Abstract
The ranks higher than the species in the prokaryotic taxonomy are primarily designated based on phylogenetic analysis of the 16S rRNA gene sequences, but no definite standards exist for the absolute relatedness (measured by 16S rRNA or other means) between the ranks. Accordingly, it remains unknown how comparable the ranks are between different organisms. To gain insights into this question, we studied the relationship between shared gene content and genetic relatedness for 175 fully sequenced strains, using as a robust measure of relatedness the average amino acid identity (AAI) of the shared genes. Our results reveal that adjacent ranks (e.g., phylum versus class) frequently show extensive overlap in terms of genetic and gene content relatedness of the grouped organisms, and hence, the current system is of limited predictive power in this respect. The overlap between nonadjacent ranks (e.g., phylum versus family) is generally limited and attributable to clear inconsistencies of the taxonomy. In addition to providing means for standardizing taxonomy, our AAI-based approach provides a means to evaluate the robustness of alternative genetic markers for phylogenetic purposes. For instance, the 23S rRNA gene was found to be as good a marker as the 16S rRNA gene, while several of the widely distributed protein-coding genes, such as the RNA polymerase and gyrase subunits, show a strong phylogenetic signal, albeit less strong than the rRNA genes (0.78 > R2 > 0.69 for the protein-coding genes versus R2 = 0.84 for the rRNA genes). The AAI approach outlined here could contribute significantly to a genome-based taxonomy for all microbial organisms.
Prokaryotic taxonomy consists of three separate components: classification (i.e., the arrangement of organisms into groups or taxa), nomenclature, and identification. Although there is no official classification for prokaryotes, the classification system represented by Bergey's Manual of Systematic Bacteriology (http://www.cme.msu.edu/bergeys/) is widely accepted by the community of microbiologists and therefore is currently considered the best approximation to an official classification (2). The Bergey's classification system is based on the phylogenetic analysis of the small-subunit rRNA genes (16S rRNA), as well as on classical microscopic and biochemical observations about the relatedness of the organisms, such as G+C content deviation and DNA-DNA hybridization efficiency (2, 19, 22). This system has been valuable in describing and appreciating the breadth of prokaryotic diversity and setting the framework for the study of relationships between taxa. Further, results from new approaches enabled by the availability of whole-genome sequences, such as phylogeny based on shared content of orthologous genes (10, 14, 17, 28), indels, or signature sequences (8, 16) and concatenated alignments of many proteins (3, 11, 31), are generally congruent with the 16S rRNA gene-based phylogeny, which adds further value to the system.
It is important to realize, however, that the definition or standards for the existing taxonomic ranks are far from being well delineated, particularly for the ranks higher than the species. In fact, considerable subjectivity in designating genera, families, etc., has been allowed, which is partially attributable to the great biochemical and morphological diversity exhibited by prokaryotes that prevents the employment of the same measuring rules for all groups of organisms (2). Currently, the only major prerequisite for designating novel taxonomic ranks higher than the species rank is that clustering by 16S rRNA gene data should support such designations, but no standards exist in regard to the absolute differences between the taxonomic ranks (19). Consequently, the prokaryotic taxonomy represents, unavoidably, an artificial system, which often depends more on the intuition of individual researchers than on specific standards or knowledge of the natural history of organisms. Nonetheless, there is great comparative value in having a taxonomic system predictive of phenotypic and genetic relatedness of the grouped organisms and taxonomic ranks that are comparable, in terms of absolute differences and similarities, among lineages. It remains unclear, however, how the prokaryotic taxonomy is performing with regard to these issues, partly due to the focus on the 16S rRNA gene, which has overlooked the overall biochemical or genetic relatedness at the whole-cell level, and partly because of technological constraints in studying the differences and similarities among microorganisms.
The recent availability of complete sequences of a number of prokaryotic genomes has made it possible for the first time to study the genetic and functional relatedness between organisms at the whole-cell level, and hence, to provide novel insights into the issues described above and an independent assessment of what the 16S rRNA-based system really represents. However, genomic studies to date have mostly been focused on assessing the accuracy of phylogenetic reconstruction, particularly in the light of horizontal gene transfer (HGT), rather than the absolute differences between taxa and/or have failed to address the latter issue systematically for all prokaryotic taxa. Here, we have assessed the consistency of the taxonomic ranks for 175 fully sequenced strains and find that the whole-genome level relatedness among these strains is often inconsistent with their taxonomic relatedness and that the taxonomic ranks, as they are currently used, are not sufficiently predictive of the genetic and functional relatedness of the strains.
MATERIALS AND METHODS
Determination of conserved genes and genetic relatedness.
The genomic sequences and sequence annotation of the 175 genomes used in this study were obtained from the National Center for Biotechnology Information ftp site (ftp://ftp.ncbi.nih.gov/). Genes conserved between a pair of genomes were determined by whole-genome pairwise sequence comparisons using the BLAST algorithm release 2.2.5 (1). For these comparisons, all protein-coding sequences (CDSs) from one genome were searched against the genomic sequence of the other genome (protein query versus translated database; TBLASTN). CDSs that that had a BLAST match of at least 30% identity at the amino acid level (recalculated to an identity along the entire sequence) and an alignable region more than 70% of the length of the query CDS were considered one-way BLAST conserved genes. This cutoff is above the twilight zone of similarity searches, where inference of homology is error prone due to low similarity between aligned sequences; thus, query CDSs were presumably homologous to their matches (24-26). Further, searching against genomic sequences (as opposed to CDSs) circumvented the problem of inconsistencies in the annotation between different genomes. A reciprocal best-match approach was also employed (i.e., by extracting the matching segment from the genomic sequence and performing the reverse, BLASTX, search) to determine the presumably orthologous fraction of conserved genes between the two genomes (two-way BLAST) in an effort to achieve a more conservative estimation of functional similarity.
The genetic relatedness between a pair of genomes was measured by the average amino acid identity (AAI) of all two-way BLAST conserved genes between the two genomes as computed by the BLAST algorithm. Measuring AAI based on two-way BLAST conserved genes gives higher, but not considerably higher, values than measuring AAI based on one-way BLAST conserved genes by an average of 1.48 (standard deviation, 0.68; 10.62 maximum). Thus, the latter approach (i.e., one-way BLAST) also gives reliable results, albeit with slightly decreased accuracy compared to the former approach, particularly for larger genomes with expanded families of paralogous genes. The 16S rRNA gene or other genetic marker identity was calculated in the same way as AAI, i.e., based on BLAST searches (nucleotide level—BLASTN—for 16S and 23S rRNA and amino acid level—BLASTP—for protein-coding genes), for consistency in comparing the results.
Calibrating AAI trees.
For calibrating AAI trees, the following strategy was used. The identity of each gene conserved (two-way BLAST) between Escherichia coli strain K-12 or Bacillus subtilis and the remaining 174 genomes was calculated. The identities of all these genes, when the genes were conserved in at least 150 genomes (i.e., ∼85% of the genomes; 16 genomes are archaeal), were plotted together (191 genes in total) against the AAI between the E. coli (or B. subtilis) genome and the corresponding genome, similarly to the graphs shown (see Fig. 5) for individual genes. A plot-fitting exercise identified the logarithmic model to best describe the relationship between the identity of the widely distributed genes and AAI, and the equation y = −1,300.41 + 603.071lnx − 64.9438(lnx)2 was used to transformed the raw AAI values into calibrated AAI values.
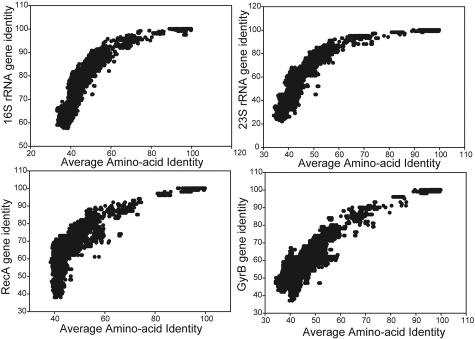
FIG. 5.
Correlations between alternative phylogenetic markers to AAI. Shown are the correspondences between the identity of a molecular marker (panel title; y axis) and AAI (x axis) for all pairs of the 175 genomes that have a clear homolog of the marker (at least 20,000 pairs for each gene) used in this study. The full-name descriptions of markers are given in Table 1.
Taxonomic information.
The taxonomic information for each of the 175 genomes was extracted from the Hierarchy Browser of the Ribosomal Database Project database, release 9 (http://rdp.cme.msu.edu/index.jsp), which implements the newer version of Bergey's taxonomy (9). The taxonomic information included all the recognized taxonomic ranks, with the exception of the subspecies rank, i.e. (from the largest to the smallest), domain, phylum, class, order, family, genus, and species (2).
RESULTS
AAI measurement of relatedness.
For our purposes, there was a need for precise measurement of the genetic relatedness between any two strains. The main limitations in performing this task universally for all prokaryotic taxa are the lack of genes that are widely distributed in all taxa (5, 27) and the still unclear effect of HGT on the inferred phylogenies (12, 21). According to our own homology-based search, there are almost no genes, including the highly conserved genes reported by Santos and Ochman (27), with detectable orthologs in all 175 genomes. Further, it is frequently not possible to identify the true ortholog of a gene when multiple matches are present or when a gene has only matches of weak similarity. For these reasons, we introduced a novel parameter, the AAI of all genes shared between two strains, to measure the genetic relatedness between the strains. By definition, the genes used in the AAI calculations are not necessarily the same in all pairwise comparisons and there are more genes conserved (and thus used to calculate AAI) between more closely related strains than between more distantly related strains. Further, the pool of conserved genes among the latter strains is more enriched toward widely distributed genes, which tend to show high degrees of sequence conservation, relative to the pool of genes conserved between the former strains, which includes many accessory genes as well. We show below, however, that these characteristics are not problematic for the comparative value of the AAI measurement because they are consistent across all lineages and introduce only a systematic effect into the AAI measurement, which can be calibrated if needed.
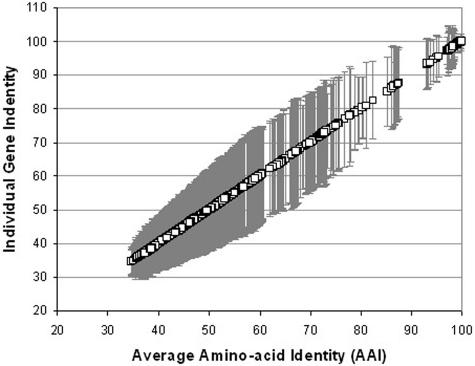
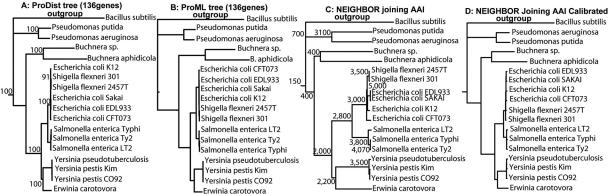
First, we have previously shown that, for short evolutionary scales, average nucleotide identity (ANI) represents a very robust measure of genetic and evolutionary relatedness between two strains because it shows strong correlation to DNA-DNA reassociation values (the classical method for species delineation in prokaryotes) and the mutation rate of the genome (18). These characteristics are applicable to AAI as well (analytical data not shown). Second, in all pairwise comparisons performed (175 genomes; 175 × 175 = 30,625 comparisons), we found that the identities of the great majority (>70%) of the genes in the genome are within ∼8.4% (STDEV = 1.85) difference from the genome average (i.e., AAI), and this is consistent regardless of the absolute genetic distance between the genomes compared, which demonstrates the power of the AAI measurement to reflect whole-genome level relatedness (Fig. 1). Finally, phylogenetic reconstruction based on AAI is very congruent in terms of tree topology, with reconstructions based on distance or maximum likelihood analysis of concatenated sequences of all genes shared between the genomes (Fig. 2, compare C with A and B). When the AAI values were calibrated based on the relationship between AAI and the degree of sequence conservation of the widely distributed genes (see Materials and Methods), the AAI tree was very congruent with the whole-genome trees in terms of branch length as well (Fig. 2, compare D with A and B). It is interesting that even the relationships among organisms with contrasting ecologies, genome sizes, and numbers of paralogous genes, such as the large-genome-size Pseudomonas (6 Mb) and the symbiotic, small-genome-size Buchnera (0.6 Mb), are accurately reconstructed on the calibrated AAI tree. These results demonstrate that the genetic distances and genome sizes of the strains compared or the varied degrees of sequence conservation of different classes of genes have little or only a systematic effect, which is not problematic, on the comparative power of AAI. Therefore, AAI represents a simple, universal, and most importantly, robust descriptor of genetic relatedness, while it avoids the problem of finding genes that are universally distributed and offers resolution at short evolutionary scales, where the widely distributed genes do not (e.g., contrast Fig. 2C with A and B for Escherichia, Salmonella, and Yersinia species).
FIG. 1.
Individual gene identity versus genome average identity. For each pair of genomes (175 genomes; 30,625 pairs), we determined the AAI, as well as the identity of each individual gene conserved (two-way BLAST; see Materials and Methods), between the two genomes. The identity of each gene was compared to the corresponding AAI value, and the variation of the identities of individual genes from the AAI, represented as 1 standard deviation from the AAI (y axis), is plotted against the corresponding AAI value (x axis). The average variation was ∼8.4 (STDEV = 1.85). These results demonstrate that the identities of the majority (>70%) of the genes conserved between two genomes are within approximately ±8.4% of the average of the genome (i.e., AAI), and this is independent of the genetic distance between the two genomes.
FIG. 2.
Phylogenetic reconstruction based on AAI versus whole-genome sequence analysis. The shared gene core between the 17 proteobacteria and Bacillus subtilis (outgroup) was determined, using a two-way BLAST approach, to be 136 genes, and these core genes were used to build the phylogenetic trees shown. (A and B) A distance and a maximum likelihood tree, respectively, built with the ProtDist and ProML algorithms of the Phylip package (13) using default settings and, as input sequence, the concatenated protein sequences of all 136 core genes aligned with the ClustalW software (6). The numbers on the nodes of the distance tree (A) indicate the statistical support of the node by 100 bootstrap replicates with ProtDist. All nodes (even the ones not shown for simplicity) have 100 bootstrap values, except for the node connecting strain K-12 to the two Shigella strains, which has 91. (C) The AAI-based tree. The numbers on the nodes of the AAI tree are rough approximations of the number of genes shared (and used in the calculations of AAI) by the genomes grouped at the node. The exact number of genes depends on the specific pair of genomes used. (D) The AAI tree calibrated as described in Materials and Methods.
Evaluation of taxonomic ranks in terms of genetic relatedness.
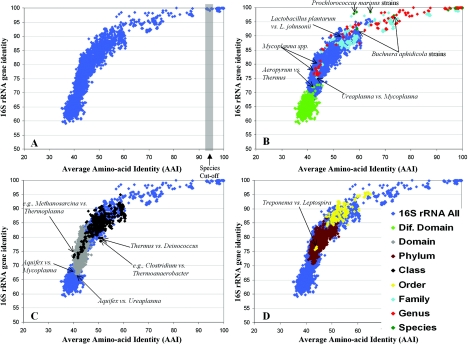
We first compared the AAI to 16S rRNA gene identity for all pairs of the 175 prokaryotic genomes used in this study (175 × 175, or 30,625 pairs in total) to gain insight into the interrelationship of these two parameters. Our results show that there is a strong correlation between 16S rRNA gene identity and AAI and that the logarithmic model best describes this correlation (R2 = 0.84; P < 0.0001) (Fig. 3A). When the analysis was restricted to pairs of genomes with higher than 87 to 90% 16S rRNA gene identity, however, there was no significant difference between the logarithmic (R2 = 0.834) and the linear (R2 = 0.825) models. These results indicate that the influence of additional mutations (presumably in the 16S rRNA gene) is offset by recurrent mutations when 16S rRNA gene sequences are less than ∼85 to 87% identical. In any case, the strong correlation observed further supports the robustness of 16S rRNA gene-based phylogeny for prokaryotes, which is consistent with other genomic approaches (11, 16, 17, 31). The 16S rRNA gene appears to have limited resolution between genomes showing higher than 80% AAI, whereas the permissible substitutions in its sequence reach saturation around 60 to 65% identity, presumably due to functional constraints.
FIG. 3.
Relationships between 16S rRNA, AAI, and taxonomic information for the 175 sequenced genomes. Each dot represents a comparison between two genomes and shows their 16S rRNA gene identity (y axes) plotted against the AAI of the genes shared between the two genomes (x axes) (A). The smallest classification rank that the two genomes of each pair (30,635 pairs in total) share has been overlaid on the graph with a color, which corresponds to the rank, in panels B, C, and D. (B to D) Pairs of genomes whose smallest shared rank is the species, genus, family, or different domain (B); the same domain or class (C); and the phylum or order (D). The ranks have been laid out in panels B, C, and D so as to avoid overlap as much as possible within the same panel. The area that corresponds to the current standards for species delineation (panel A; see the text) (18), as well as representative pairs of genomes (discussed in the text), are shown.
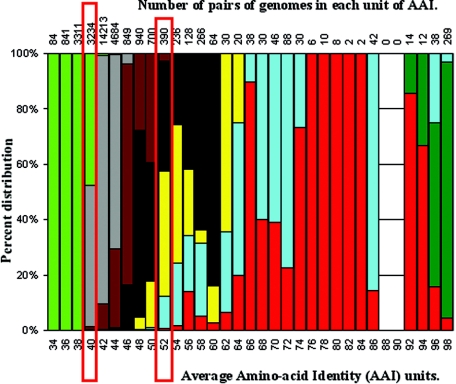
We then determined for each pair of genomes their closest taxonomic relationship, i.e., the smallest taxonomic rank they shared, and overlaid this information on the graph in Fig. 3A. The taxonomic information for each genome was extracted from Bergey's Manual (19). We found that all ranks higher than the species, with the exception of the different domains, frequently show extensive overlap (Fig. 3B, genus versus family, or C and D, same domain versus phylum). For instance, there are 390 pairs of genomes showing 52% AAI, and 39%, 43%, 17%, and 1% of these pairs have as their smallest shared rank the phylum, class, order, and family, respectively (Fig. 4). In this particular example and unit of AAI (i.e., 52%), the class appears to be the most dominant rank, representing 43% of the pairs of genomes. We found that the most dominant rank for every unit of AAI contains, on average, ∼69.3% (STDEV = 18.3) of the pairs of genomes within the particular unit; in other words, there is, on average, an ∼30.7% overlap between the ranks. The overlap is more frequent between adjacent ranks (e.g., the order and the class) than between nonadjacent ranks (e.g., the order and the phylum), which overlap, on average, 10-fold less frequently. In fact, the overlap in the latter case is limited to only a few genomes, such as between the Prochlorococcus marinus and the Buchnera aphidicola genomes (Fig. 3B) and between the Treponema and Leptospira (Fig. 3D) genomes, whose genetic relatedness is far too low, compared with the remaining data set, to justify their inclusion in the same species and order, respectively. Such cases are apparently artifacts, e.g., P. marinus strains are grouped in the same species based on their high 16S rRNA gene sequence similarity (7), and Treponema and Leptospira are assigned to the same order due to their common spirochete-like morphology (4). Finally, it is interesting that the overlap between the ranks of the taxonomy is frequently extensive in terms of 16S rRNA gene identity as well (Fig. 3).
FIG. 4.
In-depth calculation of the extent of AAI overlap between the ranks of taxonomy. We determined the number of pairs of genomes (top; x axis) related at any given unit of AAI (bottom; x axis), as well as the smallest taxonomic rank that each pair of genomes shares. The bars show the percent distribution (or overlap) of the taxonomic ranks for each unit of AAI (for an example related to the bars outlined in red, see the text). The color representation of the ranks is identical to that of Fig. 3.
Another remarkable trend revealed in our data is that several bacterial phyla and a few classes are approximately as distant from each other in terms of AAI as Bacteria is from Archaea. For instance, there are 3,234 pairs of genomes showing 40% AAI, 48% of which involve pairs of strains from different domains (i.e., an archaeon and a bacterium), whereas 51% involve pairs of strains from the same domain (Fig. 4), such as a mollicute—representing a class—and Aquifex aeolicus—representing a phylum (shown in Fig. 3C). Below 40% AAI, we found only pairs of strains from different domains; nonetheless, the difference between interdomain and interphylum AAIs is frequently too small, e.g., <2% AAI (Fig. 4). In addition, we have noted that the genetic differences between any two strains in terms of AAI correspond to comparably large functional/biochemical (gene) differences, as well (K. T. Konstantinidis and J. M. Tiedje, unpublished data); therefore, the interphylum gene content differences are also very comparable to the interdomain ones.
Evaluation of alternative markers to 16S rRNA for phylogenetic purposes.
The robustness of alternative markers to the 16S rRNA gene for phylogenetic purposes was also evaluated, using the AAI as a control in these evaluations and an approach similar to that used for the 16S rRNA gene. The results show that several of these markers, such as RNA polymerase subunits, tRNA synthetases, gyrase, and RecA protein, show considerable robustness based on the high correlation (R2 > 0.68; P < 0.0001 for all markers tested) observed between the AAI and the identity of these proteins for all pairs of genomes that have a clear homolog of the protein (Table 1 and Fig. 5). Among the protein-coding genes tested, RNA polymerase subunit B showed the highest correlation (R2 = 0.78) to AAI, and RecA protein showed the lowest (R2 = 0.68), while all protein-coding genes evaluated showed significantly lower correlation to AAI than 16S rRNA (R2 = 0.84). On the other hand, the large-subunit RNA gene (23S rRNA) showed correspondence comparable to AAI, suggesting that is a highly reliable marker (Fig. 5). A similar approach may be used to evaluate the robustness of other markers as well, targeting the full breadth of prokaryotic diversity or shorter evolutionary scales, e.g., the species level, for specific applications. For the latter case, we suggest using the ANI of the shared genes, which is more sensitive on this evolutionary scale than AAI.
TABLE 1.
Relationships of different phylogenetic markers to AAI
| Gene | R2a |
|---|---|
| 16S rRNA (small-subunit ribosomal gene) | 0.84 |
| 23S rRNA (large-subunit ribosomal gene) | 0.84 |
| RecA (DNA strand exchange and recombination protein) | 0.68 |
| RpoB (RNA polymerase; beta subunit) | 0.78 |
| GyrB (DNA gyrase subunit B) | 0.77 |
| IleS (isoleucine tRNA synthetase) | 0.72 |
| FusA (GTP-binding protein chain elongation factor EF-G) | 0.69 |
R2 is for logarithmic second-order correlation. This correlation gave among the highest R2 values from the types of correlations tested for most genes. It should be mentioned, however, that there were typically very small differences between different models (e.g., linear, power, logarithmic, and sigmoidal) in their abilities to describe the relationship between individual genes and the average of the genomes. Thus, no assumptions can be made about the underlying mechanisms of this relationship.
DISCUSSION
The whole-genome comparisons between 175 fully sequenced genomes revealed that adjacent ranks of the prokaryotic taxonomy may show, on average, ∼30% and up to ∼50% overlap in terms of genetic relatedness, meaning that for a given genetic distance between two strains, 30% of the pairs of strains belong to different ranks. In contrast, nonadjacent ranks overlap 10-fold less frequently, e.g., they constitute, on average, <3% of the total overlap (Fig. 3 and 4). Therefore, although there appears to be a coarse consistency (and a gradient) between the ranks of taxonomy, they are not always consistent with the relatedness at the whole-cell level of the grouped organisms. Kunin et al., using a whole-genome-derived measurement different from our AAI measurement, have recently reported similar trends, albeit in a considerably less systematic effort (20). These results clearly suggest that the current system requires several adjustments if the goal is to become more uniform and predictive of the genetic and biochemical relatedness of the grouped organisms.
AAI represents a convenient means to quickly identify and correct such irregularities in the classification system. AAI may also represent a powerful first step toward a genome-based taxonomy because it is a simple, robust, and pragmatic measure of relatedness for all prokaryotic taxa and computationally much easier than alternative whole-genome methods. Moreover, recent reports suggest that it may not be feasible to evaluate and/or expand the 16S rRNA-based phylogeny by including more genetic markers, due to the shortage of genes widespread in all prokaryotic taxa (5) or the difficulty in designing universal primers for widespread genes (27). One example of how AAI may be incorporated in the current taxonomy is the following: every strain, in addition to its species name, could be accompanied by its AAI value to some reference (sequenced) genomes. In this way, the classification system will gain substantial comparative value and higher accuracy while no additional confusion will be introduced. It may also be feasible to devise a new method or optimize an existing one to indirectly measure AAI, i.e., to circumvent the need for whole-genome sequencing. Multilocus sequencing typing (MLST) (23), which employs genes (not necessarily the same genes for all taxa) that evolve comparably to the genome average, may be one such approach. The methodology described here (Fig. 5) can assist the identification of good candidate genes for such an MLST-based application, and our preliminary results from seven high-draft Burkholderia genomes and seven genes used in the MLST analysis show that the MLST-based phylogenetic reconstruction is very congruent with the AAI-based one (Konstantinidis and Tiedje, unpublished).
Certainly, averaging across all genes in the genome may miss important lineage-specific information, while it is possible that, due to not comparing exactly the same genes in all pairwise comparisons, some (we believe small) error might have been introduced into the results. For these reasons, our AAI-based approach may better serve as a backbone for systematics, similar to the way the 16S rRNA gene has been used but with higher robustness and accuracy, as we have shown here (e.g., the Prochlorococcus example above), upon which finer-scale investigations would be performed. Contrary to the 16S rRNA gene, AAI (or better, ANI, as we previously showed [18]), offers better resolution between closely related species (Fig. 3). In addition, we have found that the 70% DNA-DNA reassociation threshold, the single most important criterion used since 1987 for species delineation (29, 30), corresponds to ∼95 to 96% AAI (Fig. 3A, species cutoff) (18). Therefore, AAI offers good resolving power within species as well, which is advantageous for specific applications, such as microevolution studies. Further, the effects of HGT and genome size should be less significant on AAI than on other single-gene-based and gene content-derived approaches because AAI is derived from as many genes (at least 50 and usually >500 genes in total) of the genome as possible and because of the process of amelioration of foreign DNA sequences (the prevalence of mutations toward the average nucleotide composition of the genome) that is ongoing in every cell. Consistent with these interpretations, when we compared our AAI values to the D1 genome conservation index of Kunin et al., we found generally good correlation (R2 > 0.9 for the genomes evaluated) between the two values, while our AAI generally provided a better measurement of evolutionary (and genetic) relatedness in ambiguous cases. For example, the D1 value for comparisons between the E. coli and Buchnera aphidicola genomes is 54 to 57, and that between E. coli and Yersinia pestis is 46 to 47. Our AAIs are ∼58% and ∼72% for the same pairs, suggesting that E. coli is more closely related to Yersinia than Buchnera, which is consistent with the whole-genome trees, as well (Fig. 2).
We have not fully investigated whether the sequenced strains used in this study represent “nontype material,” i.e., whether they represent strains that have been assigned to a species without a comparison to the type strain of the species, and hence, their species designation is ambiguous. Such nontype material might have confounded our results with respect to the extent of overlap between the ranks of taxonomy. We expect, however, that the overlap due to nontype material is relevant only for the lower ranks of the taxonomy, i.e., the species and genus ranks, given that the classification of strains almost always employs comparisons of 16S rRNA gene sequences and the 16S rRNA gene has good resolution at the family level or higher. Further, many strains whose histories can be easily tracked down, including strains causing overlap between nonadjacent ranks, such as the Prochlorococcus strains (6), represent the type strain of the species or have been compared to the type strain. In any case, we anticipate that the overlap due to nontype material is narrow, probably much narrower than the overlap between nonadjacent ranks caused by clear inconsistencies in classification, and our approach identifies the genomes (nontype or not) whose classification needs to be reevaluated.
The genomic comparisons also revealed that there is probably a continuum of genetic diversity in the prokaryotic world as opposed to clear boundaries that separate organisms into specific groups or ranks (Fig. 3). Although Fig. 3 and 4 clearly show that there are many fewer genomes that are highly or moderately related (e.g., showing 60 to 90% AAI), our more detailed evaluation of the γ-Proteobacteria and the Firmicutes, the phyla that are best represented with genomic sequences, suggest that this is presumably a sampling bias rather than evidence of clear boundaries of relatedness. Therefore, the art of setting standards or cutoffs for designating the ranks of taxonomy will always be somewhat arbitrary, even with the availability of whole-genome sequences for all living organisms. Nonetheless, there is great comparative value in making the classification consistent, and the whole-genome-derived approach outlined here can significantly contribute to this goal.
Among the most interesting irregularities we noted in the current classification system is that the differences in terms of genetic distance between several of the bacterial or the archaeal phyla are comparable (or only slightly smaller) than the differences between Archaea and Bacteria. This is consistent with recent studies on shared gene content trees, as well as our own unpublished results that show many bacterial and archaeal phyla to be very deeply branching and close to the root between Archaea and Bacteria (15). In our own data set, only 16S rRNA gene data clearly support the idea that the interdomain differences are larger than the interphylum differences (Fig. 3). Although the possibility that the 16S rRNA gene has better resolution at the domain level than the genome average (e.g., AAI or gene content trees) cannot be excluded at this point, the relationship between 16S rRNA gene identity and AAI (Fig. 3), as well as the extensive genetic and biochemical distinctiveness of organisms related at this level, which presumably imposes varied functional constraints and selection pressures on the 16S rRNA gene, raise serious concerns as to how quantifiable 16S rRNA gene differences are at this level of relatedness. In other words, the differences we noted in terms of phenotype and genetic relatedness at the whole-cell level are not consistent with clear distinctions between (even) the higher ranks of the taxonomy, i.e., the domain and the phylum ranks.
Acknowledgments
We thank George Garrity, James Cole, and Joel Klappenbach, as well as two anonymous reviewers, for helpful discussions regarding the manuscript.
This work was supported by the Bouyoukos Fellowship Program (K.T.K.), the Department of Energy's Genomics:GtL Program, the Ribosomal Database Project (supported by the Department of Energy, the National Science Foundation, and the National Institutes of Health), and the Center for Microbial Ecology.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, D., J. Staley, and N. Krieg. 2000. Classification of prokaryotic organisms and the concept of bacterial speciation, p. 27-31. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 3.Brown, J. R., C. J. Douady, M. J. Italia, W. E. Marshall, and M. J. Stanhope. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281-285. [DOI] [PubMed] [Google Scholar]
- 4.Canale-Parola, E. 1984. Order I: Spirochaetales Buchanan 1917, 163AL, p. 38-39. In N. R. Krieg, N. and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. William and Wilkins, Baltimore, Md. [Google Scholar]
- 5.Charlebois, R. L., and W. F. Doolittle. 2004. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chisholm, S., S. Frankel, R. Goericke, R. Olson, B. Palenik, B. Waterbury, L. West-Johnrud, and E. Zettler. 1992. Prochlorococcus marinus nov. gen. sp.: an oxyphototrophic prokaryote containing divinyl chrolophyll a and b. Arch. Microbiol. 157:297-300. [Google Scholar]
- 8.Coenye, T., and P. Vandamme. 2004. Use of the genomic signature in bacterial classification and identification. Syst. Appl. Microbiol. 27:175-185. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubin, V., M. Gouy, and G. Perriere. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daubin, V., N. A. Moran, and H. Ochman. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829-832. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2129. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- 14.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gophna, U., W. F. Doolittle, and R. L. Charlebois. 2005. Weighted genome trees: refinements and applications. J. Bacteriol. 187:1305-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta, R. S., and E. Griffiths. 2002. Critical issues in bacterial phylogeny. Theor. Popul. Biol. 61:423-434. [DOI] [PubMed] [Google Scholar]
- 17.Hong, S. H., T. Y. Kim, and S. Y. Lee. 2004. Phylogenetic analysis based on genome-scale metabolic pathway reaction content. Appl. Microbiol. Biotechnol. 65:203-210. [DOI] [PubMed] [Google Scholar]
- 18.Konstantinidis, K. T., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krieg, N., and G. Garrity. 2000. On using the manual, p. 15-19. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 20.Kunin, V., D. Ahren, L. Goldovsky, P. Janssen, and C. A. Ouzounis. 2005. Measuring genome conservation across taxa: divided strains and united kingdoms. Nucleic Acids Res. 33:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence, J. G., and H. Hendrickson. 2003. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50:739-749. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, W., and H.-P. Klenk. 2000. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics, p. 49-65. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 23.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasko, D. A., G. S. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12:85-94. [DOI] [PubMed] [Google Scholar]
- 26.Sander, C., and R. Schneider. 1991. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9:56-68. [DOI] [PubMed] [Google Scholar]
- 27.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 28.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 29.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the Ad Hoc Committee for the Re-evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 30.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 31.Wolf, Y. I., I. B. Rogozin, N. V. Grishin, R. L. Tatusov, and E. V. Koonin. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]