRL, IRR, and the fimA promoter (pA). The thin arrow indicates the direction of transcription. The Rm mutations constructed in this study (1 to 11) are listed, and the regions altered by these mutations are indicated by a line in the upper portion of the diagram. Rm2 (dotted line) was constructed by replacing Lrp site 3 with site 2, whereas all of the other mutations contain heterologous sequences. The wild-type nucleotide sequence, with Lrp site 3 and potential Lrp site 4 underlined, together with base changes made in each mutant, are shown in the lower half of the figure. “4?” denotes that Lrp has not been shown to bind to the fourth potential binding site.
Lrp controls a regulon that comprises numerous genes in E. coli, many of which are involved in amino acid biosynthesis, transport, and degradation (10, 26, 38). In addition, Lrp is involved in the regulation of several types of fimbriae, including fim, pap, sfa, daa, fan, and clp (5, 9, 35, 47). In all of the systems described to date, Lrp binds cooperatively to multiple sites to bring about its regulatory functions. Lrp can act as either a positive or a negative regulator, and its actions may be intensified, reduced, or unaffected by the presence of exogenous leucine and, in some cases, alanine. Leucine binds to Lrp in vitro to interact with both high- and low-affinity sites, and the presence of the amino acid affects the association state of the regulatory protein (11). In the absence of leucine, the protein is found predominantly as a hexadecamer, but an octameric form is favored when the amino acid is present at mM concentrations (11, 12). Notwithstanding the results described above, leucine affects the association of Lrp with DNA, apparently as the result of two opposing effects: it exerts (i) a negative effect on Lrp binding affinity for single sites but (ii) a positive effect on the cooperativity for any Lrp bound to DNA (18, 33, 39, 49, 50).
To our knowledge, the recombination of type 1 fimbriae provides the only known example where Lrp is a positive regulator, and its effects are potentiated by leucine and alanine (5, 21). A mutation of Lrp site 3 (fimS21) that abolishes the binding of the regulatory protein to its cognate site within the fim invertible element in vitro results in a high level of recombination in vivo in the absence of isoleucine, valine, leucine, and alanine (IVLA) as well as a loss of stimulation of inversion in their presence (40). Moreover, leucine promotes the selective loss of Lrp binding to site 3 in combination with sites 1 and 2 in vitro. Thus, IVLA could stimulate the fim inversion simply by promoting the conversion of a nucleoprotein complex that is less favorable for recombination (complex 1, containing Lrp bound to sites 1 to 3) into one that facilitates the inversion (complex 2, containing Lrp bound to sites 1 and 2 only). In support of this model, we show here that a mutation that increases binding of Lrp to site 3 in vitro (i) inhibits FimB recombination in minimal medium and (ii) diminishes the stimulatory effect of the branched-chain amino acids and alanine. Further work shows that although the central portion of the fim invertible element between Lrp site 3 and IHF site II is dispensable for FimB recombination, it contains additional sequences that play both positive and negative roles in the amino acid control of the system. Thus, regulation of the fim inversion in response to IVLA is complex and likely to be affected by multiple environmental factors.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this work are listed in Table 1. All plasmids used for mutagenesis in this work were derivatives of pIB445 (19). All bacterial strains are derivatives of E. coli K-12. Strain AAEC189 (Δfim) was used as the host strain for recombinant plasmids to ensure that the invertible element was maintained in the on or off orientation and that the DNA was suitably methylated to allow subsequent transformation of the strain MG1655 (6).
TABLE 1.
Strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| MG1655 | K-12 wild type; λ− F− Fim+ | E. coli Genetic Stock Center; 24 |
| AAEC189 | YMC9 (λ− F−supE44 hsdR17 mcrA mcrB endA1 thi-1 ΔargF-lac-205) ΔfimB-H ΔrecA | 6 |
| BGEC144 | MG1655, ΔlacZYA Δ′fimE-fimA′Ω(sacB-Kanr) fimE′(Am)18 fimA′-lacZYA | 20 |
| BGEC378 | MG1655, ΔlacZYA fimA′-RNase III cleavage site-lacZYA fimE-(Am)18 | 19 |
| BGEC560 | BGEC378 fimS21 | 40 |
| BGEC771 | BGEC378 Rm2 | This study |
| KCEC047 | BGEC378 Rm3 | This study |
| KCEC049 | BGEC378 Rm1 | This study |
| KCEC261 | BGEC378 fimS21 Rm1 | This study |
| KCEC300 | BGEC378 Rm6 | This study |
| KCEC302 | BGEC378 Rm7 | This study |
| KCEC304 | BGEC378 Rm8 | This study |
| KCEC306 | BGEC378 Rm9 | This study |
| KCEC308 | BGEC378 Rm4 | This study |
| KCEC310 | BGEC378 Rm5 | This study |
| KCEC312 | BGEC378 Rm10 | This study |
| KCEC314 | BGEC378 Rm11 | This study |
Construction of strains.
Allelic exchange to replace the wild-type fim sequence was carried out as described previously (7). Intermediate strain BGEC144, containing deletions of the switch region replaced by a sacB-Kanr cassette, was transformed with fim derivatives of the temperature-sensitive plasmid pMAK705 (20, 25).
Media and growth conditions.
The media included L broth (5 g of sodium chloride, 5 g of yeast extract, and 10 g of tryptone per liter [Difco]) and L agar (L broth with 1.5% agar [Difco]). Sucrose agar, used to select recombinant bacteria (7), is L agar supplemented with 6% sucrose in the absence of sodium chloride. Minimal MOPS [3-(N-morpholino)propanesulfonic acid] medium was prepared as described by Neidhardt et al. (37) and supplemented with 10 μM thiamine and 0.4% glucose. When required, the following amino acids were added to the minimal medium (concentrations are indicated in parentheses): isoleucine (0.4 mM), valine (0.6 mM), leucine (0.8 mM), and alanine (0.8 mM). In addition, to minimize the lag-phase period upon dilution to very low cell densities, NaHCO3 (1 mM) was added to minimal medium as described previously (37). Liquid cultures were grown aerobically at 37°C, and culture densities were monitored spectrophotometrically at 420 or 600 nm. Lactose MacConkey plates were used as an indicator medium to determine the proportion of switch-on bacteria to switch-off bacteria.
Determination of inversion frequencies.
Inversion of the fim switch was measured following growth in minimal MOPS medium with or without exogenous IVLA as described previously (21). Inversion frequencies were measured by inoculating 25 cultures with approximately 0.3 cell per tube. The ratio of on cells to off cells was estimated by plating seven replicates onto lactose-MacConkey indicator medium after approximately 22 generations of growth at 37°C with rapid aeration.
DNA manipulations.
DNA manipulations were carried out using standard protocols (3). Plasmid DNA was isolated using Mini or Midi Prep kits (Promega or QIAGEN). Restriction enzymes and DNA ligase were purchased from either Promega or New England Biolabs. DNA sequencing was performed using the dideoxy chain termination technique (with a Sequitherm EXCEL sequencing kit from Epicenter Technologies or by the Advanced Biotechnology Centre, Imperial College, United Kingdom). Oligonucleotide synthesis was performed in the DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University School of Medicine, by MWG-Biotech AG, or by QIAGEN. For electrophoretic mobility shift assays (EMSAs), DNA was amplified by PCR using Pwo polymerase (Boehringer Mannheim) as described previously (4). PCRs and radiolabeling of PCR products were carried out as described previously (40). PCR12 was carried out using primers 2774 (5′-CCTCCGCTATATGTAAAGCTAACGTTTC) and 2629-Xba (5′-GGGGGTCTAGATAAATACAAGACAATTGGGGC).
The replacement of Lrp binding site 3 with Lrp binding site 2 (Rm2), together with other small single and double replacement mutations (Rm3 to -11), was constructed using the PCR-based method of overlap extension as previously described (40). The 59-bp heterologous replacement of the fim switch (Rm1) was constructed using a similar approach. The replaced region corresponds to nucleotides 4540362 to 4540420 of the MG1655 genome, with the fim switch in the off orientation (Fig. 1). The replacement corresponds to sequences 3961 to 3999 of the vector pBR322 (45) and is flanked by PstI (IRL proximal end) and SalI (IRR proximal end) restriction endonuclease sites.
EMSA and methidiumpropyl-EDTA (MPE)-Fe(II) radical footprinting.
Reaction conditions were as described previously (40). The DNA fragment used (PCR12, encompassing all 3 Lrp binding sites) was as that described previously (Fig. 1 in reference 40). Gels were quantitated on an Ambis radioanalytic scanner (Ambis Systems). Complex formation was measured as the loss of free DNA, and dissociation constants were determined as the Lrp concentrations which resulted in 50% loss of free DNA.
Lrp binding affinity to each of the individual sites within the fim switch and site 3 replaced with site 2 was measured relative to the central portion (29/50 bp) of a control fragment described by Cui et al. in 1995 (13). Oligonucleotides for these experiments were purified by polyacrylamide gel electrophoresis. Each binding site was constructed by annealing a 5′-end-labeled oligonucleotide (29 bp) with its complement (3). The Lrp binding sites were positioned in the centers of the oligonucleotides, with 7 bp of adjacent sequence extending on either side. The EMSA was carried out as described previously (40), except that the reaction mixtures contained 5 nM annealed DNA and 30 nM Lrp and were fractionated by electrophoresis through 8% (wt/vol) polyacrylamide (40:1 acrylamide/bisacrylamide). Binding affinity was calculated as the ratio of protein-bound DNA to free DNA. Relative binding was determined by dividing the binding affinity for a particular construct by the binding affinity for the control fragment determined in the same experiment and multiplying by 100. MPE-Fe(II) footprinting was carried out as described previously (40).
RESULTS
The central portion of the fim invertible element is dispensable for FimB recombination but is required for control in response to IVLA.
According to our current model, IHF bound to site II and Lrp occupying sites 1 and 2 (complex 2) stimulate the fim inversion, whereas Lrp complex 1 (Lrp bound to sites 1 to 3) produces a nucleoprotein structure that is less favorable for recombination (4, 40). We suppose that the branched-chain amino acids (leucine in particular) and alanine stimulate inversion because they promote the selective loss of Lrp binding to site 3 (40). An assumption implicit in this model is that IHF and Lrp alone are required for high levels of FimB recombination and that Lrp sites 1 to 3 are sufficient for full control in response to IVLA. To test these hypotheses further, we replaced the central portion of the fim invertible element (defined as the sequences between IHF site II and Lrp site 3) (Fig. 1) with heterologous sequences of identical length (Rm1; part of the coding region of the ampicillin resistance gene of pBR322; strain KCEC049), and the effect of the mutation on FimB recombination was characterized. The replacement mutation, which is 59 bp in length, extends to a position 8 bp from Lrp site 3 (40).
The results of this experiment showed that the central portion of the fim invertible element can be replaced without the loss of FimB recombination (Table 2). However, as was observed for an Lrp site 3 null allele (fimS21) (40), the frequency of FimB recombination was enhanced markedly (8.3-fold; Table 2) in the Rm1 mutant background in the absence of exogenous amino acids but was far less responsive (1.5-fold; Table 2) to stimulation by their inclusion. In an additional control experiment, it was found that, as expected, the fimS21 mutation had much less of an effect on FimB recombination in the absence of IVLA in the Rm1 background than it did in the wild-type background (1.8-fold compared to 6.4-fold; Table 2).
TABLE 2.
FimB recombination in Rm mutants 1 to 3, fimS21, and a fimS21 double mutant grown to exponential phase in minimal MOPS glucose medium in the absence and presence of IVLA
| Mutation(s) | IVLA | Inversion frequency (104)a | Ratio of mutant to wild type |
|---|---|---|---|
| Rm1 | − | 82.2 | 8.4 |
| + | 121.0 | 2.2 | |
| Rm2 | − | 1.0 | 0.1 |
| + | 2.9 | 0.05 | |
| Rm3 | − | 3.3 | 0.3 |
| + | 5.0 | 0.09 | |
| fimS21 | − | 56.3 | 6.4 |
| + | 97.0 | 1.2 | |
| fimS21, Rm1 | − | 104.0 | 11.7 |
| + | 156.0 | 2.0 |
Per cell per generation for inversion from the off-to-on orientation. Ninety-five-percent confidence limits, determined from at least seven values, lie within 10% of the mean.
In addition to the effect noted above, the elevation of FimB recombination in the Rm1 background (KCEC049) was slightly but reproducibly higher than that in the fimS21 mutant (BGEC560), irrespective of the presence of amino acids (Table 2). Thus, the region of the fim invertible element replaced in the Rm1 allele may also include an additional element or elements that repress FimB recombination but do not play a role in amino acid control.
Taken together, these results support a revised model in which the central portion of the fim invertible element, although dispensable for the inversion, nevertheless contains sequences that repress FimB recombination and affect its control in response to IVLA.
Lrp binding to sites 1, 2, and 3 in vitro.
We supposed that a regulatory site(s) within the central portion of the fim invertible element could suppress FimB recombination in the absence of IVLA directly, with Lrp sites 1 to 3 playing an auxiliary role in this process. Alternatively, it seemed to us that this region stabilizes the interaction of Lrp with site 3 to play a potentially regulatory role in the system. In order to investigate these alternative hypotheses in more detail, we first carried out additional experiments designed to investigate the reason why leucine affects the interaction of Lrp with site 3 specifically.
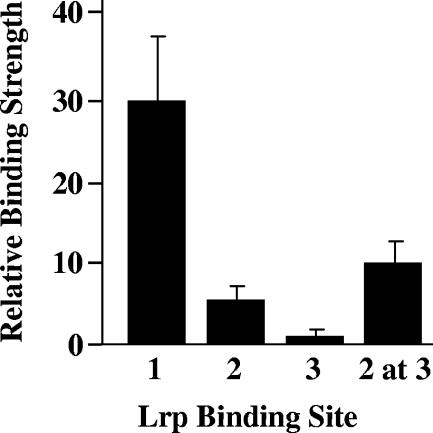
Since leucine promotes the selective dissociation of Lrp from site 3 yet has almost no effect on Lrp's affinity for sites 1 and 2 (40), we supposed that the protein binds to site 3 with an affinity lower than those for the other two sites. To test this hypothesis, we measured the interaction of the regulatory protein with each of the sites individually (the 15-bp consensus sequence with 7 bp of adjacent sequence on either side) (Fig. 2). However, even with high concentrations of Lrp (60 nM), a 50% loss of free DNA could not be attained, and thus a Kd could not be determined. Therefore, the binding affinity for each of the individual Lrp sites was measured relative to a control Lrp binding site used by Cui et al. as described previously (13) (Fig. 2). These data show that, as expected from its poor fit (9/15 bp) to the consensus for Lrp binding proposed by Cui et al. (13), Lrp does indeed bind to site 3 with affinity lower than that for either site 1 or site 2. However, we note also that although site 2 resembles the consensus for Lrp binding proposed by Cui et al. more than site 1 does (11/15 bp and 13/15 bp, respectively), site 1 actually binds the protein with higher affinity.
FIG. 2.
Quantitation of Lrp binding to each individual site in the fim switch and for site 3 replaced with site 2 (2 at 3). Binding affinity is relative to a 29-bp control fragment described in Materials and Methods. Values shown are the averages of four experiments.
Replacement of Lrp binding site 3 with Lrp binding site 2.
The analysis of individual binding site affinities described above suggested that it would be possible to prevent Lrp dissociation from site 3 by increasing the binding affinity of this site. To test this idea, we replaced Lrp site 3 with site 2 and characterized the interaction of Lrp with the mutant DNA in vitro in the absence and presence of IVLA.
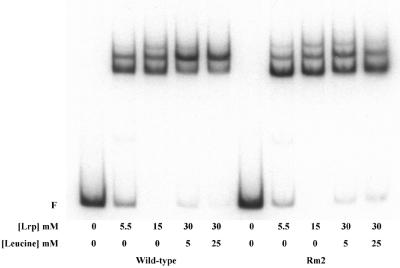
Site-directed mutagenesis was used to change the nucleotide sequence of site 3 from 5′-GAGAGAAGAATGAGG to 5′-TGGCTTAATATTCTA (changes are underlined). The effect of this mutation, Rm2, on Lrp binding to the fim switch DNA was examined by EMSA (Fig. 3) and MPE-Fe(II) footprinting (data not shown). The relative affinity of Lrp binding to site 3 replaced with site 2 (including the adjacent 7 bp on either side of site 3) was measured as described above. The relative affinity was slightly stronger than that of site 2 in its normal context, which suggests the possible involvement of sequences adjacent to the binding site in overall binding affinity (Fig. 2). In EMSAs, using a 200-bp DNA fragment that encompasses Lrp binding sites 1, 2, and 3 (PCR12, as described in reference 40) with concentrations of 5.5 nM to 30 nM Lrp, the complexes formed with the mutant template appeared virtually identical to those formed with the wild-type template (Fig. 3). However, upon the addition of 5 mM and 25 mM leucine (30 nM Lrp), the mutant template did not exhibit the pronounced enhancement of the second, more slowly migrating complex that was seen with the wild-type template. The Kd of Lrp binding to the mutant template was decreased less than twofold relative to that seen with the wild type (data not shown). MPE-Fe(II) footprinting analysis demonstrated that Lrp remained bound to site 3 even in the presence of 20 mM leucine (data not shown). While we do not understand why the loss of Lrp from binding to site 3 to form a complex with sites 1 and 2 alone produces a decrease in mobility in EMSA (40), these results show that mutation Rm2 enhances the binding of the protein to site 3 in the presence of leucine, as expected.
FIG. 3.
Electrophoretic mobility shift assay of Lrp binding to the wild type and to the Rm2 invertible element. The radiolabeled PCR products, encompassing all four known or potential Lrp binding sites (with flanking sequences extending to 17 bp beyond potential binding site 4 and 77 bp beyond binding site 1), were incubated with the specified amounts of purified Lrp and 5 mM and 25 mM leucine where indicated. Samples were separated into free (F) and bound species by polyacrylamide gel electrophoresis.
The effects of the Rm2 allele on FimB recombination in vivo.
The availability of the Rm2 mutation allowed us to address several key questions about the control of FimB recombination and to test and refine our current model. According to our model, the loss of Lrp binding to Lrp site 3 in the presence of IVLA, rather than other changes in Lrp induced by the amino acids, accounts for their stimulatory effects on FimB recombination. To test this hypothesis, the frequency of FimB recombination was measured in a mutant containing the Rm2 (BGEC771) mutation (Table 2), and, as expected, a sharp decrease in responsiveness to the amino acids (2.9-fold versus 5.7-fold) was observed. However, strain BGEC771 did retain a degree of sensitivity to IVLA stimulation, and we therefore cannot exclude the possibility that alterations in the interaction of Lrp with site 3 in the presence of the amino acids other than complete loss of binding to the element also stimulate FimB recombination.
To probe the function of the central portion of the fim invertible element, an additional mutant (KCEC047) that contains the replacement of this region (Rm1) together with the Lrp site 3-enhanced Lrp binding allele (Rm2) was constructed and characterized (Table 2). In this double mutant background (Rm3), the stimulatory effect of the Rm1 mutation was diminished in the absence of the amino acids (from 8.4-fold to 3.3-fold) considerably more than in their presence (from 2.2-fold to 1.7-fold). Thus, the central portion of the fim invertible element apparently plays a regulatory role in the response of FimB recombination to exogenous IVLA. However, this effect can be largely overcome by increasing the binding affinity of Lrp for site 3.
In addition to the results described above, a particularly interesting effect of the Rm2 mutation was that it substantially inhibited FimB recombination relative to that of the wild type (approximately 10-fold in the absence of the amino acids) (Table 2). Thus, even in the absence of exogenous IVLA, FimB recombination is likely to be restricted to cells in which Lrp site 3 is unbound by the regulator.
Analysis of a potential Lrp binding site in the central portion of the fim invertible element.
The results described above prompted us to reconsider the possibility that Lrp binds to an additional element or elements within the central portion of the fim invertible element to repress FimB recombination. The sequence was searched for potential Lrp binding sites that resemble the consensus 5′-YAGHAWATTWTDCTR (13), and the results of this analysis highlighted a possible site (5′-AAGCTAACGTTTCTG) which matches the consensus well (at the 11 positions underlined) and which lies at the same distance from Lrp site 3 as do site 3 from site 1 and site 1 from site 2 (Fig. 1).
To test the hypothesis that the loss of an interaction of Lrp with this element can explain the effect of the Rm1 mutation on the amino acid regulation of FimB recombination, the sequence was mutated by site-directed mutagenesis and its effects were characterized. The proposed element was altered at the positions underlined to 5′-ATACTAACGGACGAT (Rm4) to decrease the match from consensus by eight. When the mutation was transferred into the genome at fim by allelic exchange, it stimulated off-to-on recombination in the absence but not in the presence of the amino acids, as expected. However, the effect observed was modest (1.4- to 2.0-fold increase in recombination) (Table 3). In order to reduce the possibility that Rm4 simply failed to inhibit Lrp binding to the site by chance, a second mutation (Rm5; 5′-AAGCTAACGTTTCTG to 5′-AAGGATCCCAAAGAC) that also alters the sequence substantially was constructed and characterized. As with Rm4, Rm5 stimulated FimB recombination in the absence but not in the presence of IVLA (Table 3), but again it produced only a small (1.4- to 2.3-fold) effect.
TABLE 3.
FimB recombination in Rm mutants 4 to 11 grown to exponential phase in minimal MOPS glucose medium in the absence and presence of IVLAa
| Mutation | IVLA | Inversion frequency (104)b | Ratio of mutant to wild typeb |
|---|---|---|---|
| Rm4 | − | 36.3-42.1 | 1.4-2.0 |
| + | 59.0-61.0 | 0.8-1.1 | |
| Rm5 | − | 25.4-29.7 | 1.4-2.3 |
| + | 83.1-90.7 | 0.7-0.9 | |
| Rm6 | − | 36.8-39.2 | 1.2-1.4 |
| + | 114.0-137.0 | 1.4-1.7 | |
| Rm7 | − | 18.5-25.1 | 0.7-0.9 |
| + | 78.8-131.0 | 1.1-1.3 | |
| Rm8 | − | 5.5-10.8 | 0.2-0.4 |
| + | 62.5-97.7 | 0.9-1.0 | |
| Rm9 | − | 27.0-40.7 | 1.4 |
| + | 52.0-122.0 | 1.0-1.6 | |
| Rm10 | − | 5.8-8.26 | 0.3-0.5 |
| + | 77.2-88.4 | 0.9-0.7 | |
| Rm11 | − | 31.5-45.3 | 1.9-2.2 |
| + | 149-210 | 1.5 |
Values obtained in a duplicate experiment are shown.
Per cell per generation for inversion from the off-to-on orientation. Ninety-five-percent confidence limits, determined from at least seven independent cultures, lie within 10% of the mean.
The results described above indicate that Lrp may bind to the additional element identified here, even though this seems unlikely to account fully for the effect that the Rm1 mutation has on the amino acid regulation of FimB recombination. In previous studies, EMSAs were used to examine Lrp binding to sites 1, 2, and 3 together, and it was determined that the Kd for purified Lrp binding (determined as the Lrp concentration that resulted in a 50% loss of free DNA) to these sites was 2.4 ± 0.3 nM (40). We note that we were unable to demonstrate an interaction of Lrp with the region replaced in Rm1 in vitro previously (21, 40). Furthermore, the Rm5 mutation had no detectable effect on the interaction of Lrp with a PCR product in EMSA (PCR12 [40]) that contains the putative binding site together with Lrp sites 1 to 3 either in the presence or in the absence of IVLA (data not shown). Thus, if Lrp does bind to the potential novel site identified here, the interaction is likely to be of a low affinity and is not readily detected in vitro.
Scanning and additional replacement mutagenesis of the central portion of the fim invertible element.
To search for the additional sequence or sequences within the central portion of the fim invertible element involved in controlling FimB recombination, four smaller adjacent replacement mutations (Rm6 to -9) that collectively span the sequences replaced in the 59-bp Rm1 mutation were constructed and characterized (Fig. 1 and Table 3).
In agreement with the results obtained from the directed mutagenesis of the potential Lrp binding site described above, both the Rm8 and Rm9 mutations mostly produced a specific effect on FimB recombination in the absence of IVLA. However, the effect produced was again relatively small. Surprisingly, Rm8 (and to a lesser extent, Rm7) actually decreased FimB recombination specifically in the absence of the amino acids. Thus, in the Rm8 mutant strain, the stimulatory effect of IVLA on recombination was increased substantially (from 4.7-fold to over 15-fold).
To define the regions within the replacements that control FimB recombination in more detail, an additional mutation (Rm10) that spans the junction between Rm7 and Rm8 (5′-TATATGT to 5′-ATAGGCA) was constructed and analyzed. The results of this experiment showed that the mutation produced an effect comparable to that of Rm7, resulting in an inhibition of FimB recombination that was more pronounced in the absence of IVLA (Table 3). We note that although mutation Rm7 does extend into the near-consensus sequence for Lrp binding described above, Rm10 does not.
To account for the results described above, we suggest that the Lrp site 3-proximal half of the central portion of the fim invertible element contains multiple elements that participate in the response of FimB to IVLA. According to this hypothesis, two or more alternative negative and functionally redundant elements (possibly including the putative fourth Lrp site) act in concert with Lrp site 3 to inhibit recombination, and a positive element (mutation in Rm7 and Rm10) antagonizes this effect partially under the conditions studied. In support of this concept, it was found that the inhibitory effect of Rm10 on FimB recombination in the absence of IVLA was suppressed completely when this mutation was combined with Rm5 (Rm11) (Table 3).
In contrast to the effects described above, mutation Rm6 showed a small but reproducible increase in FimB inversion irrespective of the presence of IVLA. Thus, it appears that the IHF site II-proximal part of the region, replaced in Rm1, contains sequences that repress FimB recombination irrespective of the presence of IVLA. We note that the loss of this region would explain why the replacement of the entire central portion of the fim invertible element also increases FimB recombination in the presence of IVLA.
DISCUSSION
Recombination of the fim invertible element is inhibited in an lrp mutant (5). Lrp binds with high affinity (Kd of 2.4 ± 0.3 nM) to the invertible region in vitro (20), and mutations that diminish binding to two sites within this region (sites 1 and 2) produce a parallel decrease in recombination in vivo (20). Leucine decreases Lrp's affinity for DNA (18, 33, 39, 50), but, as expected, it has little effect on the Kd of Lrp binding to fim (40). Despite this, the branched-chain amino acids (particularly leucine) and alanine stimulate the fim inversions (21). Further work reveals that Lrp binds cooperatively to a third site within the invertible region (site 3, forming complex 1) and that leucine promotes the selective loss of the protein from this element in vitro (forming complex 2 [40]). Since a mutation (fimS21) that blocks binding to site 3 leads both to a loss of IVLA stimulation of recombination and to a high constitutive level of recombination in the absence of the amino acids, it was proposed that the dissociation of Lrp from site 3 in vivo explains how the amino acids stimulate the fim inversion (40).
It was suggested previously that Lrp and IHF bound within the fim invertible element serve together to loop the DNA to bring the sites of recombination into a juxtaposition to favor productive synapses (4). Within the confines of the invertible region of only approximately 314 bp in length, the protein-induced bends should impose a tight topological constraint on the system. In line with this model, we suppose that Lrp bound to sites 1 and 2 (complex 2) forms a nucleoprotein structure that promotes recombination, whereas interaction of the protein with all three sites (complex 1) is less favorable for inversion.
A significant finding of this study is that the replacement of the entire central portion of the invertible element (a 59-bp region between Lrp site 3 and IHF site II) with heterologous DNA still permits high levels of FimB recombination. However, the replacement mutation leads to an almost complete loss of IVLA stimulation of FimB recombination, indicating to us that either (i) Lrp complex 1 is necessary but not sufficient for the inhibition of inversion in the absence of the amino acids or (ii) the central portion of the fim invertible element contains sequences that enhance the interaction of Lrp with site 3 within complex 1.
An unresolved question addressed here is why leucine should exert a specific effect on Lrp bound to site 3. All of the Lrp binding sites at fim are separated from their neighbors by an identical spacing of around two helical turns (20, 40), and although leucine decreases binding to individual sites, it increases cooperativity between sites. Two lines of evidence presented in this study suggest that the effect of leucine on Lrp bound to site 3 can be attributed simply to the low affinity of this site for the DNA binding protein. First, when the relative binding affinities for the individual sites were measured, it was found that site 3 was the weakest of the three. Second, replacing Lrp site 3 with site 2 (Rm2) inhibited the ability of leucine to promote the selective dissociation of Lrp from site 3 in vitro.
The availability of the Rm2 mutation allowed us to test further our current model for how IVLA controls the fim recombination. As expected, we found that the mutation lead to a decrease in amino acid stimulation of the inversion. However, the recombination reaction was still affected by IVLA in the mutant background, and we therefore cannot rule out the possibility that changes in the activity of Lrp bound to site 3, in addition to a loss of binding to the element, contribute to the control. We note that leucine alters the oligomeric state of Lrp, and changes in the activity of the regulator other than DNA binding have been implicated in the control of the dadAX operon in response to alanine and leucine (11, 12, 51).
In addition to the results discussed above, we found that the effect of replacing the central portion of the fim invertible element on FimB recombination can be largely suppressed by the Rm2 mutation. These data support the hypothesis that the region between IHF site II and Lrp site 3 participates in the regulation of recombination in response to IVLA in an auxiliary role, presumably enhancing Lrp binding to site 3 rather than being an indispensable component of a nucleoprotein complex that inhibits recombination. However, further work to map and characterize the control elements within the central portion of the fim invertible region, although showing that the IRL-proximal (Lrp site 3-proximal) half is involved in the response of FimB to IVLA, highlights the apparent complexity of the system. We first considered the idea that the interaction of Lrp with a fourth binding site (5′-AAGCTAACGTTTCTG) accounts fully for the effects of the larger replacement mutation on FimB recombination. However, although most of the mutations in this sequence did affect FimB recombination as expected, the effects observed were always comparatively modest. Moreover, it remains unclear whether Lrp interacts with this sequence or not, as we have been unable to show binding to the putative fourth site in vitro, either alone or in a complex with sites 1 to 3 (21, 40; also data not shown). Thus, it seems unlikely to us that an interaction of Lrp with the potential fourth site studied here accounts fully for the effects that the larger Rm1 mutation has on the control of FimB recombination in response to IVLA. We suggest instead that the region contains at least two alternative control elements that function in concert with Lrp site 3 to repress FimB recombination.
A surprising result of this study was that mutations near the center of the invertible region (mutations Rm8 and Rm10) and over 30 bp from Lrp site 3 inhibit FimB recombination specifically in the absence of the amino acids. As Rm8 alters the putative fourth Lrp site, we suppose that, by chance, it could increase the Lrp binding to this element. Likewise, we have not ruled out the possibility that Rm10 has a similar effect, even though it does not alter the putative consensuslike sequence itself. However, these possibilities seem unlikely to us, and to account for the results obtained we suggest that the action of one (or more) negative control element(s) is antagonized partially under the conditions studied by an additional regulatory site mutated in both Rm8 and Rm10.
From results published previously, we were unable to tell whether recombination in the absence of the exogenous amino acids utilizes complex 1 (Lrp bound to sites 1 to 3) or 2 (Lrp bound to sites 1 and 2 alone) (40). An important finding reported here is that the mutation that enhances Lrp binding to site 3 (Rm2) decreased FimB recombination substantially (10-fold) even in the absence of exogenous IVLA. Thus, FimB recombination is likely to be restricted mostly to cells in which Lrp site 3 is unbound by the regulator even in the absence of exogenous IVLA. lrp expression is increased under less-favorable growth conditions (32), and thus our results lead us to predict that Lrp site 3 and IVLA will have a more substantial effect on the fim inversion at growth rates lower than those tested here (glucose minimal MOPS medium [37]). Furthermore, starvation for IVLA should increase occupancy of site 3 and hence inhibit FimB recombination.
The interaction of Lrp with the GATCdist binding sites in the papBA control region is stimulated by PapI (28). The results presented here raise the possibility that the activity of Lrp as a repressor of recombination in complex 1 could also be controlled by one or more additional factors that interact with the adjacent portion of the fim invertible region. Thus, the effects of IVLA on fim phase variation may themselves be regulated in response to additional environmental conditions. Although the physiological function of the regulation of fim phase variation by IVLA is unknown, it was proposed recently that the control of FimB by sialic acid and by N-acetylglucosamine decreases the expression of the adhesin in response to the activation of host defenses (17, 44). Type 1 fimbriae mediate proinflammatory effects by stimulating the release of interleukin-6, interleukin-8, and tumor necrosis factor alpha and act synergistically with other bacterial products, such as Lps and formylated peptides (22, 23, 34, 42, 43) The biosynthesis of the branched-chain amino acids is severely restricted by the superoxide anion, since dihydroxyacid dehydratase is inhibited by this agent (27). Thus, the control of fim phase variation in response to low levels of the branched-chain amino acids could limit the adhesins' expression in the course of inflammation to help to maintain a balanced interaction between E. coli and its hosts.
Acknowledgments
We thank Alice Bonnen, Nadja Dreesbeimdieke, Sammia El-Labany, Simon Friar, Sara Sanchez-Perales, and Shadi Shafei for technical assistance. We also thank Eric Roesch and the DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University.
The DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University was supported in part by NIH grant CA-12197. I.C.B. was supported by NIH grant GM50406 and by grant 96/P12206 from the BBSRC. P.L.R. was supported in part by NIH Training Grant T32 AI07401 as a predoctoral student, and M.L. was supported by grant 96/P12206 from the BBSRC.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljundquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. Narayana, and L. S. Pierson III. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R, Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Blomfield, I. C., D. H. Kulasekara, and B. I. Eisenstein. 1997. Integration host factor stimulates both FimB- and FimE-mediated site specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol. Microbiol. 23:705-717. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. MacClain, and B. I. Eisenstein. 1993. Lrp stimulates the phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., M. S. McClain, and B. I. Eisenstein. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439-1445. [DOI] [PubMed] [Google Scholar]
- 7.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 8.Blomfield, I. C., and M. van der Woude. 2002. Regulation and function of phase variation in Escherichia coli, p. 89-113. In M. Wilson (ed.), Advances in molecular and cellular biology, vol. 1. Bacterial adhesion to host tissues: mechanisms and consequences. Cambridge University Press, Cambridge, England. [Google Scholar]
- 9.Braaten, B. A., J. V. Platko, M. W. van der Woude, B. H. Simons, F. K. de Graaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein controls the expression of both pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S., and J. M. Calvo. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318:1031-1042. [DOI] [PubMed] [Google Scholar]
- 12.Chen, S., M. H. Rosner, and J. M. Calvo. 2001. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312:625-635. [DOI] [PubMed] [Google Scholar]
- 13.Cui, Y., Q. Wang, G. D. Stormo, and J. M. Calvo. 1995. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 177:4872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman, C. J., and C. F. Higgins. 1987. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J. Bacteriol. 169:3840-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dybvig, K. 1993. DNA rearrangements and phenotypic switching in prokaryotes. Mol. Microbiol. 10:465-471. [DOI] [PubMed] [Google Scholar]
- 16.Eisenstein, B. I., D. S. Sweet, V. Vaughn, and D. I. Friedman. 1987. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:6506-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49:1109-1118. [DOI] [PubMed] [Google Scholar]
- 18.Ernsting, B. R., J. W. Denninger, R. M. Blumenthal, and R. G. Matthews. 1993. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J. Bacteriol. 175:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gally, D. L., J. Leathart, and I. C. Blomfield. 1996. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 21:725-738. [DOI] [PubMed] [Google Scholar]
- 20.Gally, D. L., T. J. Rucker, and I. C. Blomfield. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godaly, G., B. Frendeus, A. Proudfoot, M. Svensson, P. Klemm, and C. Svanborg. 1998. Role of fimbriae-mediated adherence for neutrophil migration across Escherichia coli-infected epithelial cell layers. Mol. Microbiol. 30:725-735. [DOI] [PubMed] [Google Scholar]
- 23.Goetz, M. B. 1989. Priming of polymorphonuclear neutrophilic leukocyte oxidative activity by type 1 pili from Escherichia coli. J. Infect. Dis. 159:533-542. [DOI] [PubMed] [Google Scholar]
- 24.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 27.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 28.Kaltenbach, L. S., B. A. Braaten, and D. A. Low. 1995. Specific binding of PapI to Lrp-pap DNA complexes. J. Bacteriol. 177:6449-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemm, P. 1984. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Eur. J. Biochem. 143:395-399. [DOI] [PubMed] [Google Scholar]
- 30.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosturko, L. D., E. Daub, and H. Murialdo. 1989. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 17:317-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgraf, J. R., J. Wu., and J. M. Calvo. 1996. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 178:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, R., B. Ernsting, I. N. Hirshfield, R. G. Matthews, F. C. Neidhardt, R. L. Clark, and E. B. Newman. 1992. The lrp gene product regulates expression of lysU in Escherichia coli K-12. J. Bacteriol. 174:2779-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malaviya, R., T. Ikeda, E. Ross, and S. N. Abraham. 1996. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature (London) 381:77-80. [DOI] [PubMed] [Google Scholar]
- 35.Martin, C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and l-alanine at the transcriptional level. Mol. Microbiol. 21:281-292. [DOI] [PubMed] [Google Scholar]
- 36.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308-53014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture media for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman, E. B., and R. Lin. 1995. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 49:747-775. [DOI] [PubMed] [Google Scholar]
- 39.Ricca, E., D. A. Aker, and J. M. Calvo. 1989. A protein that binds to the regulatory region of the Escherichia coli ilvIH operon. J. Bacteriol. 171:1658-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roesch, P. L., and I. C. Blomfield. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 27:751-761. [DOI] [PubMed] [Google Scholar]
- 41.Sadowski, P. D. 1993. Site-specific genetic recombination: hops, flips, and flops. FASEB J. 7:760-767. [DOI] [PubMed] [Google Scholar]
- 42.Samuelsson, P., L. Hang, B. Wullt, H. Irjala, and C. Svanborg. 2004. Toll-like receptor 4 expression and cytokine responses in the human urinary tract mucosa. Infect. Immun. 72:3179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilling, J. D., S. M. Martin, D. A. Hunstad, K. P. Patel, M. A. Mulvey, S. S. Justice, R. G. Lorenz, and S. J. Hultgren. 2003. CD14- and Toll-like receptor-dependent activation of bladder epithelial cells by lipopolysaccharide and type 1 piliated Escherichia coli. Infect. Immun. 71:1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohanpal, B. K., S. El-Labany, M. Lahooti, J. A. Plumbridge, and I. C. Blomfield. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 101:16322-16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutcliffe, J. G. 1979. Complete nucleotide sequence of Escherichia coli plasmid pBR322. Cold Spring Harbor Symp. Quant. Biol. 43:77-90. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: application to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Woude, M. W., and D. A. Low. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the E. coli sfa and daa pili operons. Mol. Microbiol. 11:605-618. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q., and J. M. Calvo. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 12:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiese, D. E., B. R. Ernsting, R. M. Blumenthal, and R. G. Matthews. 1997. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J. Mol. Biol. 270:152-168. [DOI] [PubMed] [Google Scholar]
- 50.Willins, A. K., C. W. Ryan, J. V. Platko, and J. M. Calvo. 1991. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem. 266:10768-10774. [PubMed] [Google Scholar]
- 51.Zhi, J., E. Mathew, and M. Freundlich. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32:29-40. [DOI] [PubMed] [Google Scholar]