Abstract
Though careful consideration has been placed towards genetic characterization of tubercle bacillus isolates causing disease in humans, those causing disease predominantly among wild and domesticated mammals have received less attention. In contrast to Mycobacterium tuberculosis, whose host range is largely specific to humans, M. bovis and “M bovis-like” organisms infect a broad range of animal species beyond their most prominent host in cattle. To determine whether strains of variable genomic content are associated with distinct distributions of disease, the DNA contents of M. bovis or M. bovis-like isolates from a variety of hosts were investigated via Affymetrix GeneChip. Consistent with previous genomic analysis of the M. tuberculosis complex (MTC), large sequence polymorphisms of putative diagnostic and biological consequence were able to unambiguously distinguish interrogated isolates. The distribution of deleted regions indicates organisms genomically removed from M. bovis and also points to structured genomic variability within M. bovis. Certain genomic profiles spanned a variety of hosts but were clustered by geography, while others associated primarily with host type. In contrast to the prevailing assumption that M. bovis has broad host capacity, genomic profiles suggest that distinct MTC lineages differentially infect a variety of mammals. From this, a phylogenetic stratification of genotypes offers a predictive framework upon which to base future genetic and phenotypic studies of the MTC.
The Mycobacterium tuberculosis complex (MTC) is comprised of bacterial organisms that genetically share identical 16S rRNA sequences (6) and over 99.9% nucleotide identity (49). Classical MTC species, namely, Mycobacterium tuberculosis, Mycobacterium africanum, Mycobacterium microti, and Mycobacterium bovis, can be categorized according to a restricted number of laboratory phenotypes but, importantly, differ in physiological characteristics, virulence, and host range. Though it has been conventionally established that M. tuberculosis and M. africanum are isolated from humans, M. microti from voles, and M. bovis predominantly from cows, reports of MTC organisms in a variety of other domesticated and undomesticated hosts pose a challenge to this classification scheme. The host range of M. bovis is considered to be the broadest of the complex, causing disease across a variety of animals, including cattle, seals, and goats, but seldom in humans since the introduction of pasteurized dairy products (41), though in areas where tuberculosis (TB) is endemic, the exact proportion of human disease due to M. bovis is largely unknown (3). More recently, MTC organisms of goats and seals have been named, respectively, Mycobacterium caprae (1) and Mycobacterium pinnipedii (13), although the former is more commonly referred to as Mycobacterium bovis subsp. caprae and the latter was previously identified as a form of M. bovis. In contrast to the careful consideration placed towards differentiation of isolates causing TB in humans (22), those causing disease predominantly among wild and domesticated mammals have received less attention.
With the availability of genomic data from sequenced strains of the MTC (7, 11, 16, 18) and tools of comparative genomics (5, 45), it is now recognized that large sequence polymorphisms (LSPs) deleted from M. tuberculosis serve as accurate markers for diagnostic testing (23, 42) and, with the exception of mycobacteriophage DNA, as evolutionary markers (5, 8, 22, 24, 35, 36, 39, 50). As such, the presence or absence of these LSPs has been used to reveal associations between strains and host populations (22, 34, 37). By extension, genomic information about any LSPs specific to M. bovis and “M. bovis-like” organisms could guide in assessing the prevalence of the organisms through space and host type. A practical utility of this approach would be to provide a secure genomic definition for M. bovis and to help determine whether M. bovis is a single discrete genomic entity or instead a genomic continuum comprising strains that vary according to a number of properties, including (but not limited to) virulence and host preference.
We therefore selected M. bovis and M. bovis-like organisms isolated from various mammalian host types of diverse geographic origins, which present an array of genetic profiles and laboratory properties. The DNA of these organisms was then interrogated via Affymetrix GeneChip, followed by confirmatory PCR and sequencing. The results obtained reveal the previously unknown evolution of the MTC, provide markers for diagnostic testing and molecular epidemiologic assessment, and point to both geographic and host-specific forms of genomic variability.
MATERIALS AND METHODS
Bacterial isolates.
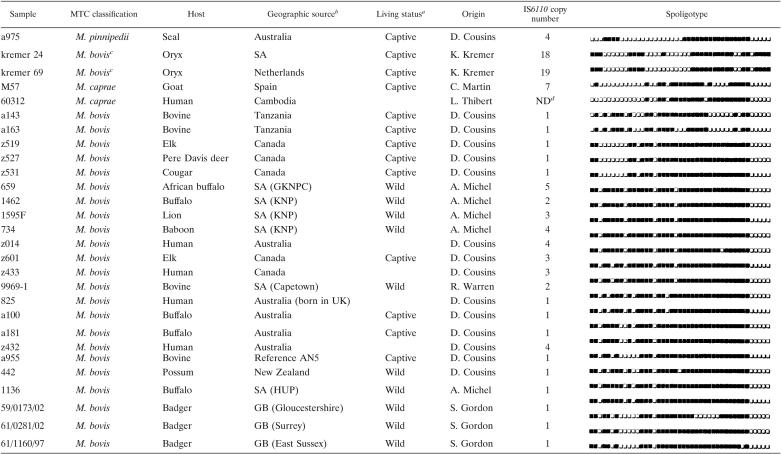
Twenty-eight MTC isolates were selected for whole-genome analysis. Isolated from a variety of mammalian hosts, their laboratory classifications and countries of origin are provided in Table 1. Beyond isolates characterized as M. bovis, also included are isolates classified as M. pinnipedii (also referred to as the “seal bacillus” [13], “oryx bacillus,” and M. caprae, because their distinction from M. bovis is still under debate [1, 13, 27]). To characterize these isolates before genomic analysis via genetic signatures undetectable to GeneChip analysis, IS6110 copy numbers (52) and spoligotypes (2) were evaluated by the original sample provider and/or specifically for this study (Table 1).
TABLE 1.
Characteristics of bacterial isolates used in this study
Captive implies farmed, domesticated, or living in a zoo, and wild implies undomesticated.
SA, South Africa; KNP, Kruger National Park; GKNPC, Greater Kruger National Park Complex; HUP, Hluhluwe-Umfolozi Park, Kwazulu-Natal; GB, Great Britain; and UK, United Kingdom.
Despite elsewhere presenting a differential genetic profile (8), “kremer 24” and “kremer 69” represent isolates originally classified as M. bovis.
ND, not determined.
In addition, 18 geographically diverse isolates of M. pinnipedii were tested via PCR for genomic deletions exclusively identified for the isolate of M. pinnipedii (a975) applied to GeneChip; Table S1 (http://www.molepi.mcgill.ca/bcg.htm) describes their origins and characterizations. (All supplemental material cited below can also be accessed at http://www.molepi.mcgill.ca/bcg.htm.) As well, 15 Spanish isolates of M. caprae presenting diverse genetic profiles were tested via PCR for genomic deletions specific for the Spanish isolate of M. caprae (M57) applied to GeneChip (Table S2 in the supplemental material). Finally, 154 isolates of M. bovis of diverse spoligotypes from cattle, badger, and deer hosts (Table S3 in the supplemental material) were tested via PCR for genomic deletions revealed from GeneChip analysis of Great Britain (GB) M. bovis isolates from a badger host (59/0173/02, 61/0281/02, and 61/1160/97).
GeneChip analysis.
The M. tuberculosis Affymetrix GeneChip is an oligonucleotide array representing sequences from the M. tuberculosis genome (45). When applied towards MTC genomic interrogation, the GeneChip is capable of identifying LSPs of hundreds of nucleotides or larger with respect to M. tuberculosis. Eight micrograms of MTC DNA was extracted, fragmented, biotin labeled, and hybridized to the GeneChip as previously described (45). Where necessary, extracted DNA was amplified using a GenomiPhi DNA amplification kit (Amersham Biosciences) to ensure that an appropriate quantity of DNA could be advanced to GeneChip and/or downstream PCR analysis. Affymetrix Microarray Suite (MAS) was used to analyze the comparative genomic data and ultimately suggest candidate deleted regions (34, 36, 37, 39).
PCR amplification and sequencing across deletions.
Candidate deleted regions inferred by GeneChip analysis were pursued as described in previous publications (34, 36, 37, 39). In brief, primers were designed to amplify regions in M. tuberculosis H37Rv flanking the putative deletion. The test strain(s) presenting the candidate deleted region, together with referent strains of M. tuberculosis and/or M. bovis BCG (included as DNA controls), was concurrently subjected to PCR amplification. PCR amplicons were run on 2% agarose gel, where products different from the expected base pair size of M. tuberculosis H37Rv could be visualized. All products obtained by amplification across a deleted region were subsequently sequenced by dideoxy terminal sequencing at the McGill University and Genome Quebec Innovation Center. Sequence results were compared by BLAST analysis to M. tuberculosis H37Rv with TubercuList (http://genolist.pasteur.fr/TubercuList/) and to M. bovis AF2122/97 with BoviList (http://genolist.pasteur.fr/BoviList/) to confirm whether the amplified MTC DNA harbored a specific deletion event. Annotation of open reading frames (ORFs) affected by deletion events was determined by using TubercuList. Nomenclature of deleted regions disclosed in this study, hereafter referred to as “new deletions,” follows previously established guidelines (7, 32, 34) and primarily denotes the MTC lineage from which strains harboring that region of difference (RD) can be specifically isolated. Nomenclature for new deletions further reflects an already published RD should it overlap the genetic region affected by a newfound RD, deleted genes for which a function has already been ascribed, the host's geographic source, and/or is numbered according to its order of discovery.
Analysis of deletions.
Primers designed to amplify new deletions are provided as supplemental material (Table S4). A genomic deletion, herein defined as a region of the ancestral genome that has been deleted only from a subgroup of clonally related organisms, is only considered a unidirectional deletion event if confirmed by PCR and sequence analysis (22, 35). For isolates applied to GeneChip, regions were called present if denoted as such via MAS. Conversely, new deletions among interrogated isolates were sequence confirmed to determine whether the exact same genomic event was being identified. The distributions of deletions already described as absent from M. bovis (5, 18, 20) were based on MAS analysis and were not pursued by further analysis. Deletion events were afterward assigned to the previously characterized bacterial isolates. Regions of difference specific to M. tuberculosis (8), namely, TbD1, RvD1, RvD2, RvD3, RvD4, and RvD5, are not available for GeneChip analysis. To confirm the presence of these regions across isolates applied to GeneChip, PCR primers designed to amplify genes internal to these deletion events were applied (Table S4).
RESULTS
Due to the close genetic relationship shared by MTC members, most GeneChip probes hybridized as present for interrogated MTC genomic DNA, whereas a marginal or absent hybridization signal was produced for regions of M. tuberculosis already described as being deleted from M. bovis (5, 18, 20) or other MTC members (32). Previously described mycobacteriophages RD3 (or phiRv1) (31) and RD11 (or phiRv2) (5, 20), which are known to have been independently deleted throughout the MTC and are thus uninformative as phylogenetic markers (8, 22, 35), were also observed as variably missing. Apart from these RDs, genomic interrogation of isolates did not reveal any deletions with junctions matching those previously described for other MTC members, including M. canetti (32; S. Mostowy and M. Behr, unpublished data), M. tuberculosis (24, 50), M. bovis (5, 20, 44, 45), M. africanum (37), M. microti (7, 17), and the “dassie bacillus” (34). With the exception of the RvD1 sequence from M. caprae M57, the sequence within deletion events specific to M. tuberculosis (8) amplified as intact for all isolates applied to Chip (data not shown). Though the absence of RvD2, RvD3, and RvD4 was initially suggested for oryx bacillus isolates (8), the absence of the sequence within these regions is not due to the same genetic event as that for isolates of M. tuberculosis but results from independent genetic events unique to the oryx bacillus described below.
A total of 29 new deletions were confirmed, affecting 152,480 bp and 164 ORFs (Table 2). The distribution of these deletions across isolates is presented in Table 3, providing distinct genomic groupings observed to correlate with spoligotypes that resembled each other (Table 1), and is independently discussed below for isolates classified as M. pinnipedii, oryx bacillus, M. caprae, and M. bovis sensu stricto (Fig. 1).
TABLE 2.
Confirmed new deletions among isolates, determined by GeneChip analysisa
| Deleted regionb | Start | End | Length (bp) | Affected ORF(s) |
|---|---|---|---|---|
| RDcap_Spain1 | 29988 | 34322 | 4,334 | Rv0026-Rv0032 |
| RDcap_Spain2 | 136718 | 138163 | 1,445 | Rv0112-Rv0114 |
| RDcap_Spain3 | 188885 | 190193 | 1,308 | Rv0160c |
| RDbovis(d)_buff1 | 484396 | 485448 | 1,052 | Rv0404 |
| RDbovis(d)_sigK | 533232 | 534213 | 981 | Rv0444c-Rv0445c |
| RDbovis(d)_1160_1 | 669913 | 683322 | 13,409 | Rv0576-Rv0585c |
| RDbovis(a)_kdp | 1151879 | 1154485 | 2,606 | Rv1028A-Rv1030 |
| RDcap_Asia1 | 1376215 | 1376864 | 649 | Rv1232c-Rv1233c |
| RDbovis(c)_Kruger | 1523189 | 1547066 | 23,877 | Rv1355c-Rv1374c |
| RDbovis(c)_wbbl2 | 1720074 | 1720719 | 645 | Rv1525 |
| RDoryx_wag22* | 1987198 | 1998793 | 11,595 | Rv1755c-Rv1765c |
| RDcap_Asia2 | 1996618 | 1999708 | 3,090 | Rv1764-Rv1765A |
| RDoryx_1* | 2038717 | 2048279 | 9,562 | Rv1799-Rv1806 |
| RDbovis(d)_0173 | 2180594 | 2187401 | 6,807 | Rv1928c-Rv1936 |
| RDcap_Spain4 | 2195377 | 2198581 | 3,204 | Rv1944c-Rv1947 |
| RDcap_Asia3 | 2359037 | 2362917 | 3,880 | Rv2100-Rv2101 |
| RD5oryx* | 2629041 | 2639541 | 10,500 | Rv2350c-Rv2356c |
| RDcap_Spain5 | 3119192 | 3120524 | 1,332 | Rv2813-Rv2814c |
| RDbovis(c)_virS | 3447448 | 3451242 | 3,794 | Rv3082c-Rv3085 |
| RD12HUP | 3477736 | 3482411 | 4,675 | Rv3109-Rv3115 |
| RD12oryx* | 3479670 | 3491252 | 11,582 | Rv3111-Rv3125c |
| RDoryx_4* | 3549075 | 3555366 | 6,291 | Rv3180c-Rv3189 |
| RDbovis(d)_1160_2 | 3823567 | 3825361 | 1,794 | Rv3403c-Rv3406 |
| RDbovis(c)_fadD18 | 3945597 | 3951329 | 5,732 | Rv3513c-Rv3515c |
| RDpin | 3967680 | 3968975 | 1,295 | Rv3530c-Rv3531c |
| RDbovis(d)_buff2 | 4142144 | 4143784 | 1,640 | Rv3699-Rv3700c |
| RDcap_Spain6 | 4194728 | 4196291 | 1,563 | Rv3743c-Rv3746c |
| RDcap_Spain7 | 4366792 | 4376362 | 9,570 | Rv3884c-Rv3894c |
| RDbovis(a)_Δpan | 4370865 | 4375133 | 4,268 | Rv3887c-Rv3892c |
| Total | 152,480 | 164 |
New deletions are from interrogated isolates relative to M. tuberculosis H37Rv. Deletions are ordered in terms of their locations within the H37Rv genome.
An asterisk indicates that an IS6110 sequence interrupts the truncation point of that deleted region.
TABLE 3.
Distribution of large sequence polymorphisms among isolatesa
| Deleted region | Sample
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a975 | kremer 24 | kremer 69 | M57 | 60312 | a163 | a143 | z531 | z519 | z527 | z014 | 659 | 1462 | 1596F | 734 | z601 | z433 | z432 | 281 | 9969-1 | 825 | a100 | a181 | a955 | 442 | 1136 | 173 | 1160 | |
| RD9 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD2seal | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDpin | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDoryx_wag22 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDoryx_1 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RD5oryx | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RD12oryx | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDoryx_4 | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RD5 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD12 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RD13 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| N-RD25 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RDcap_Spain1 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain2 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain3 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain4 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain5 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain6 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Spain7 | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Asia1 | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Asia2 | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDcap_Asia3 | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RD4 | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RDbovis(a)_kdp | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDbovis(a)_Δpan | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| N-RD17 | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RDpan | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| RDbovis(c)_virS | + | + | + | + | + | + | + | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDbovis(c)_fadD18 | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| RDbovis(c)_Kruger | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RDbovis(c)_wbbl2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | + |
| RD17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − |
| RDbovis(d)_sigK | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + |
| RDbovis(d)_buff1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + |
| RDbovis(d)_buff2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + |
| RD12HUP | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + |
| RDbovis(d)_0173 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| RDbovis(d)_1160_1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| RDbovis(d)_1160_2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| RD3 | − | − | − | + | + | + | + | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | + |
| RD11 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Distribution of regions present (+) or absent (−) among isolates applied to GeneChip interrogation. Isolates are first grouped according to the presence or absence of deletions previously employed in MTC phylogenetic analysis (8, 35) and then grouped by the presence or absence of new deletions listed in Table 2. The distribution of mycobacteriophages RD3 (31) and RD11 (5, 20), known to have been independently deleted throughout the MTC and thus uninformative as phylogenetic markers (8, 22, 35), is also provided.
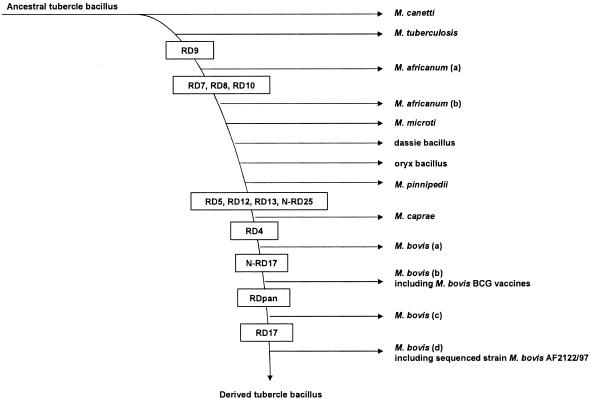
FIG. 1.
Revised deletion-based phylogeny of the MTC, based on deleted regions demonstrated through genomic analysis. The vertical axis presents the stepwise accumulation of unidirectional deletion events (RDs and N [new]-RDs) characterized among members of the MTC. Clustered along each horizontal axis are organisms for which one or more genomic deletions specific to this evolutionary branch have been revealed. Individual deletions revealed herein for M. pinnipedii, oryx bacillus, M. caprae, and multiple genomovars of M. bovis [(a) through (d)], and information about the ORFs they affect, are described in the supplemental material (Table S5). Beyond RD7 to RD10, M. microti, M. africanum (RD7 to RD10 deleted), M. pinnipedii, the dassie bacillus, and the oryx bacillus do not present a unidirectional deletion event shared with M. caprae and/or M. bovis; as a result, their respective order within this MTC phylogenetic model based on genomic deletions is arbitrary.
Deletions specific to M. pinnipedii.
In agreement with previous reports describing the seal bacillus (8, 17, 32, 35), genomic analysis of the M. pinnipedii isolate (a975) revealed the absence of RD7, RD8, RD9, RD10, and RD2seal (Table 3). MiD3 and MiD4, LSPs that involve a highly repetitive sequence in the M. tuberculosis genome (7, 17), were also observed as deleted. Regions described as characteristically deleted from M. caprae and/or M. bovis (RD5, RD12, RD13, N-RD25, and RD4) were all present in M. pinnipedii a975. In addition to these catalogued deletions, we detected in M. pinnipedii a975 one new deletion not observed in other MTC members, RDpin, affecting 1,295 bp and the Rv3530c and Rv3531c (Table 2) genes, annotated as encoding an oxireductase involved in cellular metabolism and a hypothetical protein of unknown function, respectively.
Because M. pinnipedii a975 revealed a unique genomic profile compared to those of other MTC members, deletions observed as specific to it, namely, RD2seal and RDpin, were tested for across a heterogeneous panel of isolates also characterized as M. pinnipedii (Table S1). With all isolates tested lacking both regions, these analyses revealed a common deletion profile for M. pinnipedii, irrespective of geography or pinniped host type. In all, the genomic content of M. pinnipedii isolates suggests a unique phylogenetic position within the MTC (13), genomically distinct from M. bovis (Fig. 1).
Deletions specific to oryx bacillus.
In addition to deletion events shared with other MTC members (Table 3), we detected in both oryx bacillus isolates (kremer 24 and kremer 69) five new deletions not observed in other MTC members (Table 2). Of note, all five new deletions from these isolates (which have 18 and 19 copies of IS6110 [Table 1]) present an IS6110 sequence at their deletion junction, suggesting their mediation of these deletion events (15, 33). Three deletion events specific to M. tuberculosis, RvD2, RvD3, and RvD4 (8), each overlap independent deletion events unique to oryx bacillus, RDoryx_1, RDoryx_wag22, and RD5oryx, respectively.
Deletions specific to oryx bacillus could not be tested across a heterogeneous panel of isolates also characterized as oryx bacillus, because these isolates are rarely studied. Potentially, other MTCs isolated from an oryx host could reveal a common deletion profile for oryx bacillus irrespective of geography or oryx host type, though this remains to be determined. Until then, and in agreement with previous analysis genetically differentiating it from M. bovis (8, 27), including the observation that these two isolates present the only spoligotype pattern to have retained spacers 40 to 43, the oryx bacillus appears to comprise an exclusive phylogenetic position within the MTC (Fig. 1).
Deletions specific to M. caprae.
M. caprae M57, isolated from a caprine host in Spain, was missing seven deleted regions not previously documented, here named RDcap_Spain1 through RDcap_Spain7 (Table 2). Though the region flanking RvD1 appeared intact from GeneChip analysis, primers internal to RvD1 failed to amplify in M57. Seeing that primers for this region amplified as present in all other MTC isolates tested here and elsewhere for MTC other than M. tuberculosis (8), the absence of this internal sequence as it relates to a deletion event specific to isolates of M. caprae requires further investigation. Though M57 contains seven IS6110 elements, these deletions do not appear to be generally mediated by insertion elements but involve the rearrangement of other repeat sequences in the genome, namely, PE (Pro-Glu motif) and PPE (Pro-Pro-Glu motif) genes. Functionally noteworthy among these deletions is RDcap_Spain2, which affects the genes gca (Rv0112), gmhA (Rv0113), and gmhB (Rv0114) involved in the biosynthesis of nucleotide-activated glycero-manno-heptose precursors of lipopolysaccharides (51). Also notable is RDcap_Spain7, which deletes 11 genes (Rv3884c to Rv3894c), including the mycosin mycP2 (9), and the ESAT-6-like proteins encoded by esxC and esxD (19). Genes within this region overlap two independent deletions in M. bovis described below [RDpan and RDbovis(a)_Δpan], suggesting it as a hotspot of genomic loss. M. caprae 60312, isolated from a human with TB who originally came from Cambodia, is not missing any of these seven deletions but instead lacks three genomic regions unique to it, here named RDcap_Asia1 to RDcap_Asia3 (Table 2). The specificities of these deletions from M. caprae are confirmed by the observation that they are not observed as deleted in any M. pinnipedii, oryx bacillus, or M. bovis isolates tested, but their distribution in other settings requires a greater panel of isolates. Importantly, both M. caprae M57 and M. caprae 60312 have retained the RD4 region as intact (Table 3), corroborating their phylogenetic distinction from M. bovis (1, 8, 35) (Fig. 1).
Because M. caprae M57 possessed a new set of deletions potentially specific to caprine isolates from Spain, these deletions were tested for across a diverse panel of isolates previously characterized as M. caprae from different farms across Spain to assess their geographic prevalence (Table S2). A common deletion profile for Spanish isolates of M. caprae was discovered, in that all isolates were missing RDcap_Spain2, RDcap_Spain3, RDcap_Spain5, RDcap_Spain6, and RDcap_Spain7, suggesting regional dominance of a clonal genotype.
Deletions specific to M. bovis.
In deleting RD7, RD8, RD9, RD10, RD5, RD12, RD13, N-RD25, and RD4 (Table 3), the remaining 23 isolates demonstrated the anticipated deletion profile of M. bovis (8, 35). Additionally, 13 new deletions were identified among all M. bovis isolates interrogated via GeneChip (Table 2). Assessing the distribution of these new deletions revealed four distinct genomovars of M. bovis, here referred to as M. bovis (a) through (d), characterized by the presence/absence of N-RD17 (45), a 2,405-bp deletion flanking the PAN promoter region used to identify mycobacteria (44), hereafter referred to as RDpan, and RD17 (21) (Fig. 1).
M. bovis (a).
Unlike all other isolates having deleted RD4, only M. bovis a143 and a163 have retained N-RD17 and RD17 (Table 3). Of note, both of these isolates have deleted RDbovis(a)_Δpan, an independent genetic deletion event with a different deletion location from what has been described for RDpan by Rauzier and colleagues among isolates of M. bovis (44). Furthermore, only a143 and a163 share RDbovis(a)_kdp, a deletion truncating three genes (Rv1028A to Rv1030) annotated as components of the high-affinity ATP-driven potassium transport (or KDP) system. Together, these data suggest that a143 and a163, both isolated from captive bovine hosts inTanzania, represent members of an ancestral M. bovis lineage (Fig. 1).
M. bovis (b).
Though no isolates belonging to this genomovar were directly interrogated for analysis here, M. bovis (b) represents the initial lineage lacking N-RD17. The existence of this genomovar is supported by the observation that M. bovis BCG vaccines previously interrogated via GeneChip analysis (36) share this same deletion profile, pointing to their origin from M. bovis presenting this genotype (Fig. 1).
M. bovis (c).
The deletion of RDpan was observed for a number of M. bovis isolates having one to five copies of IS6110 and infecting multiple animal species (Table 1). Though all retained RD17, isolates belonging to this intermediate genomovar of M. bovis variably presented other new deletions (Table 3).
Three isolates from different hosts in Canada with the same spoligotype pattern shared the genomic deletion of RDbovis(c)_virS affecting Rv3082c to Rv3085. This deletion includes a gene annotated as a virulence regulating transcriptional regulator (virS) that has also been independently deleted from the attenuated dassie bacillus (34). A different deletion, RDbovis(c)_Kruger, was restricted to a subset of isolates from Kruger National Park in South Africa (659, 1462, 1595F, and 734). As was observed with the Canadian deletion, the regional success of a particular clone having deleted RDbovis(c)_Kruger can be inferred from the observation that only isolates from Kruger National Park presented this deletion but that different hosts were implicated (Table 3). RDbovis(c)_Kruger was the largest deletion revealed in this study, affecting 23,877 bp and 20 ORFs, including two genes annotated as transcriptional regulators (Rv1358 and Rv1359) as well as the anti-anti sigma factor rsfA (Rv1365c) involved in regulating SigF (4). The fact that other strains from South Africa (9969-1 and 1136) have not deleted RDbovis(c)_Kruger but instead have deleted RDpan and RD17 indicates at least two genomovars of M. bovis in this region.
M. bovis (d).
This most derivative group of M. bovis was observed for 11 isolates in which RD17 was deleted (Table 3 and Fig. 1). The previously described deletion of RD17 affects treY (Rv1563c), truncating one of three known pathways for the synthesis of trehalose (14, 38). GeneChip analysis revealed seven new deletions unique to isolates missing RD17 (Tables 2 and 3). Of these, one deletion, RDbovis(d)_sigK, disrupts sigK (Rv0445c), an extracytoplasmic sigma factor recently implicated in the control of MPB70/83 production by BCG strains (10). The disruption of sigK function suggests a possible explanation for the occasional lab result where isolates of M. bovis present as negative for MPB70/83 production (29). Another deletion, RD12HUP, was only observed as deleted from a South African M. bovis isolate (1136), affecting 4,675 bp and Rv3109 to Rv3115. RD12HUP removes a cluster of genes annotated as being involved in molybdenum cofactor biosynthesis, just upstream of the RD12 region (Rv3117 to Rv3121) already missing from all isolates of M. caprae and M. bovis.
Specific to isolates of farmed East Asian water buffalo from Australia (a100 and a181), the deletion of RDbovis(d)_buff1 and RDbovis(d)_buff2 potentially indicates a strain of M. bovis that preferentially infects these hosts. Of functional interest from other studies, RDbovis(d)_buff1 deletes 1,052 bp affecting fadD30 (Rv0404), a gene annotated as involved in lipid degradation and found to be essential for in vitro growth in two independent studies (28, 46).
GeneChip analysis of GB M. bovis strains from badgers was performed on isolates from three GB-dominant spoligotype groups (48). Badger isolate 59/0173/02 from Gloucestershire showed the RDbovis(d)_0173 deletion, a lesion that removes genes involved in lipid and intermediary metabolism (Rv1928c to Rv1936) and includes one gene predicted to encode a transcriptional regulatory protein of the AraC family (Rv1931c). The East Sussex badger isolate (61/1160/97) showed a deletion, RDbovis(d)_1160_1, that removes genes encoding a predicted regulator of the ArsR family (Rv0576) and others encoding proteins of unknown function (Rv0576 to Rv0585c). Badger isolate 61/1160/97 also presented a second deletion of almost 1.8 kb that encompassed Rv3403c to Rv3406 and removes a TetR family regulator and a putative dioxygenase [RDbovis(d)_1160_2]. To determine if deletions discovered by array analysis were absent from other M. bovis strains, we then checked a panel of 154 isolates of diverse spoligotypes from cattle, badger, and deer hosts (Table S3). Irrespective of the host type from which these strains were isolated, it was found that new deletions were lost only from the subset of strains sharing the same spoligotype pattern as those applied to array analysis, thereby offering no evidence for a host-specific deletion profile. These data, in agreement with previous genetic analysis of the population structure of M. bovis in GB by spoligotyping and variable number of tandem repeats (48), provide little evidence for the appearance of host-adapted clones in GB.
DISCUSSION
Genome sequencing projects for M. tuberculosis (11, 16), M. microti (7), and M. bovis (18) have confirmed LSPs deleted from M. tuberculosis as an important medium of genomic variability among members of the MTC (8, 35). Genomic deletions have strongly contributed towards the evolutionary analysis of other clonal organisms (40), in particular, those exhibiting a species-specific host dependence (26). In the case of the MTC, these regions have proven informative both for the types of genes that vary between strains (50) and for the molecular signatures that characterize different members of the MTC (8, 35). From such studies more recently emerged the notions of geographically defined (22) and host-restricted forms (34) of the MTC. Though experimental evidence directly associating genomic deletions with in vivo niche adaptation has yet to be documented, MTC organisms with specific genomic deletion profiles appear primarily restricted to certain host types. Given that host range adaptation is not uncommon for microbial pathogens (53), it is possible that genomic deletions are at least partly responsible for the observed differences in host range among MTC variants, though one must recognize the distinct possibility that deletion profiles associated with a particular host type can be the result of other evolutionary processes, including chance. Of note, the deletion profile specific for M. pinnipedii is observed to infect a variety of pinniped species (New Zealand fur seals, Australian fur seals, South American fur seals, and sea lions) and has also been occasionally isolated from terrestrial hosts (Table S1). Thus, M. pinnipedii may not represent a host-specific MTC per se, but rather an organism that successfully infects pinnipeds of this particular ecological niche across a remarkable geographic range.
Genomic analyses have consistently described M. bovis as the most derivative MTC member (8, 35), though only in this report have LSPs been sought to study M. bovis itself. From this, a phylogenetic stratification of M. bovis genotypes (Fig. 1) offers a structure upon which to base future M. bovis study. Though it is unfortunate that isolates from the genomovar of M. bovis (b) were not analyzed here, the fact that BCG strains belong to this genomovar indicates that genetic variability between BCG strains and sequenced strains of either M. tuberculosis or M. bovis may result from polymorphisms specific to this genetic lineage and not necessarily mutations incurred during in vitro passage of BCG. The most derived genomovar of M. bovis characterized here was called M. bovis (d) and includes the sequenced strain of M. bovis AF2122/97 (18), which now de facto serves as the referent profile for M. bovis. Two practical consequences of these findings pertain to genetic and phenotypic studies of M. bovis. First, in genomic comparisons of M. tuberculosis, M. bovis, and BCG, it is noteworthy that M. bovis AF2122/97 harbors its own specific polymorphisms; therefore, the ideal referent for the genetic analysis of BCG-specific evolution should be a virulent isolate of M. bovis (b). Second, it merits reconsideration whether phenotypic properties generally ascribed to M. bovis, such as preferential growth with supplementation of pyruvate (25), constitutive production of MPB70/83 (29), and virulence in rabbits (30), are general properties of all genomovars of the organism.
A considerable number of genes disrupted through these deletions have predicted functions of potential biological impact, but their relevance for phenotypes such as virulence will clearly require targeted study. As a means of exploring the impact of these deletions, the functional classification of genes that have been deleted can be compared to their proportional distribution in the H37Rv genome (Table S6 in the supplemental material). In contrast to a null model where genetic loss is random across functional classifications, several differences are observed. As would be expected from a highly repetitive sequence, genes classified as “insertion sequences and phages” and “PE/PPE genes” are often implicated in deletion events. In contrast, genes classified as “information pathways” and “intermediary metabolism and respiration” have been relatively spared from deletion. Though deletions from M. tuberculosis sensu stricto (50) also revealed the deletion of mobile genetic elements to be more frequent than expected by chance, a high deletion rate of genes involved in intermediary metabolism and respiration is contrary to what is observed from study here. Thought to confer a selective advantage only during certain stages of infection or transmission (“intermittently essential”), the deletion of genes involved in intermediary metabolism and respiration may not present a viable strategy in the long term for derived MTC. To further explore the functional consequences of deletions, these same genes can be examined in comparison to published lists of genes essential in vitro (46) and in vivo (47) by transposon site hybridization studies (Table S5 in the supplemental material). Of genes essential in vitro, eight were implicated in the deletions characterized here (fadD30, gca, Rv1232c, Rv2813, moaA1, moaC1, moaD1, and Rv3113, where moaC1, moaD1, and Rv3113 are involved in two independent deletions), a somewhat surprising result, as the organisms typed had been isolated by culture in microbiology labs. In addition, five genes deemed essential for in vivo growth were lost in the newly described deletions (Rv1371, Rv1930c, Rv1931c, Rv2812, and Rv3114, where Rv3114 is involved in two independent deletions). Together with the loss of genes Rv1514c, Rv1563c, Rv1974, and cobL (Rv2072c) in deletions previously documented (RD4, RD17, RD7, and RD9, respectively), this indicates that a total of nine in vivo essential genes have been deleted from isolates manifestly capable of causing disease. While these data might be used to challenge the validity of the mouse model for determining genes that impact in vivo pathogenesis, one should consider that although five genes within the RD1 region deleted from BCG vaccines (Rv3871 to Rv3879c) are also considered essential in vivo (47), it is established that natural RD1 deletion mutants cause disease in voles (7) and dassies (34) yet are of attenuated virulence in a number of hosts, including humans (43). Together, these data suggest that the evolution of MTC organisms across different hosts has involved the deletion of genes elsewhere considered essential.
From the genomic comparison of M. bovis AF2122/97 to M. tuberculosis H37Rv, it was noted that the genomic evolution of MTC members appears to parallel the massive gene decay documented in the leprosy bacillus, M. leprae (12, 18). Consistent with this, examination of the ORFs in the newfound deletions reveals that the majority (98%) are absent or no longer functional in the genome of M. leprae (Table S5). The amount of genome loss between M. bovis isolates can also be contrasted with published observations on M. tuberculosis. A similar GeneChip study of 100 M. tuberculosis clones recently quantified the amount of DNA deleted from isolates with respect to the H37Rv genome and estimated the difference in gene content between two M. tuberculosis strains that are not closely related as between 40 and 100 genes (50). From this, it was concluded that the observed magnitude of genomic variability caused by LSPs within M. tuberculosis sensu stricto is similar to that found between M. tuberculosis H37Rv and M. bovis AF2122/97. LSP genomic variability among the M. bovis isolates interrogated here (restricted to samples having deleted RD4) can be quantified in a similar manner by using the sequenced strain M. bovis AF2122/97 as the referent. On average, one region affecting seven ORFs (or 8,483 bp) was deleted per isolate, with the maximum number of deleted genes being 23. Knowing that five genes are missing from regions in AF2122/97 but not in other M. bovis isolates (N-RD17, RDpan, and RD17), we can approximate that the difference in gene content between two M. bovis strains that are not closely related is on the order of 12 genes and is unlikely to exceed 28 genes. Incorporating the overall genomic variability observed across M. bovis strains, these estimates should be irrespective of the M. bovis referent genome chosen for analysis. These estimates place the diversity in gene content among M. bovis sensu stricto as less than that for M. tuberculosis sensu stricto, consistent with M. bovis being a more recent clone having evolved for less time than has M. tuberculosis (8, 35).
The approach to MTC speciation has traditionally been based upon phenotypic distinction, itself viewed as indicative of phylogenetic separation. Because of documented phenotypic convergence across the MTC, which limits the ability of in vitro tests to predict MTC genotype (37), reliance is increasingly being placed on molecular divergence from known species for diagnostic work (23, 42). With this change, standard phenotypic tests may serve as a means of describing certain physiological properties of samples, while PCR-based testing for genomic deletions such as those described here can become a simple and straightforward means of rapidly classifying organisms. With this comes the ability to address the generally held belief that, unlike the principally human-specific M. tuberculosis, M. bovis universally infects and is able to sustain disease across a broad range of animal species. Our data, from a limited number of isolates, suggests this may be partly true. In Canada and South Africa, isolates sharing the same genomic profile caused disease in a variety of hosts, pointing to geographically successful clones that are not host restricted at least in terms of establishing infection, although the capacity of M. bovis to persist in these alternative host types has been inconsistent and requires further investigation. In contrast to these cross-species events, our data confirm the genomic distinction of MTC organisms from pinniped and caprine hosts and suggest other distinct clades that may be host specific, notably, isolates from oryx and the East Asian water buffalo. Further study using these genomic markers and others across a broader sample of clinical isolates will illuminate a greater appreciation of the diversity of M. bovis organisms and, in turn, enable a more focused understanding of the challenges faced for TB control.
Acknowledgments
We thank Glyn Hewinson for his support and access to the VLA strain collection, Philip Butcher and Maryam Ehtashami for their assistance and input, and David Roquis, Corine Zotti, Pierre LePage, Daniel Vincent, Yannick Fortin, and Rob Sladek at the McGill University and Genome Quebec Innovation Center for helping with DNA sequencing and GeneChip experimentation. We also thank Eamonn Gormley (UCD) for the gift of M. bovis strains from Ireland, Rick Slayden at Colorado State University and Marc Pannunzio at Amersham Biosciences for suggestions regarding DNA amplification, and Liz Rohonczy, Chris Daborn, Alan Murray, and Amelia Bernardelli for allowing us to use DNA from local isolates from Canada, Tanzania, New Zealand, and Argentina in this study.
This work was supported by Canadian Institutes of Health Research (CIHR) grant MOP 36054 and the Department of Environment, Food, and Rural Affairs (United Kingdom). S.M. is the recipient of a studentship award from the Fonds de la Recherche en Santé du Québec. M.A.B. is a New Investigator of the CIHR.
REFERENCES
- 1.Aranaz, A., D. Cousins, A. Mateos, and L. Dominguez. 2003. Elevation of Mycobacterium tuberculosis subsp. caprae to species rank as Mycobacterium caprae comb. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 53:1785-1789. [DOI] [PubMed] [Google Scholar]
- 2.Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 34:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayele, W. Y., S. D. Neill, J. Zinsstag, M. G. Weiss, and I. Pavlik. 2004. Bovine tuberculosis: an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 8:924-937. [PubMed] [Google Scholar]
- 4.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-factor antagonists control F activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 5.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 6.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. Van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, G. D., J. A. Dave, N. C. Gey van Pittius, L. Stevens, M. R. W. Ehlers, and A. D. Beyers. 2000. The mycosins of Mycobacterium tuberculosis H37Rv: a family of subtilisin-like serine proteases. Gene 254:147-155. [DOI] [PubMed] [Google Scholar]
- 10.Charlet, D., S. Mostowy, D. Alexander, L. Sit, H. G. Wiker, and M. A. Behr. 2005. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol. Microbiol. 56:1302-1313. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 13.Cousins, D. V., R. Bastida, A. Cataldi, V. Quse, S. Redrobe, S. Dow, P. Duignan, A. Murray, C. Dupont, N. Ahmed, D. M. Collins, W. R. Butler, D. Dawson, D. Rodriguez, J. Loureiro, M. I. Romano, A. Alito, M. Zumarraga, and A. Bernardelli. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1305-1314. [DOI] [PubMed] [Google Scholar]
- 14.De Smet, K. A. L., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 15.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, J. W. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Pelayo, M. C., K. C. Caimi, J. K. Inwald, J. Hinds, F. Bigi, M. I. Romano, D. Van Soolingen, R. G. Hewinson, A. Cataldi, and S. V. Gordon. 2004. Microarray analysis of Mycobacterium microti reveals deletion of genes encoding PE-PPE proteins and ESAT-6 family antigens. Tuberculosis 84:159-166. [DOI] [PubMed] [Google Scholar]
- 18.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gey van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:research0044. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, S. V., K. Eiglmeier, T. Garnier, R. Brosch, J. Parkhill, B. Barrell, S. T. Cole, and R. G. Hewinson. 2001. Genomics of Mycobacterium bovis. Tuberculosis (Edinburgh) 81:157-163. [DOI] [PubMed] [Google Scholar]
- 22.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huard, R. C., L. C. Oliveira Lazzarini, W. R. Butler, D. Van Soolingen, and J. L. Ho. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keating, L. A., P. R. Wheeler, H. Mansoor, J. K. Inwald, J. Dale, R. G. Hewinson, and S. V. Gordon. 2005. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol. Microbiol. 56:163-174. [DOI] [PubMed] [Google Scholar]
- 26.Klasson, L., and S. G. E. Andersson. 2004. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 12:37-43. [DOI] [PubMed] [Google Scholar]
- 27.Kremer, K., D. Van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamichhane, G., M. Zignol, N. J. Blades, D. E. Geiman, A. Dougherty, J. Grosset, K. W. Broman, and W. R. Bishai. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 100:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebana, E., A. Aranaz, B. Francis, and D. Cousins. 1996. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lurie, M. B. 1928. The fate of human and bovine tubercle bacilli in various organs of the rabbit. J. Exp. Med. 48:155-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 33.McFadden, J. 1996. Recombination in mycobacteria. Mol. Microbiol. 21:205-211. [DOI] [PubMed] [Google Scholar]
- 34.Mostowy, S., D. Cousins, and M. A. Behr. 2004. Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J. Bacteriol. 186:104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 36.Mostowy, S., A. G. Tsolaki, P. M. Small, and M. A. Behr. 2003. The in vitro evolution of BCG vaccines. Vaccine 21:4270-4274. [DOI] [PubMed] [Google Scholar]
- 37.Mostowy, S., A. Onipede, S. Gagneux, S. Niemann, K. Kremer, E. P. Desmond, M. Kato-Maeda, and M. Behr. 2004. Genomic analysis distinguishes Mycobacterium africanum. J. Clin. Microbiol. 42:3594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy, H. N., G. R. Stewart, V. V. Mischenko, A. S. Apt, R. Harris, M. S. B. McAlister, P. C. Driscoll, D. B. Young, and B. D. Robertson. 2005. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 280:14524-14529. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, D., P. Brassard, D. Menzies, L. Thibert, R. Warren, S. Mostowy, and M. Behr. 2004. Genomic characterization of an endemic Mycobacterium tuberculosis strain: evolutionary and epidemiologic implications. J. Clin. Microbiol. 42:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 41.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung Dis. 76:1-46. [DOI] [PubMed] [Google Scholar]
- 42.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 44.Rauzier, J., E. Gormley, M. C. Gutierrez, E. Kassa-Kelembho, L. J. Sandall, C. Dupont, B. Gicquel, and A. Murray. 1999. A novel polymorphic genetic locus in members of the Mycobacterium tuberculosis complex. Microbiology 145:1695-1701. [DOI] [PubMed] [Google Scholar]
- 45.Salamon, H., M. Kato-Maeda, P. M. Small, J. Drenkow, and T. R. Gingeras. 2000. Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data. Genome Res. 10:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 47.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, N. H., J. Dale, J. Inwald, S. Palmer, S. V. Gordon, R. G. Hewinson, and J. M. Smith. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. USA 100:15271-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valvano, M. A., P. Messner, and P. Kosma. 2002. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology 148:1979-1989. [DOI] [PubMed] [Google Scholar]
- 52.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wren, B. W. 2000. Microbial genome analysis: insights into virulence, host adaptation and evolution. Nat. Rev. Genet. 1:30-39. [DOI] [PubMed] [Google Scholar]