Abstract
Bacterial promoters belonging to the extended −10 class contain a conserved TGn motif upstream of the −10 promoter consensus element. Open promoter complexes can be formed on some extended −10 Escherichia coli promoters at temperatures as low as 6°C, when complexes on most promoters are closed. The promoter of cspA, a gene that codes for the major cold shock protein CspA of E. coli, contains an extended −10 motif. CspA is dramatically induced upon temperature downshift from 37 to 15°C, and its cold shock induction has been attributed to transcription, translation, and mRNA stabilization effects. Here, we show that though the extended −10 motif is critical for high-level expression of cspA, it does not contribute to low-temperature expression. In fact, transcription from the wild-type cspA promoter is cold sensitive in vitro and in vivo. Thus, transcription appears to play little or no role in low-temperature induction of cspA expression.
Most Escherichia coli promoters belong to the −10/−35 class, which is characterized by the presence of two 6-bp promoter consensus elements positioned approximately 10 and 35 nucleotides upstream of the transcription initiation start point. Within the context of the RNA polymerase (RNAP) holoenzyme, the σ70 subunit conserved regions 4.2 and 2.4 recognize the −35 and the −10 promoter consensus elements, respectively, leading to promoter complex formation and transcription initiation (for reviews, see references 14 and 28). Some promoters lack the −35 promoter consensus element and instead contain a TGn extension upstream of their −10 promoter consensus element. The TGn motif of these so-called extended −10 class promoters is involved in specific interactions with an additional region of σ70, conserved region 3.0 (6, 38). This additional contact is strong enough to make promoter complex formation on extended −10 promoters independent of the σ70 region 4.2 interaction with the −35 promoter element (21).
Localized melting (opening) of promoter DNA is necessary for the synthesis of RNA. Promoter opening is temperature dependent, and promoter complexes formed on the −10/−35 class promoters close below 15°C (41). In contrast, promoter complexes on the extended −10 galP1 promoter remain open at temperatures as low as 6°C (1, 4, 5, 13). Although the mechanism of this unusual phenomenon is not fully elucidated, the extended −10 motif was shown to be critical for low-temperature opening of galP1 (13). However, this motif alone is not sufficient, and some extended −10 promoters are sensitive to low temperature (3, 5). Synthesis of the small regulatory RNA DsrA is under temperature control, and it has been shown that the sequence of the −10 box and the spacer region are the essential elements for the thermal response of this promoter (36).
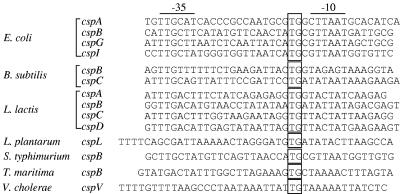
Upon the temperature downshift, the cells exhibit a cold shock response characterized by a transient arrest of cell growth, severe inhibition of general protein synthesis, and transient induction of cspA and three of its homologues, cspB, cspG, and cspI (for reviews, see references 9 and 30). Eventually, cell growth resumes but at a reduced rate. CspA homologues are widespread in bacteria and most csp genes' promoters contain the extended −10 motif (Fig. 1). For example, promoters of all of the four cold shock-inducible csp genes (cspA, cspB, cspG, and cspI) in E. coli contain a TGn motif. CspE is cold shock inducible in the absence of CspA, CspB, and CspG, and its promoter also contains TGn motif. However, this motif is not strictly conserved and is missing in promoters for genes coding for cold shock-inducible CspA homologues from, for example, Caulobacter crescentus (22), Lactobacillus bulgaricus (40), Salmonella enterica (19), and Staphylococcus aureus (18). Nonetheless, based on the published literature suggesting a link between the TGn motif and the low-temperature opening of certain E. coli promoters, it is tempting to speculate that the TGn motif contributes to transcription of E. coli cspA at cold shock conditions.
FIG. 1.
Comparison of the promoter regions of genes encoding CspA homologues. The promoter regions of E. coli cspA (43), cspB (23), cspG (29), and cspI (47), Bacillus subtilis cspB and cspC (12), Lactobacillus lactis cspA, cspB, cspC, and cspD (49), Lactobacillus plantarum cspL (25), Salmonella enterica serovar Typhimurium cspB (7), Thermotoga maritima cspB (48), and Vibrio cholerae cspV (8). The −35 and −10 regions are shown by lines, and extended −10 motifs are boxed.
It has been reported that upon temperature downshift, CspA accounts for almost 10% of the total cellular protein (17). This cold shock induction is thought to be accomplished at the level of transcription, translation, and mRNA stability (for reviews, see references 9 and 30). One of the unique features of the cspA, cspB, cspG, and cspI genes is the unusually long 5′ untranslated region. This region is responsible for both the extreme instability of csp mRNA at 37°C (a half-life of ∼12 s) and for stabilization of mRNA at low temperatures (a half-life of ∼20 min at 15°C) (26). The cspA mRNA also contains a unique sequence located 14 bases downstream of the initiation codon, termed the translation enhancing element (35). This element is also present in CspB, CspG, CspI, and CsdA and is thought to enhance translation initiation at cold shock (26). Experiments involving transcription-translation fusions indicated that the cspA promoter, which is highly active at 37°C, also contributes to cold shock induction of cspA (11, 26).
The cold shock transcription of cspA has been a topic of some discussion, the conclusion being that transcription is moderately elevated upon temperature downshift. In vivo data, such as those from primer extension, Northern blot analysis, or DNA microarray analysis, demonstrated a 4- to 5-fold increase in cspA transcript under cold shock conditions (31). However, none of these analyses can distinguish if the increase in cspA transcript was due to cold shock transcription alone, was a combined effect of transcription and mRNA stabilization upon temperature downshift, or was due solely to mRNA stabilization. In the present study, we studied low-temperature transcription of cspA through mutational analysis of the cspA promoter and in vitro transcription experiments. We show that the cspA promoter-extended −10 motif is critical for transcription at all temperatures and that its contribution to low-temperature transcription is mild. We show that transcription from the cspA promoter is strongly inhibited at cold shock conditions both in vivo and in vitro at the level of open promoter complex formation. Thus, the high-level induction of CspA upon cold shock is probably entirely due to dramatic stabilization of its mRNA and its cold-efficient translation machinery.
cspA transcription in vivo is affected by disruption of the extended −10 motif.
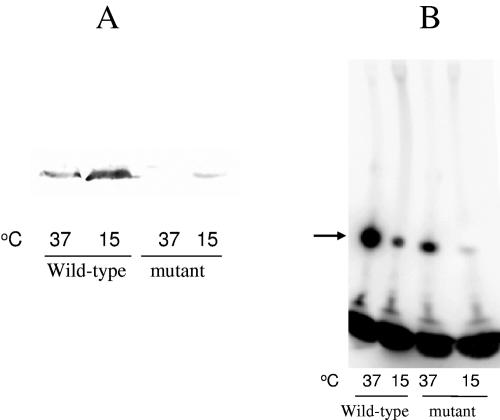
E. coli strain JM83 [F− araΔ(lac-proAB) rpsL(Strr)] (51) and its cspA deletion strain (2) were used in this study. The plasmid pJJGO2, containing entire promoter region, 5′ untranslated region, and coding region of cspA used in this study has been described previously (16). E. coli cells grown overnight in LB medium at 37°C were diluted into fresh LB medium. Cells were grown at 37°C to exponential phase (optical density at 600 nm of 0.5), and part of the cell culture was harvested and used as control. Aliquots of the cells were transferred to a prechilled LB medium at 15°C and the cells were harvested after 1 h of cold shock. The total RNA was extracted by the hot-phenol method described previously (39). The primer extension, Northern blot analysis, and the deoxyoligonucleotide used for the detection of cspA were described previously (32, 37, 50). The products were analyzed on a 6% polyacrylamide gel under denaturing conditions. The radioactive measurements of products were carried out by phosphorimaging and quantitative analysis of the bands. As seen from the primer extension (Fig. 2A) and Northern blot (Fig. 2B) analyses, the amount of the cspA transcript in wild-type E. coli cells is increased ∼5-fold upon transfer of exponentially growing E. coli culture to 15°C followed by a 60-min incubation at the same temperature. DNA microarray analysis of the cold-shocked E. coli also shows a similar increase in the cspA transcript level (31, 33).
FIG. 2.
Changes in cspA mRNA levels in response to cold shock. Total RNA was extracted by the hot-phenol method. Primer extension (A) and Northern blot analysis (B) were carried out with deoxyoligonucleotide corresponding to cspA. Control (37°C) and cold-shocked (15°C for 1 h) cells were used.
To investigate the role of the extended −10 motif in cspA transcription and cold shock induction, we substituted, by site-directed mutagenesis, the TG of the extended −10 motif by an AC. The mutant promoter, alongside the entire cspA transcription unit, was maintained on a plasmid (pJJGO2). A corresponding wild-type cspA plasmid was used as a control. Control experiments established that the copy numbers of both plasmids were the same (data not shown). First, we examined if the promoter mutation affects the cold shock induction of CspA. Plasmids carrying wild-type or mutant cspA promoters were transformed in E. coli ΔcspA cells. Cells were grown at 37°C, and the cultures were split. One half was kept at 37°C, and the other half was subjected to a 1-h cold shock at 15°C. Cell extracts were prepared and subjected to Western blot analysis (32) with anti-CspA antibodies (43). As can be seen from Fig. 3A, CspA was induced by cold shock in cells carrying the wild-type cspA promoter, as expected. Disruption of the extended −10 motif strongly affected CspA expression at both 37 and 15°C. At 37°C, the effect was not possible to quantify, since the amount of CspA present in cells carrying the mutant plasmid was beyond the detection limit. At 15°C, the amount of CspA present in cells carrying promoter mutation was increased; however, the final level was >10-fold less than that in control cells. Thus, it appears that the extended −10 motif makes a critical contribution to the overall strength of the cspA promoter but may not participate in cold shock induction.
FIG. 3.
Effect of disruption of the extended −10 motif on induction of cspA. (A) Western blot analysis of E. coli ΔcspA cells carrying pJJGO2 plasmids with either the wild-type or the mutated cspA promoter region was carried out with anti-CspA antibodies. Cell extracts were prepared from cells grown at 37°C and cells cold shocked at 15°C for 1 h. (B) Abortive transcription initiation using DNA templates containing either wild-type or mutated promoter region of cspA. The reactions were carried out at 37 or 15°C. Reaction products were separated by denaturing PAGE. The position of the synthesized product (CpApA) is indicated by an arrow.
Transcription from the cspA promoter in vitro.
To study the possible cold shock-induced transcription from the cspA promoter in the absence of RNA degradation that complicates data interpretation, we next studied transcription initiation from the wild-type and the mutant cspA promoters in vitro. Promoter complexes were prepared by using E. coli RNAP σ70 holoenzyme and linear DNA templates extending from position −100 to +50 relative to the cspA transcription start site at +1. Reactions were set up at either 15 or 37°C. Reaction mixtures were supplemented with transcription substrates, the CpA primer, and radioactive ATP, to allow the production of radioactively labeled abortive transcript CpApA. Reaction products were separated by denaturing polyacrylamide gel electrophoresis (PAGE). The results are shown in Fig. 3B. As can be seen, the cspA transcription was cold sensitive, and three times less transcript was produced from the wild-type cspA promoter at 15°C than that at 37°C. In agreement with the Western blot data, the mutant promoter was less active than the wild-type one at both temperatures. The mutant promoter appeared to be only slightly more cold sensitive than the wild type (five times less transcript produced at 15°C than at 37°C). Identical results were obtained in a runoff transcription assay, indicating that promoter clearance plays no role in cold-resistant transcription from cspA.
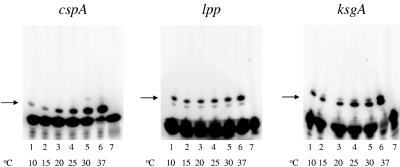
The experiment presented above assayed abortive initiation from cspA at only two temperatures, 15 and 37°C. To better understand the cspA response to temperature, we carried out abortive transcription initiation reactions from cspA at temperatures ranging from 10 to 37°C. We used, as controls, the lpp and ksgA promoters. These promoters served as negative controls, since microarray analysis suggested that the RNA synthesized from these promoters is not cold inducible (31). As can be seen from Fig. 4, decreasing reaction temperature led to decreased transcription from all three promoters. Contrary to our expectations, the cspA promoter appeared to be the most cold sensitive of the three. Therefore, despite the fact that the steady-state level of the cspA transcript increases upon cold shock, low-temperature transcription from the cspA promoter is inefficient, at least in vitro from linear DNA.
FIG. 4.
Temperature-dependent transcription profiles of cspA, lpp, and ksgA. Abortive transcription initiation using DNA templates containing promoter regions of cspA, lpp, and ksgA. The reactions were carried out at indicated temperatures. Reaction products were separated by denaturing PAGE. For lpp, CpG and [α-32P]CTP were used, and for ksgA, GpC and [α-32P]UTP were used. Lane 7 in each case is the respective [α-32P]NTP loaded on the gel as a control. The positions of the respective synthesized products are indicated by arrows.
The cspA promoter does not form open complexes at low temperature.
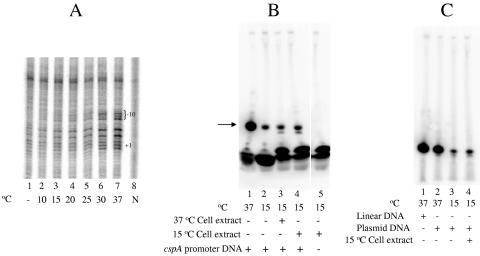
In the light of the low-temperature sensitivity of the cspA promoter in abortive transcription assays, we examined if this promoter forms open complexes at low temperature. We used KMnO4 probing (42) to monitor the cspA promoter opening at temperatures ranging from 10 to 37°C. The template, which was the same as that used for abortive transcription studies described above, was terminally labeled to visualize probing reaction products. KMnO4 modifies thymine residues that are present in single-stranded form and thus allows the following of promoter opening. In the absence of RNAP, thymine residues are present in the double-stranded form and are not accessible to KMnO4 (Fig. 5A, lane 1). The control reactions without RNAP were carried out at both 37 and 15°C and showed the same pattern. At 37°C and in the presence of RNAP, thymine residues at and around the transcription start point became susceptible to the probing agent due to open complex formation (Fig. 5A, lane 7). As the temperature of the reaction was lowered, the intensity of KMnO4-sensitive bands uniformly decreased. Little or no KMnO4 sensitivity was observed below 20°C. Thus, the cspA promoter's inability to function at low temperature in vitro is due to the fact that no open complexes form below 20°C.
FIG. 5.
Promoter opening and transcription of cspA at a low temperature. (A) Temperature-dependent open complex formation by the cspA promoter. Promoter complexes were formed at indicated temperatures by using cspA promoter-containing fragments (both strands were 32P end labeled) and RNAP holoenzyme. Complexes were probed with KMnO4 and reaction products were separated by urea-PAGE and revealed by autoradiography. Lane 1 represents a reaction without RNAP at 37°C. Lane 8 (N) represents naked DNA without KMnO4 treatment. Positions of the −10 region and transcription start sites of cspA are indicated. (B) Low-temperature transcription of cspA in the presence of cell extract. Lanes 1 and 2 represent control reactions carried out at 37 and 15°C, respectively. Lanes 3 and 4 represent reactions carried out at 15°C in the presence of cell extract prepared at 37°C and 15°C, respectively. Lane 5 represents a control reaction carried out at 15°C in the presence of 15°C cell extract, without cspA promoter DNA. The position of the synthesized product is indicated by an arrow. (C) Effect of DNA supercoiling on low-temperature transcription of cspA. Abortive transcription initiation reactions were carried out by using linear or supercoiled DNA containing the cspA promoter. Lane 1 represents a control reaction carried out at 37°C using linear DNA. Lanes 2 through 4 represent reactions carried out by using supercoiled (plasmid-based) DNA containing the cspA promoter at the indicated temperatures. Cell extract prepared from cells cold shocked at 15°C for 1 h was included in the reaction represented in lane 4.
The experiments carried out above were in a pure system devoid of any cellular factors. We considered a possibility that in vivo, the cspA promoter may be aided by some cellular factor(s) that is either induced by cold shock or that may be present even at optimum growth temperature. To explore this possibility, we prepared total cellular extracts from cells that were grown at 37°C and then subjected to a 1-h cold shock at 15°C. As control, cell extracts prepared from cells grown at 37°C were used. The cell extracts were prepared by sonicating the cell pellets followed by centrifugation to clear the cell debris. The extracts were added to abortive transcription initiation reactions from the cspA promoter with the idea that a factor(s) present in the extract may allow transcription at low temperature to occur (Fig. 5B). As can be seen from the figure, addition of cell extracts did not lead to increased transcription from cspA at low temperature. The origin of faint bands appearing below synthesized product in lanes 3 and 4 is unknown; it may suggest degradation by nucleases present in the cell extract. However, nuclease inhibitors were included in the reactions, and these minor bands did not show variation in response to different amounts of cell extract added. Inclusion of unclarified cell extracts or different amounts of cell extract in abortive transcription assays did not affect the transcription (data not shown). A control reaction (Fig. 5B, lane 5) without cspA promoter DNA shows that the cell extract does not contribute to transcription. Results of KMnO4 probing of promoter complexes formed in the presence of cell extracts were consistent with that of transcription assays (data not shown). We also considered the possibility that in vivo, the cspA promoter can become open at low temperature in the presence of all four NTPs (such a behavior was reported for a mutant RNAP that forms cold-resistant open complexes [41]). However, even in the presence of high concentration of NTPs, no transcription from and/or promoter opening of the cspA promoter was detected (data not shown). Taken together, these data do not support an idea that a specific cellular factor may aid cspA transcription during cold shock.
When cells experience cold shock, negative supercoiling of DNA transiently increases, which could favor increased transcription from some promoters (20, 27, 46). To test if the low-temperature sensitivity of the cspA promoter depends on DNA topology, we carried out abortive transcription initiation assays using supercoiled plasmid containing the cspA promoter (Fig. 5C). In lane 1, transcription products from a control reaction carried out at 37°C by using a linear cspA template are shown. Equimolar amounts of supercoiled DNA were used as templates in reactions represented in lanes 2 to 4. At 37°C, cspA transcription was not significantly influenced by template supercoiling (compare lanes 1 and 2). Moreover, supercoiling had no stimulatory effect on cspA transcription at 15°C (lane 3). The addition of cell extract prepared from cold-shocked cells to reaction mixtures containing supercoiled DNA did not allow low-temperature transcription (lane 4). We therefore conclude that supercoiling of DNA is unlikely to contribute to cold-shock induction of the cspA transcript at low temperature.
No open promoter complexes are formed on the cspA promoter in vivo.
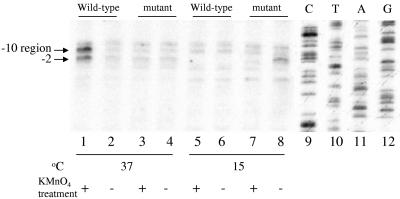
Since our in vitro analysis failed to reveal cold-resistant transcription from the cspA promoter, we next followed the cspA open promoter complex formation in E. coli cells, using in vivo KMnO4 probing. KMnO4 probing is a direct measure of open complex formation (and hence, transcription activity of a promoter). Therefore, KMnO4 probing allows one to avoid the contribution of RNA degradation and/or stabilization that influences the measurements of steady-state levels of mRNA in vivo. The in vivo KMnO4 probing was carried out as described by Marr and Roberts (24). E. coli JM83 cells were transformed with pJJGO2 plasmids carrying either wild-type or mutant cspA promoters. The cells were grown in LB broth at 37°C until exponential phase. Ten milliliters of cells was treated with KMnO4. Reactions were terminated by the addition of 11 ml STE (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA). Aliquots of 10 ml of cells were also transferred to 15°C and cold shocked for 30 to 60 min and then treated with KMnO4. Plasmids were isolated by using the QIAGEN kit. Modified thymine residues were detected by linear PCR (43) with deoxyoligonucleotide used for detection of cspA mRNA by primer extension. Equal amounts of plasmid DNA were used in the PCRs. The deoxyoligonucleotide was end labeled with [γ-32P]ATP. The reaction products were resolved by using 6% polyacrylamide-urea gels followed by autoradiography. The results are shown in Fig. 6. For each condition, a control reaction without KMnO4 treatment was carried out to assess the background sensitivity of thymine residues (compare lanes 1, 3, 5, and 7 with lanes 2, 4, 6, and 8, respectively). As expected, the wild-type cspA promoter was open at 37°C (lane 1), while the mutant promoter was closed (lane 3). At 15°C, both promoters were closed (lanes 5 and 7). A control reaction with cold-shocked cells carrying the wild-type cspA promoter and treated with rifampin prior to KMnO4 probing did not reveal any increased sensitivity of the cspA promoter thymines (data not shown). Rifampin blocks RNAP escape from the promoter and thus should have increased the level of KMnO4 sensitivity if RNAP were able to melt the cspA promoter in vivo. Based on these results, we conclude that, in agreement with our in vitro analysis, the contribution of the cspA transcription to the cold shock induction of cspA mRNA is negligible.
FIG. 6.
Temperature-dependent open complex formation on the cspA prompter in vivo. In vivo KMnO4 probing was carried out by using wild-type or mutated cspA promoters. Reaction temperatures are indicated. Lanes 2, 4, 6, and 8 represent control reactions carried out without KMnO4 treatment for lanes 1, 3, 5, and 7, respectively. The KMnO4-sensitive bands were positioned with the help of a sequencing ladder shown in lanes 9 through 12. Positions of KMnO4-sensitive thymines in the open promoter complex are indicated. The gel is representative of an experiment carried out three times.
Cold shock induction of CspA.
The cold shock induction of CspA has been a topic of extensive research over the past few years. The dramatic stabilization of the cspA mRNA and its efficient translation at low temperature by virtue of structural elements, such as translation-enhancing element contribute significantly to making CspA a major cold shock protein. On the hand, there has been a dispute regarding the importance of cold shock transcription of cspA (15). By studying the expression profiles of reporter genes such as lacZ or cat fused to the cspA promoter, two groups (11, 26) independently showed that the cspA promoter responds to temperature downshift (an ∼3-fold increase in activity at 15°C). On the other hand, Fang et al. (10) showed that CspA could be induced by cold shock independently of its promoter. However, the cold shock induction of CspA from the heterologous lpp promoter was less than that observed from the cspA promoter. In vivo data such as primer extension, Northern blot analysis or DNA microarray analysis consistently demonstrate the upshift (4- to 5-fold increase) in cspA transcript (31). However, these analyses cannot distinguish if the increase is due to cold shock transcription alone, is a combined effect of transcription and mRNA stabilization upon temperature downshift, or is solely due to message stabilization. Several expression vectors (34, 44, 45) constructed to achieve heterologous expression of proteins at low temperature under transcriptional control of cspA also did not include the cspA promoter alone but consist of many if not all of the structural elements of cspA that contribute to stabilization of its mRNA and cold efficient translation. Thus, it was not possible to adjudge the efficiency of cold shock transcription of cspA alone.
In the view of the published data that certain promoters containing the extended −10 motif stay open at low temperature, and since this motif is present in several CspA homologues, our aim in the present study was to explore the importance of this motif in cold shock induction of CspA. We initiated the study on the assumption that albeit moderate, cspA transcription is elevated upon temperature downshift. Our results showed that TGn motif is essential for efficient transcription of cspA at all temperatures and that cspA transcription is temperature sensitive. The combined results of the in vitro and in vivo experiments suggest strongly that the contribution of de novo transcription to cspA induction at cold shock is marginal. The absence of significant transcription from cspA during cold shock is also supported by the following considerations. The steady-state balance between transcription of the cspA promoter and the cspA mRNA degradation can be described by a simple mathematical formula: R = (M/t1/2) × ln2. In this equation, R is the rate of transcription, M is the steady-state level of mRNA, and t1/2 is the half-life of mRNA. Considering that the half-life of the cspA mRNA increases from 12 s at 37°C to 20 min at 15°C, as little as 5% residual transcription at 15°C (compared to transcription at 37°C) will be sufficient to account for the observed fivefold increase in the steady-state cspA mRNA level upon cold shock. Thus, the induction of CspA upon cold shock appears to be solely due to dramatic stabilization of its mRNA and its cold-resistant translation.
Acknowledgments
We thank Anirvan Sengupta for his help in mathematical modeling of the data.
This work was supported in part by the National Institutes of Health RO1 grant 64530 to K.S.
REFERENCES
- 1.Attey, A., T. Belyaeva, N. Savery, J. Hoggett, N. Fujita, A. Ishihama, and S. Busby. 1994. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the Escherichia coli galactose operon P1 promoter. Nucleic Acids Res. 22:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, W., P. G. Jones, and M. Inouye. 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 179:7081-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyaeva, T., L. Griffiths, S. Minchin, J. Cole, and S. Busby. 1993. The Escherichia coli cysG promoter belongs to the “extended −10” class of bacterial promoters. Biochem. J. 296:851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, H., and S. Minchin. 1994. Thermal energy requirement for strand separation during transcription initiation: the effect of supercoiling and extended protein DNA contacts. Nucleic Acids Res. 22:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns, H. D., T. A. Belyaeva, S. J. Busby, and S. D. Minchin. 1996. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem. J. 317:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 7.Craig, J. E., D. Boyle, K. P. Francis, and M. P. Gallagher. 1998. Expression of the cold-shock gene cspB in Salmonella typhimurium occurs below a threshold temperature. Microbiology 144:697-704. [DOI] [PubMed] [Google Scholar]
- 8.Datta, P. P., and R. K. Bhadra. 2003. Cold shock response and major cold shock proteins of Vibrio cholerae. Appl. Environ. Microbiol. 69:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ermolenko, D. N., and G. I. Makhatadze. 2002. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59:1902-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, L., W. Jiang, W. Bae, and M. Inouye. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37 degrees C by mRNA stabilization. Mol. Microbiol. 23:355-364. [DOI] [PubMed] [Google Scholar]
- 11.Goldenberg, D., I. Azar, A. B. Oppenheim, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol. Gen. Genet. 256:282-290. [DOI] [PubMed] [Google Scholar]
- 12.Graumann, P., T. M. Wendrich, M. H. Weber, K. Schroder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 13.Grimes, E., S. Busby, and S. Minchin. 1991. Different thermal energy requirement for open complex formation by Escherichia coli RNA polymerase at two related promoters. Nucleic Acids Res. 19:6113-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, C. A., C. Chan, A. Dombroski, T. Gruber, M. Sharp, J. Tupy, and B. Young. 1998. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp. Quant. Biol. 63:141-155. [DOI] [PubMed] [Google Scholar]
- 15.Gualerzi, C. O., A. M. Giuliodori, and C. L. Pon. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527-539. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, W., L. Fang, and M. Inouye. 1996. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J. Bacteriol. 178:4919-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, P. G., and M. Inouye. 1994. The cold-shock response—a hot topic. Mol. Microbiol. 11:811-818. [DOI] [PubMed] [Google Scholar]
- 18.Katzif, S., D. Danavall, S. Bowers, J. T. Balthazar, and W. M. Shafer. 2003. The major cold shock gene, cspA, is involved in the susceptibility of Staphylococcus aureus to an antimicrobial peptide of human cathepsin G. Infect. Immun. 71:4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, B. H., I. S. Bang, S. Y. Lee, S. K. Hong, S. H. Bang, I. S. Lee, and Y. K. Park. 2001. Expression of cspH, encoding the cold shock protein in Salmonella enterica serovar Typhimurium UK-1. J. Bacteriol. 183:5580-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krispin, O., and R. Allmansberger. 1995. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 134:129-135. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 22.Lang, E. A., and M. V. Marques. 2004. Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain. J. Bacteriol. 186:5603-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. J., A. Xie, W. Jiang, J. P. Etchegaray, P. G. Jones, and M. Inouye. 1994. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol. Microbiol. 11:833-839. [DOI] [PubMed] [Google Scholar]
- 24.Marr, M. T., and J. W. Roberts. 2000. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 6:1275-1285. [DOI] [PubMed] [Google Scholar]
- 25.Mayo, B., S. Derzelle, M. Fernandez, C. Leonard, T. Ferain, P. Hols, J. E. Suarez, and J. Delcour. 1997. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. J. Bacteriol. 179:3039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitta, M., L. Fang, and M. Inouye. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 26:321-335. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima, T., K. Kataoka, Y. Ogata, R. Inoue, and K. Sekimizu. 1997. Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol. Microbiol. 23:381-386. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima, K., K. Kanamaru, T. Mizuno, and K. Horikoshi. 1996. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J. Bacteriol. 178:2994-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125-136. [PubMed] [Google Scholar]
- 31.Phadtare, S., and M. Inouye. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186:7007-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154:573-580. [DOI] [PubMed] [Google Scholar]
- 34.Qing, G., L. C. Ma, A. Khorchid, G. V. Swapna, T. K. Mal, M. M. Takayama, B. Xia, S. Phadtare, H. Ke, T. Acton, G. T. Montelione, M. Ikura, and M. Inouye. 2004. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22:877-882. [DOI] [PubMed] [Google Scholar]
- 35.Qing, G., B. Xia, and M. Inouye. 2004. Enhancement of translation initiation by A/T-rich sequences downstream of the initiation codon in Escherichia coli. J. Mol. Microbiol. Biotechnol. 6:133-144. [DOI] [PubMed] [Google Scholar]
- 36.Repoila, F., and S. Gottesman. 2003. Temperature sensing by the dsrA promoter. J. Bacteriol. 185:6609-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sanderson, A., J. E. Mitchell, S. D. Minchin, and S. J. Busby. 2003. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 544:199-205. [DOI] [PubMed] [Google Scholar]
- 39.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 40.Serror, P., R. Dervyn, S. D. Ehrlich, and E. Maguin. 2003. csp-like genes of Lactobacillus delbrueckii ssp. bulgaricus and their response to cold shock. FEMS Microbiol. Lett. 226:323-330. [DOI] [PubMed] [Google Scholar]
- 41.Severinov, K., and S. A. Darst. 1997. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl. Acad. Sci. USA 94:13481-13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Severinova, E., K. Severinov, and S. A. Darst. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 279:9-18. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe, H., J. Goldstein, M. Yang, and M. Inouye. 1992. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J. Bacteriol. 174:3867-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasina, J. A., and F. Baneyx. 1997. Expression of aggregation-prone recombinant proteins at low temperatures: a comparative study of the Escherichia coli cspA and tac promoter systems. Protein Expr. Purif. 9:211-218. [DOI] [PubMed] [Google Scholar]
- 45.Vasina, J. A., and F. Baneyx. 1996. Recombinant protein expression at low temperatures under the transcriptional control of the major Escherichia coli cold shock promoter cspA. Appl. Environ. Microbiol. 62:1444-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J. Y., and M. Syvanen. 1992. DNA twist as a transcriptional sensor for environmental changes. Mol. Microbiol. 6:1861-1866. [DOI] [PubMed] [Google Scholar]
- 47.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welker, C., G. Bohm, H. Schurig, and R. Jaenicke. 1999. Cloning, overexpression, purification, and physicochemical characterization of a cold shock protein homolog from the hyperthermophilic bacterium Thermotoga maritima. Protein Sci. 8:394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wouters, J. A., J. W. Sanders, J. Kok, W. M. de Vos, O. P. Kuipers, and T. Abee. 1998. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology 144:2885-2893. [DOI] [PubMed] [Google Scholar]
- 50.Xia, B., H. Ke, W. Jiang, and M. Inouye. 2002. The Cold Box stem-loop proximal to the 5′-end of the Escherichia coli cspA gene stabilizes its mRNA at low temperature. J. Biol. Chem. 277:6005-6011. [DOI] [PubMed] [Google Scholar]
- 51.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]