Prokaryotic taxonomy is cyclically confronted with breakthroughs of methodological and conceptual innovations. The “sexiest” novelties are often met by periods of reluctance but, after some ups and downs, tend to consolidate themselves within the taxonomic framework. One of these novelties is presented in this issue of the Journal of Bacteriology, where Konstantinidis and Tiedje (10) evaluate the implementation of a genomics-based parameter to objectify and numerically confine the higher taxa (i.e., taxa above species) of the taxonomic hierarchical classification system. Their results seem promising, and one could predict that it might be an important parameter to take into account when modernizing prokaryotic taxonomy. However, at the same time, we have to be aware of what has been achieved and how the microbial classification system has evolved.
Taxonomy is an essential discipline of the biological sciences simply because it provides a framework for the scientific community to facilitate understanding and knowledge exchange. It is perhaps one of the oldest biological sciences, having been in existence for at least 2,400 years, since Aristotle devised the first hierarchy based on creationist and essentialist tenets (3). However, although the basic criteria for understanding biodiversity and evolution subsequently shifted toward abandoning essentialism, the basic hierarchical structure of how humans understand nature has remained rigid and immutable (5). Scientists take for granted that the whole of biological diversity, including prokaryotes and eukaryotes from any source, can be organized by following a single hierarchical model of categories, equally ranked for any kind of organism, which is a tendency called “monism” by philosophers (8). It is remarkable that despite all the technological and conceptual developments achieved in the last and the present centuries, the way we understand order in nature is still based on the taxonomic schema applied to higher eukaryotes devised by Linnaeus more than two centuries ago. Microbiologists adopted the system directly from the botanical and zoological taxonomies, and microbial classification was constructed following intuitive criteria of how a category could be circumscribed (14). Of course, this intuition was replaced by more solid criteria developed in parallel with the techniques that periodically appeared through technological improvements.
The taxonomic hierarchy stands on the basic category “species.” This unit is considered to be the unique real entity of the whole classification schema, whereas all other higher categories are considered to be abstract (7, 8). Species are categories motivated by the observations of recurrent patterns within the diversity of living things. However, such patterns of recurrence are necessarily different for different kinds of organisms that exhibit distinct levels of morphological and/or physiological complexity (4). This is one of the most controversial issues within the biological sciences, because it means that for these very reasons different taxonomies will have noncomparable species categories, contrary to the monistic belief (8). The search for a universal species concept has led to heated debates (8, 11), and establishing universal categories for all living organisms, including prokaryotes, has created general dissatisfaction (15). Thus, in the case of the species category, monism is a scientific tendency that hinders conceptual developments.
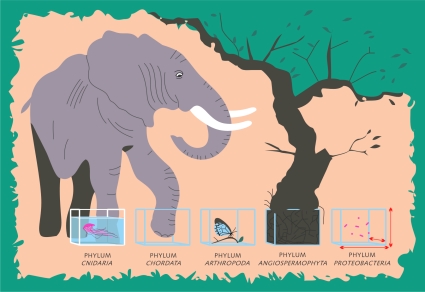
Monism is also about understanding that all living organisms can be classified within a single pattern of hierarchical categories independently of their evolutionary and ecological particularities. The whole taxonomic construct was devised by the human mind to classify nature at points where the known biological diversity was reduced to easily observable biological entities. However, after determining the extent of the real and putative biological diversity present in the biosphere, the single and rigid hierarchical system may not be appropriate for embracing all living things (Fig. 1). Actually, the hierarchical global taxonomic classification system has been severely criticized and predicted to fail (3). It is an artificial construct of the mind, intuitively devised by observations of how recurrent patterns in nature could be related. It may work, but it will remain artificial.
FIG. 1.
A monistic approach demands that a single hierarchical system be applied to describe the whole of biological diversity. However, due to the intrinsic evolutionary and ecological characteristics of each single group of living organisms, the use of the same measuring stick for category construction cannot produce a “natural classification system.” Arbitrariness of taxon circumscriptions will be unavoidable unless we accept pluralism in taxonomy. Then, the search for objective parameters to circumscribe taxa may not be a fantasy, but also, the resulting categories will no longer be comparable between taxonomies. (Drawing by David Fajardo.)
Due to the simpler nature of the prokaryotes, taxonomists understood that in the technological developments of other disciplines, there were profitable parameters that would help to reflect natural relationships among organisms. The search for and establishment of novel criteria have been recurrent themes throughout the history of prokaryote taxonomy (14). Important breakthroughs occurred, for example, when a rigid monothetic taxonomy was abandoned for a more flexible polythetic and theoretical approach based on the computerized developments of numerical taxonomy (14, 16). A little later, but almost simultaneously, came the introduction of the first genomic parameters, G+C mol% content and whole-genome hybridizations, and these were followed by the appearance of numerous techniques revealing relevant chemical components (i.e., chemotaxonomy). All such innovations were subjected to back-and-forth debates for almost 30 years before being finally recognized as necessary for classification purposes (18).
The most recent relevant innovation introduced into prokaryotic taxonomy, which has also been subjected to an almost 30-year suitability debate, was the use of gene or protein sequences, and especially that of the 16S rRNA gene, as molecular clocks (19). Since then, especially within the framework of taxonomy, there have been heated debates on the usefulness of the marker to construct organismal phylogenies, or whether analyses would provide a “more natural” framework for prokaryotic classification that could replace the one already being used, which was treated as improper because of its artificiality (6, 12, 13, 16, 20). Actually, 16S rRNA gene sequence analysis had been increasingly used for new prokaryotic classifications since its introduction, but its use as a key parameter in taxonomy has only recently been recommended (17). The most recent global prokaryotic classification is primarily based on 16S rRNA similarities and the resulting tree reconstructions (1), and the use of this parameter now seems obvious. However, a lapse of more than 30 years has been needed to convince microbiologists, and especially taxonomists, that despite its intrinsic pitfalls, the parameter is very helpful.
We have definitively entered the genomic era. The number of completely sequenced genomes is geometrically increasing with time simultaneously with the decrease in cost of such techniques. Thus, the race has begun to find those parameters that can be retrieved and that are useful for scientific taxonomic purposes using the current information data set. However, the findings must be produced before the overflow of information that is to come collapses scientists' computing and understanding capabilities. There is hope that genomics will produce tools to definitively understand “natural relationships” between microorganisms (2). The several parameters tested range from the more reductionistic ones in which a reduced set of selected functional genes are used for genealogy inferences to the more holistic ones based on the analysis of the shared content of orthologous genes (2).
In this issue of the Journal of Bacteriology, Konstantinidis and Tiedje (10) introduce a new genome-based parameter that, due to its simplicity of formulation and possible computerization, might serve as a primary criterion for delineation of higher taxa in the midterm period. The average amino acid identity (AAI) and the already formulated average nucleotide identity (ANI) (9) are two clear parameters resulting from pairwise genome comparisons and averaging the sequence identities of shared orthologous genes (amino acid or nucleotide, respectively). Both achieve, to a great extent, the goal that whole-genome DNA hybridizations (DDH) pursued and that has been especially determinant for the circumscription of prokaryotic species. Such pairwise comparisons have two main advantages over DDH. First, they permit in silico analyses and the construction of cumulative databases, which are pitfalls severely criticized in DDH (17). Second, due to the nature of the constraints of the macromolecules, the ANI and AAI comparisons provide two levels of relatedness signals, the former for close relationships and the latter for more distant relationships. ANI may directly reflect DDH experiments, and it has been shown to correlate nicely with the results accumulated over many years, which are measurable because of the availability of whole-genome sequences of closely related strains (9). A different question, which will not be discussed here, is whether the “size” of the prokaryotic species circumscribed after DDH is conservative. Both AAI and ANI provide, however, an undoubtedly holistic approach to compare genomes, with the advantage of a simple theoretical background that facilitates interpretation.
Due to the intrinsic characteristics of amino acid identity comparisons, AAI may be suitable for comparisons of distant genomes rather than ANI, because the latter may contain more evolutionary noise. However, distantly related genomes have the disadvantage that the absolute and shared gene contents between a given pair of strains may differ tremendously. This is indeed an important point when trying to standardize criteria for taxon circumscription. There will surely be thresholds of, for instance, minimum shared gene content, or minimum amino acid identity values, that hamper attempts to establish “natural relationships.” Nevertheless, it is still too soon to evaluate the potential pitfalls, since important efforts in sequencing and establishing comparisons still need to be made.
The main question in applying the AAI approach to prokaryotic taxonomy is whether ranks above species can be numerically (for some understood as “objectively”) confined. Indeed, the complaint is that the most modern view of hierarchic classification has been based on 16S rRNA gene analyses (1), although classifications are still subject to considerable subjectivity. So, the question here is whether AAI will provide an objective framework to encapsulate taxa within other taxa, like metaphorical Russian dolls. The main conceptual obstacle to an “objective” achievement is that the system is artificial per se and demands a fractal distribution of the taxon hierarchies. This means that, for all living organisms, identical evolutionary rhythms are necessary. A “natural classification” may not be achievable if we do not permit some freedom in how the system appears to our own eyes when new approaches are used. It is, however, very important to note that if we succeed in numerically circumscribing higher taxa with parameters such as AAI, the resulting criteria used for the definitions will necessarily be different from equivalent comparisons among eukaryotes. If AAI is to be applied, then higher taxa will be no more than abstract entities equally comparable among taxonomies, and we should be aware of this before starting a (to me unfruitful) debate, such as the ongoing one concerning prokaryotic species and their incomparability with those of eukaryotes.
The use of AAI has brought interesting results that seem promising for circumscribing higher taxa. Nevertheless, it is also true that the currently available genome database is strongly biased toward microorganisms with clinical importance, which may constitute an insignificant portion of the prokaryotic world. Preliminary results indicate that in general, it is possible to numerically confine the already classified higher taxa. However, as Konstantinidis and Tiedje mention in this issue (10), precisely because the taxa are arbitrary categories, and due to the microbial diversity still to be revealed, the objective cutoffs now acceptable may be inadequate in the future, thus challenging the supposed objectivity of the hierarchic system.
We might have to wait for a third 30-year-lapse cycle to ascertain whether AAI or ANI is a successful parameter to construct/restore prokaryotic taxonomy. However, for the foreseeable future, a period of disagreement, with back-and-forth debate between scientific beliefs, is predictable.
Acknowledgments
I am grateful to Rudolf Amann, Josefa Antón, Rafael Bosch, Jürgen Busse, Gorazd Avgustin, Peter Kämpfer, Jorge Lalucat, Silvia Marqués, Raúl Rivas, and Brian Tindall for critically reading the manuscript; to Chris Rodgers for correcting the English style; and to David Fajardo for drawing the allegory of the artificial taxonomic system.
My work is supported by grants BOS-2003-05198-C02-01 and -02 from the Spanish Ministry of Science and Education and by the European Network of Excellence Marine Genomics Europe (GOCE-CT-505403) through the sixth framework program.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Brenner, D., J. Staley, and N. Krieg. 2000. Classification of procaryotic organisms and the concept of bacterial speciation, p. 27-38. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer Verlag, New York, N.Y. [Google Scholar]
- 2.Coenye, T., D. Gevers, Y. Van de Peer, P. Vandamme, and J. Swings. 2005. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 29:147-167. [DOI] [PubMed] [Google Scholar]
- 3.Ereshefsky, M. 1994. Some problems with the Linnaean hierarchy. Phyl. Sci. 61:186-205. [Google Scholar]
- 4.Hey, J. 2001. Genes, categories and species. Oxford University Press Inc. New York, N.Y.
- 5.Hull, D. L. 1965. The effect of essentialism on taxonomy—two thousand years of stasis. Br. J. Phil. Sci. 15:314-326, 16: 1-18. [Google Scholar]
- 6.Hull, D. L. 1970. Contemporary systematic philosophies. Annu. Rev. Ecol. Syst. 1:19-54. [Google Scholar]
- 7.Hull, D. L. 1976. Are species really individuals? Syst. Zool. 25:174-191. [Google Scholar]
- 8.Hull, D. L. 1997. The ideal species concept—and why we can't get it, p. 357-380. In M. F. Claridge, H. A. Dawah, and M. R. Wilson (ed.), Species: the units of biodiversity. Chapman & Hall, London, United Kingdom.
- 9.Konstantinidis, K., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konstantinidis, K., and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187:6258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayden, R. L. 1997. A hierarchy of species concepts: the denouement in the saga of the species problem, p. 381-424. In M. F. Claridge, H. A. Dawah, and M. R. Wilson (ed.), Species: the units of biodiversity. Chapman & Hall, London, United Kingdom.
- 12.Mayr, E. 1998. Two empires or three? Proc. Natl. Acad. Sci. USA 95:9720-9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palleroni, N. 2003. Prokaryote taxonomy of the 20th century and the impact of studies on the genus Pseudomonas: a personal view. Microbiology 149:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 15.Rosselló-Mora, R. 2003. The species problem, can we achieve a universal concept? Syst. Appl. Microbiol. 26:323-326. [DOI] [PubMed] [Google Scholar]
- 16.Sneath, P. H. A. 1995. Thirty years of numerical taxonomy. Syst. Biol. 44:281-298. [Google Scholar]
- 17.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kämpfer, M. C. J. Maiden, X. Nesme, R. Rosselló-Mora, J. Swings, H. G. Trüper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the Ad Hoc Committee for the Re-Evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 18.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, L. Krichevsky, L. H. Moore, W. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 19.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woese, C. R. 1998. Default taxonomy: Ernst Mayr's view of the microbial world. Proc. Natl. Acad. Sci. USA 95:11043-11046. [DOI] [PMC free article] [PubMed] [Google Scholar]