Abstract
The global regulators AbrB, Abh, and SpoVT are paralogous proteins showing their most extensive sequence homologies in the DNA-binding amino-terminal regions (about 50 residues). The carboxyl-terminal portion of AbrB has been hypothesized to be a multimerization domain with little if any role in DNA-binding recognition or specificity. To investigate the multimerization potentials of the carboxyl-terminal portions of AbrB, Abh, and SpoVT we utilized an in vivo multimerization assay system based upon fusion of the domains to the DNA binding domain of the λ cI repressor protein. The results indicate that the N and C domains of all three paralogues are independent dimerization modules and that the intact Abh and SpoVT proteins are most probably tetramers. Chimeric proteins consisting of the AbrB N-terminal DNA-binding domain fused to the C domain of either Abh or SpoVT are indistinguishable from wild-type AbrB in their ability to regulate an AbrB target promoter in vivo.
The Bacillus subtilis AbrB protein is known to regulate the expression of well over 100 genes that are involved in adaptation to nutrient-limiting environments. In Bacillus species, AbrB plays a crucial regulatory role in the production of antibiotics (14, 19, 27), the formation of biofilms and fruiting bodies (5, 10), the development of competence (9), stationary phase survival (19, 24), sporulation gene expression (16, 34), the production of extracellular proteases and other degradative enzymes (19), cannibalistic behavior (8), and, in B. anthracis, the expression of toxin genes (20). Critical for AbrB's global regulatory properties is its ability to recognize and specifically bind a diverse, yet finite, subset of differing base sequences located in the promoter regions of genes it controls (26). Although optimal binding sequences converging to a consensus can be selected in vitro, examination of known chromosomal binding sites cannot be readily characterized by derivation of a base sequence consensus (31). It is believed that AbrB-DNA binding interactions entail recognition of specific stretches of three-dimensional DNA helix configurations that are characterized by a subset of allowable structural parameters (e.g., minor groove width, degree of propeller twisting, etc.) with which the protein can productively interact (4, 23).
Initial observations had suggested that the native B. subtilis AbrB protein was a hexamer of identical 96-amino-acid monomeric subunits (25, 29), but it is now known that it is actually a homotetramer of 94-amino-acid residue subunits (3, 28). On the basis of a study of abrB mutations and mutant proteins it was hypothesized that the monomeric subunit could be divided into separable domains with the N-terminal portion possessing most, if not all, of the determinants required for DNA recognition and specificity (29). The role of the C-terminal domain was posited to be primarily in multimer formation. Analysis of purified N-terminal domain peptides (residues 1 to 53 and 1 to 55) confirmed that DNA-binding specificity resided therein, and nuclear magnetic resonance (NMR) analysis of these domains revealed that they form a stable dimeric structure in solution (28, 30). Genes encoding AbrB orthologues (primarily having highest homology in the N-terminal DNA-binding domain) have been discovered in a wide variety of gram-positive, gram-negative, and archaeal species. Two AbrB paralogues are present in B. subtilis (1, 13) and other Bacillus and Clostridium species that have been sequenced. These paralogues, Abh and SpoVT (Fig. 1), exhibit a very high degree of amino acid sequence identity with AbrB over the first 50 residues (i.e., the DNA-binding domain). The C-terminal domains are significantly more diverse in sequence and, in the case of SpoVT, size. Recently, Abh has been shown to be a DNA-binding global regulator of gene expression (M. A. Strauch, unpublished data). Although less is known about SpoVT, it has been shown to regulate expression of at least 15 genes, presumably via DNA-binding interactions with target promoters (1).
FIG. 1.
Sequence alignment of the B. subtilis AbrB, Abh, and SpoVT proteins. An asterisk indicates positions of amino acid identity in all three paralogues; a period denotes positions where two of the three proteins have identical amino acids at that position and the third protein has a related amino acid. The pound sign (#) denotes the conserved proline at position 50 in AbrB and Abh (position 51 of SpoVT).
The discovery of the dimerization ability of truncated AbrB N domains, the overall tetrameric structure of the intact protein, and analysis of additional abrB mutations led us to hypothesize that the C domains of AbrB (and its paralogues) are independent dimerization modules required for formation of the functional tetramer. To test this hypothesis, we utilized an in vivo multimerization assay system based upon fusion of putative multimerization domains to the DNA-binding domain of the λ cI repressor protein (15). Additionally, we constructed chimeric proteins having the AbrB N-terminal DNA-binding domain fused to the C-terminal domains of Abh and SpoVT and assayed the ability of the chimeras to regulate an AbrB-dependent target promoter in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage strains.
Strains and plasmids used, or constructed, in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Description, relevant properties, or use | Reference and/or source |
|---|---|---|
| E. coli | ||
| DH5α F′lacIQ | General-use host strain for cloning plasmids | Invitrogen |
| AG1688 | MC1061 F′128 lacIqlacZ::Tn5 | J. Hu (15) |
| JH372 | AG1688(λ202) = λPROR(OR2−)-lacZ reporter strain | J. Hu (15) |
| JH607 | AG1688(λ112OsPs) = λOsPs-lacZ reporter strain | J. Hu (15) |
| XZ980 | AG1688(λXZ970) = λOSPS(OR1434)-lacZ reporter | J. Hu (15) |
| TP611 | pcn mutant for low-copy-number cloning | K. Devine |
| B. subtilis | ||
| SWV994 | trpC2 spo0A12 tolB24 ΔthrC::abrB8-lacZ spcr | Lab strain |
| SWV1030 | SWV994 ΔamyE::pSpac-abrB, cmr | Lab strain |
| FYS75 | SWV994 ΔamyE::(pFY80) | This study |
| FYS76 | SWV994 ΔamyE::(pFY81) | This study |
| Plasmids | ||
| pJH391 | Cloning vector for cI DBD fusion constructs | J. Hu (15) |
| pFG157 | pJH391 with intact λcI | J. Hu (15) |
| pKH101 | pJH391 with cI DBD only | J. Hu (15) |
| pJH370 | pJH391 with cI-GCN4 (dimeric) | J. Hu (15) |
| pJH622 | pJH391 with cI-GCN4 (tetrameric) | J. Hu (15) |
| pFY7 | cI DBD fused to AbrB 48-94 | This study |
| pFY8 | cI DBD fused to SpoVT 50-178 | This study |
| pFY12 | cI DBD fused to AbrB 48-80 | This study |
| pFY13 | cI DBD fused to AbrB 48-94 L67P | This study |
| pFY15 | cI DBD fused to Abh 50-92 | This study |
| pFY16 | cI DBD fused to AbrB 27-94 | This study |
| pFY17 | cI DBD fused to SpoVT 92-178 | This study |
| pFY18 | cI DBD fused to AbrB 67-94 | This study |
| pFY20 | cI DBD fused to SpoVT 50-131 | This study |
| pFY21 | cI DBD fused to SpoVT 92-131 | This study |
| pFY22 | cI DBD fused to SpoVT 124-178 | This study |
| pFY28 | cI DBD fused to AbrB 4-53 | This study |
| pFY31 | cI DBD fused to AbrB 55-94 | This study |
| pFY32 | cI DBD fused to AbrB 4-94 | This study |
| pFY34 | cI DBD fused to SpoVT 59-178 | This study |
| pFY36 | cI DBD fused to SpoVT 59-131 | This study |
| pFY37 | cI DBD fused to SpoVT 4-160 | This study |
| pFY38 | cI DBD fused to SpoVT 59-160 | This study |
| pFY39 | cI DBD fused to Abh 55-92 | This study |
| pFY46 | cI DBD fused to AbrB 4-53 R24S | This study |
| pFY47 | cI DBD fused to AbrB 4-53 L34H | This study |
| pFY48 | cI DBD fused to AbrB 51-94 | This study |
| pFY49 | cI DBD fused to AbrB 53-94 | This study |
| pFY50 | cI DBD fused to AbrB 48-88 | This study |
| pFY51 | cI DBD fused to SpoVT 4-54 | This study |
| pFY52 | cI DBD fused to SpoVT 4-178 | This study |
| pFY53 | cI DBD fused to AbrB 4-53 E35G | This study |
| pFY54 | cI DBD fused to AbrB 4-53 R23S | This study |
| pFY55 | cI DBD fused to AbrB 48-94 N64I | This study |
| pFY56 | cI DBD fused to AbrB 4-94 R23S | This study |
| pFY58 | cI DBD fused to AbrB 48-94 C54Y | This study |
| pFY59 | cI DBD fused to AbrB 48-94 C54S | This study |
| pFY60 | cI DBD fused to AbrB 48-94 C54W | This study |
| pFY61 | cI DBD fused to AbrB 4-53 I43S | This study |
| pFY64 | cI DBD fused to AbrB 48-94 C54Y | This study |
| pDR67 | pSpac, amyE integrative vector | |
| pFY66 | pSpac-(abrB 1-50) | This study |
| pFY80 | pSpac-(abrB 1-50)-gly-(spoVT 55-178) | This study |
| pFY81 | pSpac-(abrB 1-50)-gly-(abh 54-92) | This study |
| Phage-S | ||
| λKH54 | λΔcI mutant | J. Hu |
| λimm21C | Heteroimmune control | J. Hu |
| λ supervir | Supervirulent mutant of λ | D. Friedman |
| λKH54h80 | λΔcI h80 (i.e. host range of φ80) | J. Hu |
Principal and outline of the λcI repressor fusion assay for multimerization domains.
The in vivo assay developed by James Hu and colleagues for detection of protein domains having multimerization activity was used in this study. Methods for constructing and assaying the fusions were according to previously published protocols and information supplied by the Hu Laboratory as part of their generous package of vectors, strains, and phages required for the technique (15, 32, 33). The principle of the assay is that the DNA-binding N-terminal domain (DBD) of the λcI repressor is unable to bind its cognate operator sequences unless fused to a peptide segment that can self-associate to form dimers or higher-order multimeric complexes. When the DBD is fused to a segment capable of self-multimerization, Escherichia coli cells expressing the fusion are immune to infection by lambdoid phages having the λ immunity region but are sensitive to phages expressing heteroimmune regions or to virulent mutants of λ. Chimeric genes encoding the λcI DBD fused at its C terminal to the peptide segment to be assayed are cloned into a plasmid vector downstream of a lacUV5 promoter. An initial assessment of a segment's capacity for self-association is resistance to λ infection of E. coli cells harboring the fusion vector. A semiquantitative confirmation can then be obtained by placing the fusion plasmid into different reporter strains having a lacZ gene under control of various λ promoter-operator constructs (see below).
Construction of cI fusion proteins.
Specific DNA fragments encoding various portions of the abrB, abh, and spoVT genes were cloned into the cI-DBD fusion vector pJH391 at the latter's SalI-BamHI sites as has been described. Oligonucleotide primers (IDT) for PCR generation of the fragments were designed to produce the desired in-frame fusion of the coding segments when joined to the SalI site of pJH391. When necessary, translation stop codons were engineered into the design of the oligonucleotides to truncate the fused segment at a desired point. The sequences of all oligonucleotides used in this study are available upon request. The fusion plasmids were initially introduced into DH5α lacIq for selection and propagation. Plasmid preparations for transformation of reporter strains were made using kits purchased from QIAGEN. When necessary, DNA sequencing to confirm the proper nature of the fusion point, or the presence of a desired abrB mutation in a segment, was performed at the Biopolymer/Genomics Core Facility of the Department of Microbiology and Immunology at the University of Maryland at Baltimore.
Construction of AbrBN′-C′SpoVT and AbrBN′-C′Abh fusion proteins.
The construction of genes encoding chimeric AbrBN′-C′SpoVT and AbrBN′-C′Abh fusion proteins was performed in two separate cloning steps. First, PCR was used to generate a DNA fragment corresponding to the ribosome binding site through residue 50 (proline) of the AbrB gene. The oligonucleotides were designed to contain a HindIII site at the 5′ upstream end and a SmaI site at the 3′ downstream end (the CCC of the SmaI site corresponding to a codon for the proline residue at position 50). This fragment was cloned into HindIII-SmaI-digested pDR67 (12) to generate pFY66. Next, PCR was used to obtain DNA fragments corresponding to codons 54 to 92 of the abh gene and codons 55 to 178 of the spoVT gene (each followed by its naturally occurring stop codons). These C-terminal domain fragments were engineered to have a SmaI site at their 5′ upstream ends and an XbaI site at their downstream 3′ ends. The fragments were then cloned into SmaI-XbaI-digested pFY66 to generate pFY80 and pFY81 (TP611 as an E. coli transformation host). The chimeric fusion protein encoded by pFY80 thus contains AbrB residues 1 to 50, followed by a glycine “linker” residue, followed by residues 55 to 178 of SpoVT; that encoded on pFY81 is (AbrB 1-50)-gly-(Abh 54-92).
Genetic manipulations.
Competent cells of E. coli strains AG1688, JH372, JH607, and XZ980 were prepared using the CaCl2 technique and transformed with plasmids (selection for resistance to 100 μg/ml ampicillin on Luria-Bertani [LB] plates) by use of standard protocols (21). DH5αF′lacIq competent cells were purchased from Invitrogen (Carlsbad, CA) and transformed according to the manufacturer's instructions. B. subtilis strains were constructed using standard transformation protocols and media (11).
Phage immunity test.
Phages λKH54 (cI−) and λimmP21c were painted as lines on agar plates, and AG1688 transformants harboring the fusion protein plasmid constructs were struck perpendicularly across the two lines of phage. A construct encoding a fusion protein capable of multimerization is immune to infection by λKH54 but sensitive to λimmP21c infection and lysis. Cells with fusions incapable of multimerization would be sensitive to infection and lysis by λKH54. To confirm that resistance to λKH54 was not due to the cells having acquired a mutation affecting phage host range, the cross-streak procedure was repeated using λKH54 h80 as the initial discriminator and a virulent mutant of λ (λsupervir) as final arbitrator.
E. coli in vivo multimerization assays.
JH372 is lysogenic for a λ prophage containing a lacZ reporter gene driven by the λPR promoter under the control of a λOR operator with a mutation in the OR2 portion. Proteins having the λcI DBD fused to a peptide segment capable of self-assembly are able to repress PR by binding at the OR2 operator variant (at the levels attained from the lacUV5 promoter driving fusion protein expression under noninducing conditions); cI DBD fusions to peptides incapable of self-assembly will not repress transcription of the reporter under these conditions. JH372 transformants of the various fusion protein plasmids were grown in Luria-Bertani (LB) medium, and samples were taken (in quadruplicate) during mid-logarithmic growth (optical density at 600 nm, 0.5 to 1.0) for assaying. β-Galactosidase assays were performed as has been described, and activity was expressed in Miller units (21). Each strain was assayed by this procedure on at least two independent occasions.
The reporter present in JH372 cannot be used to discriminate between dimerization and other higher-order forms of multimerization (15). To accomplish this, comparative β-galactosidase assays were performed using JH607 and XZ980 reporter strains harboring the fusion constructs. The properties of the cI-repressible operators present upstream of lacZ reporters in these phage have been previously described (2, 15, 33). Briefly, any cI-DBD fusion construct capable of dimerization will lead to repression of these reporters, but in the case of the JH607 reporter the formation of trimers or higher will give a greater degree of repression (i.e., lower activity) than dimer formation. The XZ980 reporter shows little, if any, difference between repressed activities brought about by dimers or higher multimerization states. In practice, the β-galactosidase activities produced by the two strains harboring a particular fusion are compared. If the repressed activity of the JH607 reporter is equal to or greater than the XZ980 reporter then the dimer state is indicated; a repressed activity in JH607 lower than that in XZ980 indicates formation of an oligomer state greater than dimerization. The fusion constructs we tested in the JH607 and XZ980 reporters were grown and assayed for β-galactosidase activity as described in the preceding paragraph.
Assay of chimeric fusions in B. subtilis.
B. subtilis strains containing the wild-type abrB gene under pSpac control (SWV1030) and the chimeric abrBN′-C′spoVT and abrBN′-C′abh fusion constructs (FYS75 and FYS76, respectively) were grown in Schaeffer's medium (22) with and without 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cultures were sampled at various times during growth, and β-galactosidase assays were performed as previously described (7).
A note on the NMR structure of AbrB N domains.
An NMR structure of AbrB N-terminal DNA-binding domain dimers has appeared in print (28). However, this structure contains errors which have now been corrected (a correction notice appears in Nat. Struct. Biol. [vol. 12, no. 4, p. 380; 1 April 2005, posting date]). The revised structure is available from the Protein Data Bank, entry 1Z0R. Interested readers are referred to this revised structure and warned against consulting the now-obsolete entry 1EKT.
RESULTS AND DISCUSSION
AbrB multimerization domains.
We cloned 20 distinct fragments of the abrB gene to create cI DBD fusion proteins containing the AbrB N-terminal domain (residues 4 to 53), the N-domain segment with missense mutations, various segments from the AbrB C-terminal domain, a C-domain segment (residues 48 to 94) containing different missense mutations, and an essentially intact abrB segment (residues 4 to 94) with and without the R23S missense mutation. The fusion constructs were introduced into E. coli strains (AG1688, DH5αlacIq, and JH372), and the transformants were tested for phage immunity as an initial indication of the propensity of the fused segments to self-multimerize. Transformants of JH372 were then assayed for β-galactosidase activity as described in Materials and Methods. The results of these experiments are given in Fig. 2 (which also includes results of our tests of SpoVT and Abh domains; see below). We observed an exact correlation between the immunity test results and the level of LacZ expression in the JH372 reporter strain. When the presence of a fusion conferred immunity to λ infection, significantly less than 750 Miller units of β-galactosidase activity were formed by the JH372 strain harboring the fusion. When the presence of the fusion did not confer λ immunity, 2,000 Miller units or more were formed in the reporter strain.
FIG. 2.
Summary of assay results for fusion constructs present in the E. coli JH372 reporter strain. The results for each of the λcI DBD fusions we constructed and examined in the JH372 reporter strain are depicted. The extent of each segment from AbrB, Abh, and SpoVT fused to the λcI DBD is denoted numerically (e.g., 4-94 indicates fusion of amino acid residues spanning position 4 to position 94). The AbrB monomer is 94 residues in length, Abh is 92 residues, and SpoVT is 178 residues. Fused fragments containing a missense mutation are indicated by the nature of the amino acid change (one-letter code) at the residue position relative to the native, intact protein monomer (e.g., C54Y indicates a mutation changing cysteine to tyrosine at the 54th residue relative to the native monomer start point). The results of both phage immunity tests and β-galactosidase assays are given. S, sensitive to λ infection (i.e., no multimerization brought about by the fused segment); I, immune to λ infection (i.e., the fused segment can self-assemble into a multimeric state). The dimeric and tetrameric forms of the GCN4 protein fusions used as controls are described in references 15 and 33. (Figures 3, 5, and 6 present cartoons summarizing different aspects of these results along with the results shown in Fig. 4.)
Two of the AbrB segments we tested encompassed residues Thr4-Thr53 and Thr53-Lys94. Each of these segments gave results indicating that they contained a self-assembling domain (Fig. 2 and 3). Previous NMR experiments have shown that a truncated AbrB protein encompassing residues 1 to 53 forms a stable homodimer in solution (28). Our results with the T53-K94 segment indicate that the carboxyl-terminal portion of the monomer also possesses a multimerization domain (most probably a dimerization domain; see below) and that its self-assembly is independent of the dimerization activity of the N-terminal domain.
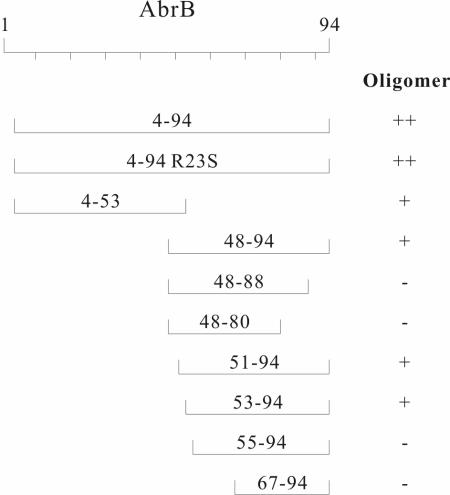
FIG. 3.
Summary of multimerization abilities of portions of the AbrB protein. The oligomeric natures of (λcI DBD)-(AbrB segments) fusion proteins, as determined on the basis of the results shown in Fig. 2 and 4, are indicated. +, dimer; −, monomer; ++, greater than dimers (i.e., tetramers).
The nature of the molecular interactions between residues in the AbrB N domain that are responsible for dimerization are known from the NMR structure of AbrBN′(1-53) dimers. In an effort to delimit the region in the carboxyl domain responsible for its multimerization capability, we examined different truncated forms of the domain. As shown by the results in Fig. 2 and Fig. 3, elimination of the last six residues of the domain abolishes self-assembly (i.e., residues 48 to 94 multimerize, residues 48 to 88 do not). The amino-proximal extent necessary for multimerization must include residue Cys54 (the lone cysteine in the monomer) and possibly Thr53. Thus, the integrity of almost the entire C-terminal portion of the protein is required for its independent multimerization activity.
The immunity test assay and the semiquantitative β-galactosidase assays using the reporter construct in JH372 are not capable of distinguishing between dimerization and higher-order multimerization states. However, comparative assays of β-galactosidase expression by use of the two different promoter/operator-lacZ constructs present in strains JH607 and XZ980 can be employed to make this type of distinction (see Materials and Methods). The results presented in Fig. 4 indicate that segments containing residues 4 to 53 self-assemble into dimers (in agreement with the NMR results mentioned previously); segments containing residues 48 to 94 also self assemble into dimers, while the essentially intact protein (residues 4 to 94) assembles into a higher-order multimerization state. A variety of studies have demonstrated that native AbrB is a tetramer in solution (3, 28). Thus, AbrB tetramer formation requires the action of two independent dimerization domains, one present in the amino-terminal portion of the monomer and the other located in the carboxyl-terminal portion.
FIG. 4.
Assessment of the ability of select AbrB and SpoVT domains to self-assemble into oligomer states greater than dimers. The results of β-galactosidase assays for various fusions present in the differential reporter strains XZ980 and JH607 are shown. Segments fused to the λcI DBD are denoted as given in the legend to Fig. 2. For an indicated fusion, the ratio of activity in XZ980 versus JH607 is indicated numerically. A XZ980/JH607 ratio of less than 1 is indicative of dimer formation; a ratio greater than 1 indicates formation of trimers or greater (see text for description and discussion). w.t., wild type.
Effect of AbrB mutations on multimerization.
Previous studies of purified mutant forms of AbrB had indicated that the R23S mutant protein is tetrameric, L67P is probably a dimer, and C54Y may have a reduced, but not abolished, capacity to form tetramers (18, 29). To examine these assessments further and to investigate the multimerization defect of other missense mutations, we constructed a variety of cI DBD fusions to segments of AbrB containing the mutations. The results are given in Fig. 2 and summarized in Fig. 5.
FIG. 5.
Effects of single amino acid changes on the multimerization ability of the AbrB N and C domains. The oligomeric natures of (λcI DBD)-(AbrB segments containing missense mutations) fusion proteins, as determined on the basis of the results shown in Fig. 2 and 4, are indicated. +, dimer; −, monomer.
The results obtained for the 4-53 (Fig. 5) and 4-94 (Fig. 4) segments containing the R23S mutation support the conclusion that this mutation affects neither dimerization of the N domain nor tetramerization of the intact protein. The arginine residue (R24) adjacent to R23 in the putative DNA-binding α-helical portion of the N domain likewise does not appear to play a role in multimerization, as shown by the dimerization capacity of the 4-53(R24S) segment. A critical feature of the dimerization of N domains, as revealed by the NMR structure of AbrBN53, is a four-stranded intermolecular β-sheet interface (residues 34 to 39 and 42 to 48 of each monomer). Strong intermolecular hydrophobic interactions forming the interface occur between Ile43 from one monomer and Leu45 in the other. Mutation of I43 would thus be expected to abrogate dimerization, and this is the result we observed for the 4-53(I43S) segment. The β-sheet regions of each monomer also form a scaffold for hydrophobic packing interactions defining the monomer structure. Some of the intramolecular hydrophobic packing interactions involve the Leu34 residue. Mutation of Leu34 to a histidine residue abolishes dimer formation (Fig. 5), indicating the critical relationship between overall monomer structure and the ability to form dimers. The E35G mutation does not affect dimerization, but it does severely impair AbrB regulatory function in vivo (K. Zoller and M. A. Strauch, unpublished data). This seemingly indicates that the E35 residue plays a crucial role in DNA binding. Given its position relative to the putative DNA-binding surface including arginine residues critical for DNA interactions (R8, R15, R23, R24), we do not believe E35 is involved in direct contact with DNA moieties, although this cannot be entirely ruled out. Rather, we favor a view that E35 is responsible for proper spatial configuration of the actual DNA-binding surfaces relative to the β scaffolding of the N-domain dimers, due to a likely role for E35 in charge-packing interactions with residues K47 and K49 in the monomer.
The result observed for the 48-94 segment containing L67P supports the idea of a dimerization defect in the C domain which would have no effect upon N-domain-mediated dimerization, in keeping with the previous assessment that purified AbrB L67P is a dimer. When present in the AbrB 48-94 segment, the N64I mutation also causes a dimerization defect in the C domain. The role played by the lone cysteine residue (Cys54) of the AbrB monomer is enigmatic. The C54Y, C54S, and C54W mutations all result in an AbrB mutant phenotype in B. subtilis (17, 18, 29). The defect caused by the absence of a cysteine at this position cannot be due to elimination of disulfide bond formation between subunits, as it has been shown that the sulfhydryls of Cys54 residues are present in the reduced state in active AbrB tetramers (18). When present in the AbrB 48-94 segment fused to the λcI DBD, the C54S and C54W mutations prevent dimerization but the C54Y mutation does not (Fig. 2 and 5). Previous observations had indicated that purified C54Y mutant AbrB proteins were capable of forming tetramers in vitro but that these tetramers behaved as though the multimeric interactions were somehow weaker than in the wild-type protein (25, 29). Purified C54S appeared to be a dimer in these in vitro assays. (C54W was not obtained in sufficiently pure form to allow characterization.) Although there is some evidence that the sulfhydryl groups of the Cys54 residues have altered solvent accessibility upon binding DNA (18), other lines of evidence strongly imply that free Cys54 sulfhydryl groups are not involved in direct DNA contact (30). On the basis of the observations, we believe that Cys54 residues in their reduced state must play a crucial role in the stability of the tetrameric configuration needed for stable DNA binding. An explanation accounting for the ability of C54Y C domains to dimerize (and C54Y mutant proteins to form in vitro tetramers, albeit with reduced stability) could be that the substituted tyrosines are capable of functional interactions with some portion of the protein that partially mimic the Cys54 interactions.
SpoVT multimerization domains.
Preliminary NMR studies indicate that N-terminal truncations of the SpoVT protein (residues 1 to 54) appear to form dimers in much the same manner as occurs for the AbrB and Abh N domains (D. Sullivan and J. Cavanagh, personal communication). Compared to the AbrB and Abh proteins, the SpoVT protein has a much larger peptide chain extending beyond the highly conserved N-terminal domain (Fig. 1). We sought to determine, using λcI DBD fusions, what portion, if any, of the SpoVT C domain might be a multimerization domain. As the actual multimerization state of SpoVT had not yet been conclusively determined (6), we also examined λcI DBD fusions to the N domain (residues 4 to 54) and to the (essentially) entire SpoVT peptide (residues 4 to 178).
As shown in Fig. 2 and summarized in Fig. 6, SpoVT is comprised of two separable multimerization domains: one present between residues 4 and 54 (N-domain), the other between 59 and 178 (C domain). Furthermore, each of these independent domains must specify dimerization only, as shown by the results presented in Fig. 4. Just as is the case for AbrB, the (essentially) full-length SpoVT protein (residues 4 to 178) results in a multimerization state greater than dimerization when fused to the λcI DBD (Fig. 4). While the data do not afford conclusive proof as to the exact multimer state, on the basis of the AbrB model it seems logical to assume that the SpoVT protein is also homotetrameric (two independent dimerization domains acting to form a tetramer).
FIG. 6.
Summary of multimerization abilities of portions of the SpoVT protein. The oligomeric natures of (λcI DBD)-(SpoVT segments) fusion proteins, as determined on the basis of the results shown in Fig. 2 and 4, are indicated. +, dimer; −, monomer; ++, greater than dimers (probably tetramers).
On the basis of the limited number of truncations in the C domain we examined, residues from 60 to 91 and from 161 to 178 appear necessary for dimerization (Fig. 2 and 6). It is tempting to speculate that either these subregions form part of the dimerization interface or that one interacts with the other in an intermonomeric fashion, but other explanations cannot be ruled out.
Abh multimerization domains.
Truncated versions of the Abh protein encompassing amino-proximal residues 1 to 53 exist as dimers in solution, as revealed by NMR structural determinations (B. Bobay and J. Cavanagh, personal communication). The results observed when the Abh carboxyl-terminal residues from positions 50 to 92 are fused to the λcI DBD indicate that dimerization takes place (Fig. 2). Although we have not dissected the C domain of Abh in as much detail as the AbrB and SpoVT domains, evidence presented below indicates that a truncated segment containing Abh residues 54 to 92 is sufficient for dimerization.
Interchangeability of C domains for AbrB in vivo functioning.
Given our results indicating that the C domains of AbrB, Abh, and SpoVT are independent dimerization modules and our hypothesis that the sole function of the AbrB C domain is related to multimerization, we next tested the ability of the C-domain modules of Abh and SpoVT to substitute for the AbrB C domain in the creation of active chimeric proteins in B. subtilis. We created chimeric genes fusing the AbrB N-terminal domain to the C domains specified by the abh and spoVT genes (see Materials and Methods) and placed the constructs under control of the IPTG-inducible pSpac promoter. The reporter for AbrB activity was an abrB8-lacZ fusion integrated at the thrC locus in a strain with knockout mutations in the chromosomal copies of the spo0A and abrB genes. As shown in Fig. 7, expression of each of the chimeric fusions repressed the abrB8-lacZ reporter in a manner indistinguishable from that seen when the wild-type intact AbrB protein was expressed. Expression of only the N domain of AbrB (residues 1 to 55) under these conditions did not have any regulatory effect upon the reporter (data not shown). Thus, the C domains of Abh and SpoVT are capable of supplying the multimerization activity needed to form active AbrB tetramers in vivo. Additionally, the presence of a “foreign” C-domain dimerization module had no apparent effect upon the level of AbrB-mediated regulatory activity seen at the target promoter used in the assay.
FIG. 7.
Abh and SpoVT C domains can substitute for the AbrB C domain in vivo. Time course of β-galactosidase expression from an abrB8-lacZ reporter in strains expressing IPTG-inducible chimeric fusions of the N-terminal DNA-binding domain of AbrB to the C-terminal multimerization domains of Abh and SpoVT. Closed circles, pSpac-abrB (wild type), no IPTG; open circles, pSpac-abrB (wild type) plus 1 mM IPTG; closed squares, pSpac-abrBN′-C′abh, no IPTG; open squares, pSpac-abrBN′-C′abh plus 1 mM IPTG; closed triangles, pSpac-abrBN′-C′spoVT, no IPTG; open triangles, pSpac-abrBN′-C′spoVT plus 1 mM IPTG. Zero on the abscissa denotes the end of exponential growth and entry into stationary phase.
Concluding remarks.
The greatest degree of amino acid sequence homology between AbrB, Abh, and SpoVT lies in their N-terminal residues 50 to 51 (Fig. 1). Not surprisingly, NMR studies indicate that isolated N domains of these three paralogues have highly similar structures, with each existing in a stable dimeric state in solution (B. Bobay, D. Sullivan, J. Cavanagh, and M. A. Strauch, unpublished observations). Previous studies had indicated that the DNA-binding recognition and specificity determinants lie primarily, or solely, within the N domain of AbrB (29, 30). That the DNA-binding determinants of Abh and SpoVT also reside primarily in their N domains is a reasonable supposition. The results presented in this communication indicate that the C-terminal domains of AbrB, Abh, and SpoVT are independent dimerization modules. Intact AbrB is known to be a tetramer in solution, and the evidence presented here indicates that the most probable multimeric forms of Abh and SpoVT are also tetramers. At least in the case of the AbrB N domain, any one of the three C-domain dimerization modules can be utilized for the formation of active tetramers in vivo. We conclude that the sole role of the AbrB C domain is in formation of the active tetrameric species of the protein. Given the similarity of its size to that of the AbrB C domain, we suppose that the role of the Abh C domain is only in the formation of tetramers also. The significantly larger size of the C-terminal dimerization domain of SpoVT could indicate that it plays some additional role, possibly in directly contacting moieties of DNA targets or, perhaps more likely, in interaction with some component of the transcriptional apparatus or other regulatory factor.
Acknowledgments
We thank James Hu for providing the E. coli strains and vectors used in the construction and assay of the λcI DBD fusions, Katie Zoller for performing the B. subtilis β-galactosidase assays, and John Cavanagh for discussions regarding the NMR structure of AbrB N-domain dimers.
This work was supported in part by grant GM46000 from the National Institutes of Health, United States Public Health Service.
REFERENCES
- 1.Bagyan, I., J. Hobot, and S. Cutting. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckett, D., D. S. Burz, G. K. Ackers, and R. T. Sauer. 1993. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry 32:9073-9079. [DOI] [PubMed] [Google Scholar]
- 3.Benson, L. M., J. L. Vaughn, M. A. Strauch, B. G. Bobay, R. Thompson, S. Naylor, and J. Cavanagh. 2002. Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Anal. Biochem. 306:222-227. [DOI] [PubMed] [Google Scholar]
- 4.Bobay, B. G., L. Benson, S. Naylor, B. Feeney, A. C. Clark, M. B. Goshe, M. A. Strauch, R. Thompson, and J. Cavanagh. 2004. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry 43:16106-16118. [DOI] [PubMed] [Google Scholar]
- 5.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, T. C., S. M. Cutting, and R. J. Lewis. 2004. DNA-binding studies on the Bacillus subtilis transcriptional regulator and AbrB homologue, SpoVT. FEMS Microbiol. Lett. 233:247-256. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari, E., D. J. Henner, M. Perego, and J. A. Hoch. 1988. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J. Bacteriol. 170:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, J., M. Roggiani, and D. Dubnau. 1995. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 177:3601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305-320. [DOI] [PubMed] [Google Scholar]
- 12.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, K., K. Shoji, T. Shimizu, K. Nakano, T. Sato, and Y. Kobayashi. 1995. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J. Bacteriol. 177:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marahiel, M. A., M. M. Nakano, and P. Zuber. 1993. Regulation of peptide antibiotic production in Bacillus. Mol. Microbiol. 7:631-636. [DOI] [PubMed] [Google Scholar]
- 15.Marino-Ramirez, L., and J. C. Hu. 2002. Using λ repressor fusions to isolate and characterize self-assembling domains, p. 375-393. In E.Golemis (ed.), Protein-protein interactions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Perego, M., and J. A. Hoch. 1991. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J. Bacteriol. 173:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 18.Phillips, Z. E., and M. A. Strauch. 2001. Role of Cys54 in AbrB multimerization and DNA-binding activity. FEMS Microbiol. Lett. 203:207-210. [DOI] [PubMed] [Google Scholar]
- 19.Phillips, Z. E., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59:392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., and D. Russell. 2000. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schaeffer, P., J. Miller, and J. Aubert. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauch, M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote. J. Bacteriol. 177:6999-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauch, M. A. 1993. Regulation of Bacillus subtilis gene expression during the transition from exponential growth to stationary phase. Prog. Nucleic Acid Res. Mol. Biol. 46:121-153. [DOI] [PubMed] [Google Scholar]
- 25.Strauch, M. A., M. Perego, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 3:1203-1209. [DOI] [PubMed] [Google Scholar]
- 26.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trowsdale, J., S. M. Chen, and J. A. Hoch. 1979. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol. Gen. Genet. 173:61-70. [DOI] [PubMed] [Google Scholar]
- 28.Vaughn, J. L., V. Feher, S. Naylor, M. A. Strauch, and J. Cavanagh. 2000. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator AbrB. Nat. Struct. Biol. 7:1139-1146. [DOI] [PubMed] [Google Scholar]
- 29.Xu, K., D. Clark, and M. A. Strauch. 1996. Analysis of abrB mutations, mutant proteins, and why abrB does not utilize a perfect consensus in the −35 region of its sigma A promoter. J. Biol. Chem. 271:2621-2626. [DOI] [PubMed] [Google Scholar]
- 30.Xu, K., and M. A. Strauch. 2001. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 183:4094-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, K., and M. A. Strauch. 1996. In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures. Mol. Microbiol. 19:145-158. [DOI] [PubMed] [Google Scholar]
- 32.Zeng, X., A. M. Herndon, and J. C. Hu. 1997. Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc. Natl. Acad. Sci. USA 94:3673-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng, X., and J. C. Hu. 1997. Detection of tetramerization domains in vivo by cooperative DNA binding to tandem lambda operator sites. Gene 185:245-249. [DOI] [PubMed] [Google Scholar]
- 34.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]