Abstract
Plasmodium falciparum (Pf) is the most prevalent cause of malaria infections in humans. Due to the development of resistant strains, newer drugs, or drugs acting at novel targets are constantly being sought. Here, we report the design and preparation of 48 new 2,4-diaminopyrimidine derivatives targeting the Pf protein kinome. Bioinformatics methods have been used to identify the most probable target(s). Cheminformatics and molecular modelling have been used to guide the structural modifications. Our primary goal was to enhance the antimalarial activity of the series, reduce mammalian cytotoxicity, and increase aqueous solubility. The antimalarial activity of all 48 compounds has been assessed in chloroquine-resistant Pf3D7 strain and for their mammalian cytotoxicity in HepG2 cell lines. Phosphate buffer solubility, MDCK permeability, and metabolic clearance in human and rat microsomes were also assessed. Compounds 68 and 69 demonstrated good antimalarial activity (Pf IC50) of 0.05 and 0.06 μM, respectively, and good selectivity over the mammalian cell line (SI >100 fold). The compounds also demonstrated much improved aqueous solubilities of 989.7 and 1573 μg mL−1, respectively, along with moderate intrinsic clearance (∼3 mL min−1 g−1) and permeability (>60 nm s−1).
Cheminformatics-guided optimization of the antimalarial activity and physicochemical properties of 2,4-diaminopyrimidines.
Introduction
According to the World Health Organization (WHO), there were an estimated 249 million malaria cases globally in 2022, resulting in 608 000 deaths.1 The majority of these deaths are caused by the Plasmodium falciparum (Pf) species of the parasite. Artemisinin-based combination therapy (ACT) is the gold standard in treating malaria as resistance to earlier generations of medications, including chloroquine and mefloquine, has increased. Unfortunately, resistance to newer artemisinin-based therapies has now also been reported.2 This has prompted a search for new drug classes with novel mechanisms of action to tackle the resistance problem.3
Protein kinases are a family of enzymes that play a crucial role in regulating various cellular processes. Activation or deactivation of cell signalling pathways involving the almost 500 enzymes found in human can be highly effective in treating a variety of diseases. Over 80 marketed kinase drugs exist for cancer, and inflammatory indications4 and their applicability to other therapeutic conditions including cardiovascular and neurological are being actively explored.5–7 Additionally, kinases inhibitors have been extensively investigated as potential antimalarial therapies since the Pf genome encodes over 80 putative protein kinases,8 several of which are essential for the parasite's survival.9–12
Several potential Pf kinases have been investigated to date, including PfCDPKs and PfPKs. PfCDPK1 and PfCDPK2 have been identified in the asexual blood of late and mature schizont stages, respectively,13,14 while PfPK6 has been associated with asexual blood stage development.9 However, no single ideal target has been identified. Indeed researchers have considered the potential benefits of employing a polypharmacology strategy or targeting host cell kinases.10,12
Aminopyrimidines have shown potential as antimalarials targeting Pf protein kinases including PfCDPK1,15PfCDPK2,15,16PfPKB,16PfNEK3,16 and PfPK6 (ref. 15 and 17) (Fig. 1). Compounds 1 and 2, identified initially by phenotypic screening,18 and were later found to display significant inhibitory effects at PfPK6 and PfCDPK1, respectively.15 Furthermore, 3 demonstrated inhibitory effects against PfCDPK2 and PfPKB,164 at PfPKB and PfNEK319 and 5 against PfPK6.17
Fig. 1. Structures of 2,4-diamino-pyrimidine and related derivatives previously confirmed as either Pf or human kinase inhibitors.
Herein, we report the preparation of forty-eight new 2,4-diaminopyrimidines compounds in our efforts to improve upon the properties of 6, a potent anti-parasitic agent identified in our previous studies on this chemotype.21–23 The molecule displays good activity in a phenotypic-based 3D7 Pf model with an IC50 of 0.093 μM. Unfortunately, subsequent testing demonstrated relatively low solubility (0.33 μg mL−1) as well as moderate metabolic stability in human and rat microsomes (5.0 and 8.8 mL min−1 g−1, respectively).21
We undertook further optimization of the series using a computationally enabled approach with a particular focus on increasing the low solubility and clearance of the series, while also trying to increase or maintain Pf activity. To this end we performed datamining of ChEMBL Pf activity data to identify general trends, and more specific quantitative structure activity relationship (QSAR) models on our existing data in this series. This data was then used to prioritize a set of structural changes with the potential to increase the Pf activity and improve their overall physicochemical properties.
The suitability of the proposed designs to the putative Pf kinase targets, identified from bioinformatics, were assessed using molecular docking prior to their initial preparation (Fig. 2a). Designs that were capable of binding to the ATP site consistent with other amino pyrimidines to related Pf (i.e. PK6, PDB ID 9CMZ) and human targets (i.e. PDB ID CDK2, 3UNK24) were prioritized. The antimalarial activity of the 48 new compounds were then assessed against the 3D7 strain of Pf parasite. The cytotoxicity of the compounds in mammalian HepG2 cells was also determined. The physiochemical properties of the most promising compounds were then measured. Finally, the inhibition profile of the most potent and selective molecules was assessed against 10 diverse human kinases at 1 μM to assess their potential for host-kinome interaction.
Fig. 2. (a) Docking of exemplar 68, prepared in this study, to the active sites of X-ray structures of 4MVF (PfCDPK2, blue), 9CMZ (PfPK6, green), and homology models of PfCDPK1 (gray) and PfPKB (pink) generated using AlphaFold.20 Hydrogen bond represented as a yellow dash line (b) 2D representation of the interaction of 68 within the kinase active site (green line for PfPK6 conformation), (c) amino acids sequence alignment for residues within 5 Å of ATP binding site (blue indicates conserved, white non conserved), and (d) similarity of residues within 5 Å of ATP binding site.
Result and discussion
Cheminformatics analyses
Incorporation of an acidic or basic moiety into our neutral derivatives21–23 would be expected to favourably impact solubility.25,26 However, such a modification would also lead to a decrease in lipophilicity and this could have effects on other parameters such as the activity,27 permeability, intrinsic clearance etc.25,26 To understand the impact on antimalarial activity, approximately 21 000 unique compounds with Pf IC50s were extracted from the ChEMBL database and classified as; basic, neutral, acidic, or zwitterionic (Fig. 3a). Analysis of the variance in the activities between these groups was then performed, in the form −log[IC50] or pIC50. For this diverse set it is apparent that the incorporation of a basic center results in a statistically significant (Table S1) increase in activity, of almost 0.3 log units, compared to neutral molecules. In contrast, incorporation of an acidic group results in an approximately 0.4 log unit drop-in activity. Indeed, the majority of antimalarial drugs (52.2%) contain a basic center, with only 26.1% being neutral, 17.4% being acidic, and 4.3% being zwitterionic (Fig. 3b). These observations suggest a good strategy for our series would be to incorporate a basic center27 to improve both the anti-parasitic activity and solubility.
Fig. 3. The effect of ionization state on the (a) the antimalarial activity of compounds reported in ChEMBL with Pf IC50s and (b) the ionization state of approved antimalarial drug. Boxes represent interquartile range and whiskers 5–95 percentiles.
The impact of physicochemical properties on anti-malarial activity was explored further by generating a diamino-pyrimidine specific partial least squares (PLS) QSAR model using 54 diamino-pyrimidine compounds of ours previously reported with 3D7 pIC50 data (Fig. 4a & Table S4).21,22 The PLS model was built using 15 common atomic and molecular descriptors (see the SI), fitting 2 components in total. The model demonstrated an R2 of 0.65 for the training set (Fig. 5b) which can be considered acceptable given the complexity of the phenotypic endpoint being investigated.
Fig. 4. (a) Plot of observed vs. predicted Pf pIC50 for the training and test set, (b) molecular descriptor roles for antimalarial activity.
Fig. 5. Design scope of compounds 22–69.
Even modestly predictive models can offer useful insights during the design of new series modifications. Indeed, analysis of Fig. 4b shows that lipophilicity (i.e. clog P and clog D) drives increased Pf activity. However, this is generally contraindicated for improving solubility.25,26 From an analysis of the coefficients, we observe evidence to support an increase in basic pKa (ref. 27) will lead to increased activity. In contrast decreasing the acidic pKa is preferred. That being said, care needs to be taken in the latter case since the training set contains only 5 compounds with an acidic pKa ≥ 7 and only 1 compound with an acidic pKa < 7.
We would subsequently validate the PLS model using the newly prepared compounds (22–69, Fig. 5). The agreement between the observed and predicted values for this test set was reasonable (r2 = 0.40) given the complexity of the endpoint being predicted. While modest, the r2 value indicates a useful degree of predictive performance that could be useful to pre-screen molecules for synthesis. Indeed, if only the top 50% of predicted compounds were prepared, all compounds with pIC50s > 6.5 would have been identified.
Design
We decided on a 2-phase strategy to try and maximize the solubility of our most promising hit to date (6)21 while also maintaining or increasing the anti-malarial activity. Unfortunately, a lipophilicity decreases for example would be expected to improve the former but lead to a deterioration in the latter parameter. Thus, one subset of prepared compounds would focus on increasing polarity21 and reducing the aromatic character/disrupting planarity28 of the molecules to improve solubility and a second subset would primarily focus on the incorporation of a basic center (Fig. 5).
New 2,4-diaminopyrimidines were subsequently identified through isosteric modifications at the 2-position and 4-position and 5-position of the pyrimidine. Modifications at the 2-position were limited to 3 aromatic heterocycles investigated as part of our previous study.29 Modifications at the 4-position involved the incorporation of a range of polar or basic moieties. These were selected based on an analysis of bioisosteres present in related potent anti-malarials (i.e.Fig. 1) or inhibitors of human kinases with structurally similar Pf equivalents (Tables S2 and S3). We also investigated the effect of –H substitution with –Cl at the 5-position of pyrimidine as this corresponds to the ATP binding site gatekeeper position.30
Docking of exemplars (i.e.68) into putative Pf kinases targets suggested there were several acidic residues towards the mouth of the ATP pocket with the potential to interact with H-bond acceptors or a basic center of our compounds (Fig. 2a). Comparison of the sequences for Pf kinases confirmed as targets to related compounds (Fig. 1) shows that the ATP binding sites of PfCDPK1, PfCDPK2, PfPK6, and PfPKB are highly similar and are potential targets of the molecules here (Fig. 2c and d). The SAR at the 2-position was therefore the most extensively investigated here. Several linkers were incorporated and these were connected to a variety of solubilizing moieties.
Prior to synthesis, all proposed molecules were evaluated in the Pf QSAR model. We did not exclude any of the compounds at this stage as we were interested in (a) assessing how the model performed in real world prediction and (b) we did not want to solely prepare the most potent predicted molecules as these would potentially be the higher lipophilicity, lower solubility molecules that we were interested in moving away from. Initially, twenty-eight compounds containing sulfonyl and urea groups at the 2-position of pyrimidine were prepared. SAR at the 4-position was investigated using indole, benzaxolone, and phenylureas (Table 1). In addition, we explored the impact of incorporating –Cl at the 5-position in place of -H. The initial SAR demonstrated that sulfonyl groups unfortunately reduced Pf activity, 1H-indole was preferred at 4-position (Fig. S3 and Table S7) and –Cl at the 5 position was not advantageous. Therefore, a further 20 compounds were then prepared that incorporated a basic moieties (Table 2).
Table 1. Compounds 22–49 with sulfone and urea linker at R3.

| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | R1 | R2 | R3 (para) | R3 (meta) | MW | clog Da | LEb | LLEc | Pf3D7 IC50 (μM) | HepG2 IC50 (μM) | SI |
| Dihydroartemisinin | 284.4 | 2.84 | 0.55 | 5.26 | 0.008 ± 0.001 | 57.6± 33.5 | 7198 | ||||
| Mefloquine | 378.3 | 1.71 | 0.39 | 5.69 | 0.040 ± 0.004 | 4.68 ± 0.2 | 117 | ||||
| Hydroxychloroquine | 335.9 | −0.26 | 0.43 | 7.53 | 0.059 ± 0.034 | 12.07 ± 1.6 | 205 | ||||
| Palbociclib (hCDK4/6 inhibitor) | 447.5 | 1.35 | 0.19 | 3.25 | >25 | 33.5 ± 15.8 | <1.3 | ||||
| Seliciclib (hCDK2/7/9 inhibitor) | 354.5 | 2.89 | 0.24 | 1.71 | >25 | 18.0 ± 5.5 | <0.72 | ||||
| 6 | H |

|
H |

|
435.5 | 5.45 | 0.28 | 1.58 | 0.093 ± 0.018 | 5.0 ± 0.4 | 54 |
| 22 | H |

|

|
H | 440.5 | 2.29 | 0.24 | 3.04 | 6.6 ± 5.8 | >100 | >15 |
| 23 | H |

|

|
H | 440.5 | 2.29 | 0.24 | 3.04 | 6.6 ± 5.8 | >100 | >15 |
| 24 | H |

|

|
H | 411.4 | 2.16 | 0.24 | 3.00 | 7.6 ± 4.1 | >100 | >13.2 |
| 25 | H |

|
H |

|
393.5 | 2.90 | 0.27 | 2.55 | 3.7 ± 1.5 | 15.1 ± 0.5 | 4.1 |
| 26 | H |

|
H |

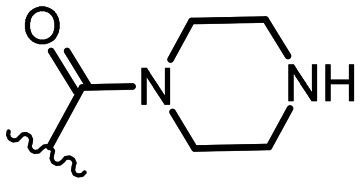
|
440.5 | 2.29 | 0.23 | 2.93 | 7.0 ± 5.0 | >100 | >14.3 |
| 27 | H |

|
H |

|
411.4 | 2.16 | 0.24 | 2.91 | 8.6 ± 2.0 | >100 | >11.6 |
| 28 | H |

|

|
H | 394.5 | 2.67 | 0.26 | 2.70 | 4.4 ± 1.6 | 11.5 ± 0.4 | 2.62 |
| 29 | H |

|

|
H | 441.5 | 2.06 | 0.23 | 3.13 | 6.7 ± 2.5 | >100 | >15 |
| 30 | H |

|

|
H | 412.4 | 1.93 | 0.24 | 3.05 | 11.9 ± 8.1 | >100 | >8.4 |
| 31 | H |

|
H |

|
394.5 | 2.67 | 0.27 | 2.75 | 4.2 ± 2.5 | 17.6 ± 0.9 | 4.2 |
| 32 | H |

|
H |

|
441.5 | 2.06 | 0.25 | 3.54 | 3.9 ± 1.5 | >100 | >25.8 |
| 33 | H |

|
H |

|
412.4 | 1.93 | 0.27 | 3.69 | 5.4 ± 3.2 | >100 | >18.6 |
| 34 | H |

|

|
H | 387.5 | 3.79 | 0.27 | 1.90 | 2.1 ± 0.8 | 26.4 ± 2.6 | 12.4 |
| 35 | H |

|
H |

|
387.5 | 3.79 | 0.30 | 2.52 | 0.51 ± 0.2 | 30.1 ± 8.7 | 59.5 |
| 36 | Cl |

|

|
H | 427.9 | 3.51 | 0.25 | 1.86 | 4.7 ± 2.8 | 4.7 ± 0.5 | 0.99 |
| 37 | Cl |

|

|
H | 475.0 | 2.89 | 0.24 | 2.61 | 3.4 ± 1.7 | 24.4 ± 6.3 | 7.3 |
| 38 | Cl |

|

|
H | 445.9 | 2.77 | 0.25 | 2.61 | 4.6 ± 2.7 | 6.3 ± 1.3 | 1.4 |
| 39 | Cl |

|
H |

|
427.9 | 3.51 | 0.27 | 2.25 | 1.8 ± 0.4 | 4.6 + 0.0 | 2.6 |
| 40 | Cl |

|
H |

|
475.0 | 2.89 | 0.23 | 2.46 | 4.5 ± 0.7 | 4.9 ± 0.5 | 1.1 |
| 41 | Cl |

|
H |

|
445.9 | 2.77 | 0.24 | 2.53 | 5.4 ± 2.5 | 5.3 ± 0.3 | 0.99 |
| 42 | Cl |

|

|
H | 428.9 | 3.28 | 0.25 | 2.08 | 4.7 ± 2.6 | 3.6 + 0.0 | 0.77 |
| 43 | Cl |

|

|
H | 476.0 | 2.66 | 0.20 | 1.94 | 25.1 ± 0.0 | 15.4 ± 1.7 | 0.61 |
| 44 | Cl |

|

|
H | 446.9 | 2.54 | 0.26 | 3.11 | 2.4 ± 1.4 | >100 | >40.1 |
| 45 | Cl |

|
H |

|
428.9 | 3.28 | 0.27 | 2.34 | 4.6 ± 4.9 | 9.8 ± 1.3 | 2.2 |
| 46 | Cl |

|
H |

|
476.0 | 2.66 | 0.23 | 2.67 | 4.7 ± 1.1 | 7.5 ± 1.1 | 1.6 |
| 47 | Cl |

|
H |

|
446.9 | 2.54 | 0.24 | 2.79 | 4.7 ± 0.8 | 10.8 ± 0.2 | 2.3 |
| 48 | Cl |

|

|
H | 421.9 | 4.39 | 0.24 | 0.94 | 4.7 ± 0.3 | 4.4 ± 0.2 | 0.94 |
| 49 | Cl |

|
H |

|
421.9 | 4.39 | 0.24 | 0.85 | 5.8 ± 1.5 | 7.5 ± 0.1 | 1.3 |
ChemAxon JChem 24.1.2.
Ligand efficiency (LE) = (1.37 × Pf pIC50)/(heavy atoms).
Ligand lipophilic efficiency (LLE) = (Pf pIC50 − clog D).
Table 2. Compound 50–69 with amide linker at R3.

| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | R3 (para) | R3 (meta) | MW | clog Da | LEb | LLEc | Pf3D7 IC50 (μM) | HepG2 IC50 (μM) | SI |
| 50 |

|
H | 427.5 | 3.50 | 0.29 | 3.38 | 0.14 ± 0.05 | 6.4 ± 2.4 | 46.5 |
| 51 | H |

|
427.5 | 3.52 | 0.29 | 3.15 | 0.22 ± 0.09 | 10.6 ± 6.0 | 47.5 |
| 52 |

|
H | 456.6 | 2.71 | 0.27 | 4.09 | 0.16 ± 0.04 | 11.5 ± 1.5 | 70.9 |
| 53 | H |

|
456.6 | 2.71 | 0.25 | 3.52 | 0.60 ± 0.04 | 18.1 ± 2.6 | 30.4 |
| 54 |

|
H | 510.7 | 1.24 | 0.24 | 5.44 | 0.22 ± 0.07 | 13.6 ± 2.6 | 62.6 |
| 55 | H |

|
510.7 | 1.24 | 0.24 | 5.54 | 0.17 ± 0.001 | 13.9 ± 1.8 | 82.6 |
| 56 |

|
H | 415.5 | 2.25 | 0.28 | 4.01 | 0.56 ± 0.1 | 8.9 ± 0.9 | 15.9 |
| 57 | H |

|
415.5 | 2.25 | 0.27 | 3.89 | 0.73 ± 0.2 | 21.7 ± 2.5 | 29.6 |
| 58 |

|
H | 427.5 | 3.09 | 0.25 | 2.72 | 1.6 ± 0.09 | 18.8 ± 0.3 | 12.1 |
| 59 | H |

|
427.5 | 3.10 | 0.25 | 2.84 | 1.2 ± 0.2 | 20.4 ± 0.7 | 17.6 |
| 60 |

|
H | 413.5 | 2.50 | 0.27 | 3.69 | 0.65 ± 0.06 | 48.2 ± 13.2 | 73.9 |
| 61 | H |
|
413.5 | 2.50 | 0.27 | 3.62 | 0.77 ± 0.2 | 23.7 ± 5.4 | 30.9 |
| 62 |

|
H | 414.5 | 3.38 | 0.23 | 1.73 | 8.2 ± 3.4 | 28.7 ± 4.2 | 3.5 |
| 63 | H |

|
414.5 | 3.38 | 0.24 | 2.15 | 3.0 ± 0.9 | 54.4 ± 23.2 | 17.9 |
| 64 |

|
H | 481.6 | 1.98 | 0.25 | 4.52 | 0.32 ± 0.03 | 24.5 ± 7.1 | 77.7 |
| 65 | H |

|
481.6 | 2.01 | 0.23 | 4.14 | 0.71 ± 0.2 | 23.6 ± 8.3 | 33.2 |
| 66 |

|
H | 399.5 | 1.89 | 0.28 | 4.32 | 0.63 ± 0.2 | 23.5 ± 7.5 | 37.1 |
| 67 | H |

|
399.5 | 1.89 | 0.28 | 4.28 | 0.69 ± 0.2 | 11.7 ± 3.4 | 17 |
| 68 |

|
H | 467.6 | 2.00 | 0.28 | 5.19 | 0.060 ± 0.004 | 8.7 ± 0.3 | 135.7 |
| 69 | H |

|
467.6 | 2.03 | 0.29 | 5.26 | 0.050 ± 0.004 | 5.2 ± 0.8 | 101.2 |
ChemAxon JChem 24.1.2.
LE = (1.37 × Pf pIC50)/(heavy atoms).
LLE = (Pf pIC50 − clog D).
Chemistry
Intermediates were prepared according to Scheme 1. All compounds were synthesized using nitroaromatic derivatives. Substituted ureas 7a–b were prepared using commercially available isocyanate nitrobenzene and ethylamine in DCM at room temperature in yields of 85 and 88%.31 Intermediates 8a–d were synthesized from substituted bromomethyl nitrobenzene and secondary amines in DCM at room temperature affording yields ranging from 85–93%. Amide substituted intermediates 9a–j were prepared from nitro benzoyl chloride and appropriate amines in DCM at 0 °C and TEA resulting in yields of 49–93%.
Scheme 1. a) DCM, rt, overnight, b) DCM, rt, 2–4 h, c) DCM, 0 °C, 2 h, (d) THF, rt, 2 h, (e) i-PrOH, 85 °C, overnight, (f) Pd/C, H2, 4–7 h, (g) SnCl2, EtOAc, rt, overnight, (h) TEA or NaHCO3, EtOH, 90 °C, overnight, (i) TFA, i-PrOH, 100 °C, 16 h, (j) TFA, DCM, rt, overnight.
Intermediates 11a–f were prepared from 10a–b according to the general procedure from the literature.32,33 by reacting under reflux conditions with appropriate secondary amines in isopropanol to obtain intermediates 11a–f. Aniline products were obtained by hydrogenation reaction of all substituted nitro intermediates using Pd/C in a hydrogen atmosphere or SnCl2 with yields of approximately 90%.23,34,35
2,4-Diamino-pyrimidine derivatives 16–21 were prepared from 4-Cl of pyrimidine and anilines with NaHCO3 or TEA in ethanol resulting in yields 18–92%. Final products were prepared by substitution of the 2-Cl of intermediates 16–21 and appropriate anilines with acid catalyst to afford compounds 22–69 in yields of 6–92%. Compounds 60, 61, 66, and 67 required an additional in situ Boc-deprotecting step with TFA/DCM at room temperature to give the corresponding amine in yields of 7–17%.
Antimalarial activity
Forty-eight compounds in total were evaluated for in vitro antimalarial activity against the chloroquine-resistant 3D7 strain of P. falciparum (Tables 1 and 2). Compounds 22–69 displayed antimalarial IC50 values ranging from 25 to 0.05 μM. The top compounds identified compared favourably to marketed drugs Hydroxychloroquine and Mefloquine with IC50s of 0.059 and 0.04 μM, respectively. However, the activity was still considerably lower than the gold standard treatment Dihydroartemisinin at 0.008 μM.
Despite the similarity between 22–69 and human CDK inhbitors Palbociclib and Seliciclib (Fig. 6), the former display dramatically better activity against Pf. Palbociclib and seliciclib are both inactive against the malaria parasite (IC50s > 25 μM). This suggests that despite targeting quite similar Pf homologs of human CDKs (Tables S2 and S3), our derivatives show a distinct preference for the former.
Fig. 6. Selected anti-malarial and CDK drug standards used for comparison.
Focusing first on the neutral chemotypes, 22–49 demonstrated somewhat lower Pf3D7 IC50s overall, ranging from 25–0.5 μM. The lower activity of this set is consistent with earlier observations that compounds that incorporate a basic center generally have improved anti-malarial activity over neutral chemotypes (Fig. 3).
Analysis of the SAR shows that replacement of –H with –Cl at the R1-position did not show a statistically significant effect on the Pf activity (Fig. S3). The most active compound of the set was found to be 35, (IC50 = 0.51 μM). This compound has a –H at R1 compared to 5.9 μM for the equivalent possessing a –Cl (49). Thus, as a result of the higher log D, and lower ligand lipophilic efficiency (LLE) of the –Cl substituted compounds, we decided to focus on –H substitution at the R1 position for later compounds.
Modifications at R2 included the previously studied 1H-indole ring,21 as well as new isosteres N-ethyl-N-phenylurea and benzoxazolone. Unfortunately, compounds containing benzoxazolone (11.9–2.4 μM) and urea substituted anilines (3.4–7.0 μM) showed generally poor Pf activity. Maintaining the 1H-indole at R2 led to better Pf activity as exemplified by compounds 34, 35, and 39 with IC50s of 2.1, 0.51, and 1.8 μM, respectively. 1H-Indole was found to be the most preferable, in line with our previous SAR studies21 (Fig. 7d). At the R3 position, there was no statistically significant between (a) the sulfone-substituted anilines and the urea-substituted anilines investigated, and (b) whether substituents were located at the meta or para position. The most potent urea compound was meta substituted (35) and had an IC50 of 0.51 μM. The most potent sulfone (39) was also meta substituted and had an IC50 of 1.78 μM.
Fig. 7. Effect of R group substitution on Pf activity, HepG2, and clog D at R1 (a–c) and R2 (d–f). Box represents interquartile range; whisker (5–95 percentile); *(significant). Statistical data is shown in Tables S8 and S10.
We subsequently limited our SAR investigation by fixing the R1 as –H, the R2 as 1H-indole and varying only the R3 position (Table 2). Several aliphatic ring systems containing basic nitrogens were incorporated at either the meta or para position of the anilino ring attached to the 2-position of pyrimidine (50–69). The Pf activity of these compounds were considerably improved compared to the neutral chemotypes (22–49) (Fig. 7a). Pf3D7 IC50s ranging from 6.2–0.05 μM for the former. Compounds 68 and 69 demonstrated the most potent Pf activity of all prepared here with IC50 = 0.060 and 0.050 μM. These contained 4-(pyrrolidin-1-yl)piperidine at the 4- and 3-positions respectively. Addition of a basic center was found to improve the activity of the chemotype to a range equivalent to hydroxychloroquine (0.059 μM) and mefloquine (0.040 μM).
It can be concluded from Table 2 that the addition of a basic center, as exemplified by 60–61, 64–66, leads to a dramatic increase in potency, irrespective of the basic moiety used (pyrrolidine, piperazine, or pyrrolidinyl piperidine) or the point of attachments to aniline (meta or para). To confirm the importance of the basic center, structurally similar but neutral morpholino derivatives (62 & 63), were prepared and these showed >4-fold poorer activity compared to their piperazine counterparts (60 & 62). This leads us to conclude that a basic moiety is critical for activity in this series.36,37
Finally, we explored the SAR associated with the amide and urea linker by preparing new derivatives (66–69). Removing the carbonyl group generally improved the antimalarial activity (i.e.64vs.68 and 65vs.69). We postulate that removal of the carbonyl H-bond acceptor may result in an improved interaction between the basic center and acidic residues commonly located towards the mouth of the target kinase (Fig. 2b).38
HepG2 cytotoxicity SAR
The selectivity of compounds was also assessed by counter screening for cytotoxicity against a mammalian liver cell line (HepG2). The top compounds identified against Pf (68 & 69) showed the greatest selectivity of all compounds with SI values of 135-fold and 101-fold, respectively. These SI values are comparable to hydroxychloroquine (205) and mefloquine (117), but lower than the gold standard dihydroartemisinin at 7198. Interestingly, the SI values are dramatically better than the human kinase inhibitors Palbociclib and Seliciclib. These compounds in fact have greater activity against HepG2 cells than Pf parasites.
At the R1 position, replacing the 5-pyrimidine hydrogen with a –Cl substituent leads to a sizeable increase in toxicity, 10-fold higher on average (Fig. S3). At the R2 position, benzoxazolone was found to be marginally less toxic than 1H-indole (Fig. 7e); however, the key exemplars such as 68 and 69 defied this trend.
The SAR at R2 and R3 positions was rather flat. Only a small difference in activity between the indole and benzoxazoles were found to be statistically significant. The mean pIC50 being 0.42 log units lower in the latter than the former.
In summary, we identified compounds with good Pf activity and selectivity versus HepG2 cell (i.e. >50 fold). This included compounds 68 and 69. Thus, these two compounds along with several other exemplars were selected for further physicochemical profiling.
Physicochemical properties and intrinsic clearance
The experimental log D7.4 and phosphate buffer solubility of the most promising compounds were evaluated (Table 3). These compounds showed log D7.4 values in an acceptable range (2.33 ± 0.54),39 considerably lower than the best exemplar identified in previous studies.21 Additionally, the lower log D7.4 of the basic exemplars contributes to a significantly lower overall LLE of 2.55 ± 0.65. This compares to 3.88 ± 1.05 for the neutral chemotypes.
Table 3. Physicochemical properties of selected compounds.
| ID | Pf IC50 (μM) | SIa | Clearance (mL min−1 g−1) | MDCK Papp | log D7.4 | Solubility pH 7.4 | |
|---|---|---|---|---|---|---|---|
| Human Clint | Rat Clint | nm s−1 | μg mL−1 (μM) | ||||
| 6 (ref. 21) | 0.093 ± 0.02 | 54 | 5.0 | — | — | >4.0 | 0.33 (0.76) |
| 25 | 3.7 ± 1.46 | 4.1 | — | — | — | 2.7 | 5.8 (14.8) |
| 35 | 0.51 ± 0.16 | 59.5 | — | — | — | 2.96 | 0.68 (1.75) |
| 39 | 1.8 ± 0.40 | 2.6 | — | — | — | 3.1 | 3.9 (9.16) |
| 50 | 0.1 ± 0.05 | 46.5 | — | — | — | 3.0 | 5.9 (13.77) |
| 51 | 0.22 ± 0.09 | 47.5 | — | — | — | 1.94 | 65.3 (152.8) |
| 52 | 0.16 ± 0.04 | 70.9 | 1.63 | 3.36 | 43 | 1.4 | 800.2 (1752) |
| 54 | 0.22 ± 0.07 | 62.6 | — | — | — | 2.1 | 337.6 (661.2) |
| 55 | 0.17 ± 0.001 | 82.6 | — | — | — | 2.1 | 475.6 (931.4) |
| 64 | 0.32 ± 0.03 | 77.7 | — | — | — | 1.98 | 1203 (2498) |
| 68 | 0.06 ± 0.004 | 135.7 | 3.50 | 2.44 | 62 | 2.39 | 989.7 (2116) |
| 69 | 0.05 ± 0.004 | 101.2 | 3.12 | 6.21 | 73 | 2.0 | 1573 (3365) |
| Palbociclib | >25 | <1.3 | — | — | — | 1.80 | 48.01 (107.2) |
| Seliciclib | >25 | <0.72 | — | — | — | 3.45 | 409.8 (1156) |
SI = Pf IC50/HepG2 IC50.
The phosphate buffer solubility of the exemplar compounds has also dramatically improved over 6, particularly for the basic chemotypes, and to a lesser degree than neutral exemplars (i.e.6, 25, 35 & 39). The most active compounds (68 and 69) showed much improved solubilities at 989 and 1573 μg mL−1, respectively. Compounds 52 and 64, with moderate Pf activity, had reasonable solubilities of 800.2 and 1203 μg mL−1, respectively. These results compared very favourably to our the previously identified compound 6 (0.33 μg mL−1). For comparison, the most soluble of the neutral chemotypes prepared was 25 at 5.8 μg mL−1.
The permeability of the most potent, selective, and soluble molecules (52, 68 & 69) was assessed in a Madin–Darby canine kidney (MDCK) permeability assay. Compounds 68 and 69 demonstrated good permeability (62 and 73 nm s−1). Compound 52 showed a permeability of 43 nm s−1. While only moderate in magnitude, the permeability rates compare well to the marketed antimalarial drugs Mefloquine and Quinidine, which have reported MDCK permeabilities <40 nm s−1.40,41
The metabolic stability of compounds 52, 68 & 69 were assessed in human and rat liver microsomes. Compounds were found to be marginally more stable in human than the original lead (6), albeit this was obtained under slightly different experimental conditions.21 Compound 52 showed the highest stability in human (1.63 mL min−1 g−1) while 68 and 69 showed moderate stability at ∼3 mL min−1 g−1. Compound 68 showed the best balance in clearance with moderate values in both human (3.50 mL min−1 g−1) and rat (2.44 mL min−1 g−1). Comparable antichagasic molecules with Clint of 2.2 and 3.0 mL min−1 g−1 have progressed to further in vivo studies.42,43 Nevertheless, further optimization is likely required to improve the metabolic stability of the series.
Kinase inhibition profile
Finally, we sought to probe the potential for human host effects by determining inhibitory activity. We selected the two most promising compounds for testing against an in vitro panel of 10 structurally diverse human kinases at a concentration of 1 μM (Table 4). The human CDK2/7/9 inhibitor Seliciclib was also assessed for the purpose of comparison.
Table 4. Activities (% inhibition at 1 μM) of compounds 68 and 69 and seliciclib at a panel of 10 kinases. Standard deviation reported in parenthesis.
| hKinase | 68 (%) | 69 (%) | Seliciclib (%) | Active similarity to Pf kinasesa |
|---|---|---|---|---|
| hPIM1 (P11309) | 107.4 ± 0.5 | 86.9 ± 4.3 | 138.5 ± 1.4 | PfPK2 (65.7%), PfCDPK2 (62.9%)b |
| hCDK2 (P24941) | 111.6 ± 7.2 | 104.3 ± 0.2 | 28.1 ± 0.2 | PfPK5 (90.9%), PfPK6 (75.8%)b |
| hCDK9 (P50750) | 95.8 ± 0.3 | 97.8 ± 14.2 | 91.2 ± 9.3 | PfCRK3 (84.8%), PfPK6 (69.7%) |
| hERK1 (P27361) | 109.2 ± 8.4 | 105.0 ± 5.1 | 115.6 ± 8.4 | PfMAPK1 (86.7%), PfMAPK2 (76.7%) |
| hJAK3 (P52333) | 98.0 ± 10.9 | 111.5 ± 21.2 | 98.6 ± 16.7 | PfPK5 (61.8%), PfGSK3 (61.8%)b |
| hMEKK1 (Q13233) | 76.3 ± 10.2 | 73.7 ± 6.9 | 69.9 ± 6.3 | PfCDPK7 (74.3%), PfPK2 (68.6%) |
| hHER4 (Q15303) | 38.9 ± 8.2 | 52.5 ± 7.3 | 29.0 ± 5.3 | PfGSK3 (64.7%), PfMRK (58.8%) |
| hTAK1 (O43318) | 64.0 ± 2.1 | 57.5 ± 0.1 | 90.0 ± 3.7 | PfPK2 (71.4%), PfNIMA4 (70.6%) |
| hAURA (O14965) | 82.2 ± 3.0 | 71.3 ± 2.7 | 109.6 ± 8.4 | PfARK2 (82.9%), PfPKB (77.1%)b |
| hVEGFR1 (P17948) | 88.7 ± 11.0 | 72.3 ± 1.2 | 89.1 ± 3.5 | PfGSK3 (61.8%), PfCDPK5 (51.4%) |
Similarity within 5 Å of their ATP binding sites. Sites defined from ligands of PDB structures 9CMZ (PfPK6), 4MVF (PfCDPK2), 8ERD (hCDK2), 4I6Q (hJAK3).
Related Pfkinases target reported previously with 2,4-diaminopyrimidines scaffold.
Compounds 68 and 69 demonstrated significant inhibition against hPIM1, hCDK2, hCDK9, hERK1, and hJAK3 at the relatively high concentration assessed. The activity profile is qualitatively similar to Seliciclib in terms of both the magnitude and individual kinases targeted. This is unsurprising given that these compounds are postulated to target Pf variants structural similar to CDK kinases.15–17 Compounds 68 and 69 exhibited good activity against hPIM1, hCDK2, hCDK9, which share sequence similarity of >60% at the ATP binding site level to confirmed Pf kinase targets PfPK6 (ref. 16) and PfCDPK2.17
It is worth noting that while we found that 68 and 69 were considerably more potent in the blood stage 3D7 assay when compared to human CDK inhibitors Seliciclib and Palbociclib, Arang et al.44 reported Seliciclib as an effective liver stage Pf inhibitor. It was found to clear >85% of liver-stage Plasmodium parasite burden at 500 nM after 24 h. Potentially 68 and 69 could also function in the latter stage also although testing would be require to prove this.
In summary, further work is needed to fully understand the potential risks and benefits of treating Pf with protein kinases inhibitors. A critical component of that will be to validate the true target or targets of the lead molecules and the potential for undesirable host effects. This is a non-trivial task given that it has been postulated that targeting of host kinases, rather than Pf kinases may contribute to the phenotypic response observed.45–48
Conclusions
In this study we have designed and synthesized 48 new 2,4-diaminopyrimidines anti-malarial agents targeting the chloroquine-sensitive Pf3D7 Pf strain. Our goals were to enhance or maintain the antimalarial activity of the series, reduce mammalian cytotoxicity and increase aqueous solubility of our previously reported lead compound 6.21
Bioinformatics was used to identify the most probable Pf kinase targets for our compounds. Molecular docking to key exemplars was then used to guide our structural modifications. Indeed, a recent target-based study to develop antimalarial agents acting at PfCDPK4 did identify potent in vitro inhibitors (IC50 = 0.21 μM). However, these unfortunately did not translate to potent anti-malarial agents in the phenotypic assays (IC50 = 18.7 μM).49 This challenge in part led us to take a phenotypic approach here with secondary focus on the precise targets involved.
In conclusion, we have successfully identified new antimalarials by incorporating a basic moiety into our previously reported 2,4-diaminopyrimidine scaffold. This has led to improved Pf activity and cytotoxic selectivity against a HepG2 mammalian cell line. In addition, the compounds showed markedly improved solubility. Compounds 68 and 69 demonstrated the best antimalarial activity at Pf (0.06 and 0.05 μM). Both demonstrated much-improved solubility (∼1000 μg mL−1), as well as gains in permeability (60–70 nm s−1) and reductions in human microsomal clearance (∼3 mL min−1 g−1).
Experimental procedures
General methods and instruments
All reagents and solvents were purchased from commercial suppliers and were used without further purification. Thin-layer chromatography (TLC) was performed on TLC aluminum/silica gel plate (Silicycle UltraPure Silica gels) with UV light visualization. Purification of the compounds by column chromatography was performed using silica gel G60 (60–200 μm). 1H NMR and 13C NMR spectra were recorded on a Jeol spectrometer model: ECZ500/S1 at 500 MHz (1H NMR), 125 MHz (13C NMR). Chemical shifts (δ) and coupling constants are quoted in parts per million (ppm) and hertz (Hz), respectively, with the residual DMSO-d6 solvent line acting as the internal standard. Mass spectra were obtained from an LC-MSD-Trap mass spectrometer, ion source type ESI(+) mode. High-resolution mass spectra (HRMS) were determined on Bruker Daltonics – micrOTOF-Q III in source ESI(+) mode. Melting points were determined on a digital melting point meter (KRÜSS M3000). The purity of compounds was confirmed ≥95% based on a UV absorbance HPLC chromatogram (Agilent 1100 system, Sunshell C8 column using a mobile phase consisting of (a) 0.1% formic acid in water and (b) acetonitrile: 99 : 1% to 1 : 99% gradient run in 5 and 8 minutes).
All final compounds were characterized by 1H and 13C-NMR. Compound purity was assessed using HPLC-UV and molecular weight/formula confirmed by HRMS. For example, compound 22 exhibited three distinct singlet signals corresponding to –NH protons from the 1H-indole moiety and two substitution sites (2- and 4-pyrimidine NHs), appearing between δ 11.02 and 9.13 ppm. Aromatic protons were observed in the δ 7.95–6.16 ppm region. A methylene (–CH2) appeared as a singlet at δ 4.37 ppm, and a methyl (–CH3) was observed as a singlet at δ 2.87 ppm. Purity was confirmed to be greater than 95% by HPLC, and the observed mass by HRMS matched the calculated [M + H]+ value. All other compounds were characterized in the same manner.
General synthetic procedure for the preparation of intermediate compounds 7a–b
A mixture of nitrophenyl isocyanate (200 mg, 1.22 mmol, 1.0 equiv.) and ethyl amine (82.46 mg, 1.83 mmol, 1.5 equiv.) in DCM (5 mL) was stirred at room temperature for 12 hours. The completion of the reaction was checked with TLC. The mixture was concentrated under reduced pressure to obtain the desired compound 7a as a yellow solid, which was directly used in the next step without further purification.
1-Ethyl-3-(4-nitrophenyl)urea (7a)
Yellow solid, 203.8 mg, 79.9% yields. 1H NMR (500 MHz, DMSO-d6) δ 9.35 (s, 1H), 8.12–8.06 (m, 2H), 7.62–7.55 (m, 2H), 6.46 (t, J = 5.5 Hz, 1H), 3.09 (qd, J = 7.2, 5.6 Hz, 2H), 1.02 (t, J = 7.2 Hz, 3H). ESI-HRMS: calcd for C9H12N3O3 [M + H]+: 210.0873; found: 210.0873.
1-Ethyl-3-(3-nitrophenyl)urea (7b)
Yellow solid, 240.8 mg, 94.4% yields. 1H NMR (500 MHz, DMSO-d6) δ 8.97 (s, 1H), 8.47 (t, J = 2.2 Hz, 1H), 7.69 (ddd, J = 8.1, 2.3, 0.9 Hz, 1H), 7.60 (ddd, J = 8.1, 2.2, 1.0 Hz, 1H), 7.45 (t, J = 8.1 Hz, 1H), 6.28 (t, J = 5.6 Hz, 1H), 3.09 (qd, J = 7.2, 5.6 Hz, 2H), 1.02 (t, J = 7.2 Hz, 3H). ESI-HRMS: calcd for C9H12N3O3 [M + H]+: 210.0873; found: 210.0874.
General synthetic procedure for the preparation of intermediate compounds 8a–d
4-Nitrobenzylbromide (100 mg, 0.47 mmol, 1 equiv.) and secondary amine (0.93 mmol, 2 equiv.) were dissolved in DCM (5 mL). The mixture was stirred for 2–4 hours and monitored with TLC. The precipitation was filtered and washed with DCM, or the mixture was extracted with DCM/water. The organic phase was dried and evaporated to obtain the product.
tert-Butyl 4-(4-nitrobenzyl)piperazine-1-carboxylate (8a)
White solid, 133.3 mg, 89.2% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.18–8.13 (m, 2H), 7.59–7.53 (m, 2H), 3.58 (s, 2H), 3.28 (t, J = 6.2 Hz, 4H), 2.29 (t, J = 5.1 Hz, 4H), 1.34 (s, 9H). ESI-HRMS: calcd for C16H24N3O4 [M + H]+: 322.1761; found: 322.1744.
tert-Butyl 4-(3-nitrobenzyl)piperazine-1-carboxylate (8b)
White solid, 127.4 mg, 85.3% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.14–8.06 (m, 2H), 7.76–7.70 (m, 1H), 7.59 (t, J = 7.9 Hz, 1H), 3.58 (s, 2H), 3.28 (d, J = 5.2 Hz, 4H), 2.30 (t, J = 5.0 Hz, 4H), 1.34 (s, 9H). ESI-HRMS: calcd for C16H24N3O4 [M + H]+: 322.1761; found: 322.1743.
1-(4-Nitrobenzyl)-4-(pyrrolidin-1-yl)piperidine (8c)
Yellow solid, 115.5 mg, 85.8% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.19–8.11 (m, 2H), 7.58–7.50 (m, 2H), 3.53 (s, 2H), 2.69 (dd, J = 11.6, 4.2 Hz, 2H), 2.43–2.38 (m, 4H), 2.00–1.83 (m, 3H), 1.78–1.70 (m, 2H), 1.64–1.57 (m, 4H), 1.41–1.30 (m, 2H). ESI-HRMS: calcd for C16H24N3O2 [M + H]+: 290.1863; found: 290.1832.
1-(3-Nitrobenzyl)-4-(pyrrolidin-1-yl)piperidine (8d)
Yellow solid, 125.1 mg, 93.0% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.11–8.04 (m, 2H), 7.71 (d, J = 7.6 Hz, 1H), 7.58 (t, J = 7.8 Hz, 1H), 3.53 (s, 2H), 2.71 (dd, J = 11.3, 4.1 Hz, 2H), 2.40 (h, J = 3.5 Hz, 4H), 2.01–1.81 (m, 3H), 1.78–1.71 (m, 2H), 1.66–1.55 (m, 4H), 1.41–1.30 (m, 2H). ESI-HRMS: calcd for C16H24N3O2 [M + H]+: 290.1863; found: 290.1835.
General synthetic procedure for the preparation of intermediate compounds 9a–j
A mixture of 3- and 4-nitrobenzoyl chloride (100 mg, 0.54 mmol, 1.0 equiv.) and appropriate amine (0.65 mmol, 1.2 equiv.) in DCM (5 mL) was stirred at 0 °C for 2 hours. Triethylamine (TEA, 150.6 μL, 1.08 mmol, 2 equiv.) was added dropwise. The completion of the reaction was checked with TLC. The mixture was concentrated under reduced pressure, and the desired compound 9a was directly used in the next step without further purification.
tert-Butyl 4-(4-nitrobenzoyl)piperazine-1-carboxylate (9a)
White solid, 158.3 mg, 87.4% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.29–8.23 (m, 2H), 7.69–7.63 (m, 2H), 3.58 (t, J = 5.2 Hz, 2H), 3.42–3.38 (m, 2H), 3.30 (d, J = 1.1 Hz, 2H), 3.23–3.18 (m, 2H), 1.37 (s, 9H). ESI-HRMS: calcd for C11H14N3O3 [M+H]+-Boc: 236.1030; found: 236.1005.
tert-Butyl 4-(3-nitrobenzoyl)piperazine-1-carboxylate (9b)
White solid, 169.9 mg, 93.8% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.27 (dd, J = 8.3, 2.4 Hz, 1H), 8.19 (t, J = 1.9 Hz, 1H), 7.83 (d, J = 7.6 Hz, 1H), 7.71 (t, J = 7.9 Hz, 1H), 3.58 (s, 2H), 3.40 (s, 2H), 3.30 (s, 4H), 1.37 (s, 9H). ESI-HRMS: calcd for C11H14N3O3 [M + H]+-Boc: 236.1028; found: 236.1005.
(4-Methylpiperazin-1-yl)(4-nitrophenyl)methanone (9c)
Yellow solid, 103.5 mg, 76.8% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.27–8.21 (m, 2H), 7.65–7.59 (m, 2H), 3.60 (t, J = 5.0 Hz, 2H), 3.21 (t, J = 5.1 Hz, 2H), 2.34 (t, J = 5.2 Hz, 2H), 2.22 (q, J = 5.7 Hz, 2H), 2.15 (s, 3H). ESI-HRMS: calcd for C12H16N3O3 [M + H]+: 250.1186; found: 250.1189.
(4-Methylpiperazin-1-yl)(3-nitrophenyl)methanone (9d)
Yellow solid, 95.5 mg, 70.9% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.26 (ddd, J = 8.2, 2.4, 1.1 Hz, 1H), 8.14 (dd, J = 2.3, 1.6 Hz, 1H), 7.81 (dt, J = 7.6, 1.3 Hz, 1H), 7.70 (t, J = 7.9 Hz, 1H), 3.53 (d, J = 65.8 Hz, 4H), 2.34 (s, 2H), 2.23 (s, 2H), 2.15 (s, 3H). ESI-HRMS: calcd for C12H16N3O3 [M + H]+: 250.1186; found: 250.1188.
Morpholino(4-nitrophenyl)methanone (9e)
Yellow solid, 81.5 mg, 63.8% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.26–8.23 (m, 2H), 7.66–7.64 (m, 2H), 3.65–3.59 (m, 4H), 3.50 (d, J = 5.4 Hz, 2H), 3.24 (t, J = 4.7 Hz, 2H). ESI-HRMS: calcd for C11H13N2O4 [M + H]+: 237.0870; found: 237.0868.
Morpholino(3-nitrophenyl)methanone (9f)
Yellow solid, 95.1 mg, 74.5% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.26 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H), 8.18 (t, J = 1.9 Hz, 1H), 7.84 (dt, J = 7.6, 1.3 Hz, 1H), 7.71 (t, J = 7.9 Hz, 1H), 3.61 (s, 4H), 3.54–3.48 (m, 2H), 3.31–3.25 (m, 2H). ESI-HRMS: calcd for C11H13N2O4 [M + H]+: 237.0870; found: 237.0869.
N-(2-(Dimethylamino)ethyl)-4-nitrobenzamide (9g)
Yellow solid, 98.1 mg, 76.5% yield. 1H NMR (500 MHz, DMSO-d6) δ 9.24 (t, J = 5.6 Hz, 1H), 8.33–8.25 (m, 2H), 8.18–8.09 (m, 2H), 3.63 (q, J = 5.8 Hz, 2H), 3.25 (t, J = 6.0 Hz, 2H), 2.78 (s, 6H). ESI-HRMS: calcd for C11H16N3O3 [M + H]+: 238.1193; found: 238.1190.
N-(2-(Dimethylamino)ethyl)-3-nitrobenzamide (9h)
Yellow solid, 64 mg, 49.9% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.47 (s, 1H), 9.29 (q, J = 5.0 Hz, 1H), 8.69 (t, J = 2.1 Hz, 1H), 8.42–8.32 (m, 1H), 7.76 (td, J = 8.0, 1.5 Hz, 1H), 3.65 (q, J = 5.9 Hz, 2H), 3.26 (t, J = 6.0 Hz, 2H), 2.78 (s, 6H). ESI-HRMS: calcd for C11H16N3O3 [M + H]+: 238.1186; found: 238.1189.
(4-Nitrophenyl)(4-(pyrrolidin-1-yl)piperidin-1-yl)methanone (9i)
White solid, 131.8 mg, 80.4% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.29–8.23 (m, 2H), 7.66–7.60 (m, 2H), 4.51 (d, J = 13.2 Hz, 1H), 3.48 (d, J = 14.0 Hz, 1H), 3.21–3.01 (m, 4H), 2.80 (t, J = 12.7 Hz, 1H), 2.12 (d, J = 12.0 Hz, 1H), 1.97–1.90 (m, 1H), 1.86 (s, 4H), 1.64 (dd, J = 28.9, 13.9 Hz, 2H). ESI-HRMS: calcd for C16H22N3O3 [M + H]+: 304.1656; found: 304.1623.
(3-Nitrophenyl)(4-(pyrrolidin-1-yl)piperidin-1-yl)methanone (9j)
White solid, 113.6 mg, 69.3% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.31–8.21 (m, 1H), 8.14 (t, J = 2.0 Hz, 1H), 7.81 (dt, J = 7.6, 1.4 Hz, 1H), 7.70 (t, J = 7.9 Hz, 1H), 4.26–4.19 (m, 1H), 3.45 (dd, J = 11.9, 6.1 Hz, 2H), 3.03 (dt, J = 35.0, 12.1 Hz, 2H), 2.50 (dd, J = 3.8, 1.9 Hz, 2H), 2.29–2.18 (m, 1H), 1.94–1.81 (m, 1H), 1.81–1.68 (m, 1H), 1.68–1.58 (m, 4H), 1.44–1.38 (m, 1H), 1.36 (s, 1H), 1.19 (s, 2H). ESI-HRMS: calcd for C16H22N3O3 [M + H]+: 304.1656; found: 304.1627.
General synthetic procedure for the preparation of intermediate compounds 11a–f
Compound 10a–b was prepared by reacting 4-nitroaniline or 3-nitroaniline with chloroacetyl chloride according to general step in literatures.32,33 Subsequently, each product (100 mg, 0.47 mmol, 1 equiv.) was dissolved in isopropanol (5 mL). Appropriate secondary amines (0.94 mmol, 2 equiv.) were added and refluxed for 16 h. The mixture was cooled to room temperature and extracted between EtOAc and sat. NaHCO3 (1 : 1). The organic phase was dried over anhydrous Na2SO4 to afford the desired product.
N-(4-Nitrophenyl)-2-(pyrrolidin-1-yl)acetamide (11a)
Yellow solid, 88.4 mg, 76.2% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.34 (s, 1H), 8.20–8.13 (m, 2H), 7.92–7.84 (m, 2H), 3.27 (d, J = 0.9 Hz, 2H), 2.54 (dddd, J = 8.3, 7.1, 3.3, 2.1 Hz, 4H), 1.70 (tq, J = 4.1, 1.2 Hz, 4H). ESI-HRMS: calcd for C12H16N3O3 [M + H]+: 250.1186; found: 250.1158.
N-(3-Nitrophenyl)-2-(pyrrolidin-1-yl)acetamide (11b)
Yellow solid, 98.2 mg, 84.6% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.19 (s, 1H), 8.66 (t, J = 2.2 Hz, 1H), 7.98 (ddd, J = 8.2, 2.1, 1.0 Hz, 1H), 7.86 (ddd, J = 8.2, 2.3, 0.9 Hz, 1H), 7.54 (t, J = 8.2 Hz, 1H), 3.25 (s, 2H), 2.55 (ddt, J = 7.2, 5.7, 2.2 Hz, 4H), 1.76–1.64 (m, 4H). HRMS: calcd for C12H16N3O3 [M + H]+: 250.1186; found: 250.1157.
2-(4-Methylpiperazin-1-yl)-N-(4-nitrophenyl)acetamide (11c)
Yellow solid, 88.4 mg, 68.2% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.29 (s, 1H), 8.22–8.14 (m, 2H), 7.89–7.83 (m, 2H), 3.14 (s, 2H), 2.56–2.47 (m, 2H), 2.44 (s, 2H), 2.39–2.27 (m, 4H), 2.12 (s, 3H). ESI-HRMS: calcd for C13H19N4O3 [M + H]+: 279.1452; found: 279.1427.
2-(4-Methylpiperazin-1-yl)-N-(3-nitrophenyl)acetamide (11d)
Yellow solid, 95.4 mg, 73.6% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.16 (s, 1H), 8.64 (t, J = 2.2 Hz, 1H), 7.96 (ddd, J = 8.2, 2.1, 1.0 Hz, 1H), 7.88 (ddd, J = 8.2, 2.3, 1.0 Hz, 1H), 7.56 (t, J = 8.2 Hz, 1H), 3.12 (s, 2H), 2.54–2.47 (m, 2H), 2.44 (s, 2H), 2.39–2.29 (m, 4H), 2.13 (s, 3H). ESI-HRMS: calcd for C13H19N4O3 [M + H]+: 279.1452; found: 279.1426.
N-(4-Nitrophenyl)-2-(4-(pyrrolidin-1-yl)piperidin-1-yl)acetamide (11e)
Brown solid, 104.2 mg, 67.3% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.27 (s, 1H), 8.23–8.14 (m, 2H), 7.93–7.84 (m, 2H), 3.12 (s, 2H), 2.78 (dd, J = 11.2, 4.4 Hz, 2H), 2.42 (d, J = 5.8 Hz, 4H), 2.13 (td, J = 11.6, 2.5 Hz, 2H), 1.96–1.84 (m, 1H), 1.77 (dt, J = 12.7, 2.7 Hz, 2H), 1.67–1.56 (m, 4H), 1.52–1.39 (m, 2H). ESI-HRMS: calcd for C17H25N4O3 [M + H]+: 333.1921; found: 333.1887.
N-(3-Nitrophenyl)-2-(4-(pyrrolidin-1-yl)piperidin-1-yl)acetamide (11f)
Brown solid, 92 mg, 59.4% yield. 1H NMR (500 MHz, DMSO-d6) δ 10.36 (s, 1H), 8.65 (t, J = 2.3 Hz, 1H), 7.96 (dd, J = 8.1, 2.1 Hz, 1H), 7.88 (dd, J = 8.2, 2.3 Hz, 1H), 7.57 (t, J = 8.2 Hz, 1H), 3.19 (s, 2H), 3.00 (s, 2H), 2.94 (d, J = 11.7 Hz, 3H), 2.18 (t, J = 11.9 Hz, 2H), 1.96 (d, J = 12.1 Hz, 3H), 1.87 (s, 4H), 1.74 (qd, J = 12.1, 3.8 Hz, 3H). ESI-HRMS: calcd for C17H25N4O3 [M + H]+: 333.1921; found: 333.1924.
General synthetic procedure for the preparation of intermediate anilines 13a–b, 14a–d, 15a–j, and 12a–f
General procedure A for 13a–b, 14c–d, 15a–j, and 12a–f
A mixture of appropriate intermediates 7a–b, 8c–d, 9a–j, and 12a–f (1 mmol, 1 equiv.) and 10% Pd/C (0.1 equiv.) was dissolved in MeOH (20 mL) and reacted for 5 h at room temperature under an atmosphere of H2 gas (balloon). The mixture was filtered through celite plate and concentrated under vacuum. The product was used without further purification for the next step.
General procedure B for 14a–b
A mixture of appropriate intermediates 8a–b (1 mmol, 1 equiv.) and SnCl2·2H2O (5 equiv.) was dissolved in EtOAc (20 mL) and stirred overnight at room temperature. Then, sat. NaHCO3 (20 mL) was added and vigorously stirred for 1 h. The crude was filtered, and filtrate was extracted with water. Organic phase was dried over anhydrous Na2SO4 and concentrated to give white solid.34 The product was used without further purification for the next step.
General synthetic procedure for the preparation of intermediate compounds 16–21
2,4-Dichloropyrimidine derivatives (1.2 equiv.) and appropriate amine (100 mg, ca. 0.66 mmol, 1 equiv.) were dissolved in EtOH (5 mL). Sodium bicarbonate (NaHCO3, 2 equiv.) or triethylamine (TEA, 2 equiv.) was used as catalyst to prepare the intermediate. The mixture was refluxed for 6 h. After cooling to room temperature, the precipitate was filtered and washed with hot ethanol, or the resultant was added water (15 mL) and stirred at room temperature for 20 minutes. The precipitation was formed, then filtered and dried to give the crude. The crude was further purified by silica gel column chromatography to give the desired product.
N-(2-Chloropyrimidin-4-yl)-1H-indol-5-amine (16)
Brown solid, 165.4 mg, 89.4% yield. 1H NMR (500 MHz, DMSO-d6) δ 11.10 (s, 1H), 9.79 (s, 1H), 8.00 (d, J = 5.9 Hz, 1H), 7.64 (s, 1H), 7.36 (d, J = 8.6 Hz, 1H), 7.33 (t, J = 2.8 Hz, 1H), 7.07 (s, 1H), 6.57 (s, 1H), 6.39 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H). ESI-HRMS: calcd for C12H10ClN4 [M + H]+: 245.0589; found: 245.0562.
1-(4-((2-Chloropyrimidin-4-yl)amino)phenyl)-3-ethylurea (17)
Brown solid, 93.3 mg, 57.3% yield. 1H NMR (500 MHz, DMSO-d6) δ 9.80 (s, 1H), 8.37 (s, 1H), 8.04 (d, J = 5.9 Hz, 1H), 7.34 (d, J = 6.6 Hz, 3H), 6.60 (d, J = 5.9 Hz, 1H), 6.03 (t, J = 5.6 Hz, 1H), 3.06 (qd, J = 7.2, 5.6 Hz, 2H), 1.01 (t, J = 7.2 Hz, 3H). ESI-HRMS: calcd for C13H15ClN5O [M + H]+: 292.0960; found: 292.0930.
6-((2-Chloropyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (18)
Off-white solid, 127 mg, 72.6 yield. 1H NMR (500 MHz, DMSO-d6) δ 9.99 (s, 1H), 8.09 (d, J = 5.9 Hz, 1H), 7.60 (s, 1H), 7.17 (dd, J = 8.4, 1.9 Hz, 1H), 7.05 (d, J = 8.3 Hz, 1H), 6.66 (d, J = 5.9 Hz, 1H). ESI-HRMS: calcd for C11H8ClN4O2 [M + H]+: 263.0330; found: 263.0319.
N-(2,5-Dichloropyrimidin-4-yl)-1H-indol-5-amine (19)
Yellow solid, 187.7 mg, 89.2% yield. 1H NMR (500 MHz, DMSO-d6) δ 11.13 (s, 1H), 9.44 (s, 1H), 8.26 (s, 1H), 7.56 (d, J = 2.0 Hz, 1H), 7.39–7.31 (m, 2H), 7.11 (dd, J = 8.6, 2.1 Hz, 1H), 6.40 (ddd, J = 2.9, 1.9, 0.9 Hz, 1H). ESI-HRMS: calcd for C12H9Cl2N4 [M + H]+: 279.0199; found: 279.0175.
1-(4-((2,5-Dichloropyrimidin-4-yl)amino)phenyl)-3-ethylurea (20)
Brown solid, 137.5 mg, 75.8% yield. 1H NMR (500 MHz, DMSO-d6) δ 9.38 (s, 1H), 8.42 (s, 1H), 8.27 (s, 1H), 7.39–7.28 (m, 4H), 6.07 (t, J = 5.6 Hz, 1H), 3.07 (qd, J = 7.2, 5.6 Hz, 2H), 1.01 (t, J = 7.2 Hz, 3H). HRMS: calcd for C13H14Cl2N5O [M + H]+: 326.0570; found: 326.0542.
6-((2,5-Dichloropyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (21)
Off-white solid, 161.9 mg, 82.1% yield. 1H NMR (500 MHz, DMSO-d6) δ 8.30 (s, 1H), 7.39 (d, J = 2.0 Hz, 1H), 7.17 (dd, J = 8.3, 2.0 Hz, 1H), 7.00 (d, J = 8.3 Hz, 1H). HRMS: calcd for C11H7Cl2N4O2 [M + H]+: 296.9941; found: 296.9925.
General synthetic procedure for the preparation of final compounds 22–69
A mixture of 2-chloro-N-arylpyrimidin-4-amine intermediates 16–21 (100 mg, ca. 0.38 mmol, 1 equiv.), appropriate amines (1 equiv.), and trifluoroacetic acid (TFA, 0.4 mL per 100 mg of intermediate) in 2-propanol (4 mL) was heated in a sealed tube at 100 °C for 12 hours. The mixture was cooled to room temperature. The mixture was neutralized with NaOH 1 M. The precipitation was filtered and washed with hot ethanol, or the resultant was added to brine. The aqueous phase was extracted with EtOAc. The organic phase was combined and dried over anhydrous Na2SO4 and concentrated under vacuum. The crude was purified by column chromatography with a gradient elution system (methanol 5–10% in EtOAc).
N 4-(1H-Indol-5-yl)-N2-(4-((methylsulfonyl)methyl)phenyl)pyrimidine-2,4-diamine (22)
Green solid, 99 mg, 61.4% yield, mp: 107 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.02 (s, 1H), 9.17 (s, 1H), 9.13 (s, 1H), 7.95 (d, J = 5.8 Hz, 1H), 7.88 (s, 1H), 7.85–7.76 (m, 2H), 7.40–7.30 (m, 2H), 7.28–7.14 (m, 3H), 6.41 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 6.19–6.11 (m, 1H), 4.37 (s, 2H), 2.87 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.70, 161.76, 159.65, 155.35, 141.65, 133.35, 131.68, 131.54, 128.33, 126.50, 121.44, 119.38, 117.38, 111.88, 101.67, 59.68. ESI-HRMS: calcd for C20H20N5O2S [M + H]+: 394.1332; found: 394.1299.
1-Ethyl-3-(4-((2-((4-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)phenyl)urea (23)
Brown solid, 26.8 mg, 17.7% yield, mp: 173 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.20 (s, 1H), 8.40 (s, 1H), 7.93 (d, J = 5.8 Hz, 1H), 7.76–7.69 (m, 2H), 7.46 (d, J = 8.4 Hz, 2H), 7.39–7.25 (m, 2H), 7.25–7.19 (m, 2H), 6.16–6.10 (m, 2H), 4.33 (s, 2H), 3.06 (qd, J = 7.2, 5.5 Hz, 2H), 2.83 (s, 3H), 1.01 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-D6) δ 172.59, 161.13, 158.74, 158.49, 155.87, 141.46, 133.60, 131.50, 121.76, 119.50, 118.97, 118.68, 116.59, 99.09, 59.67, 34.47, 16.02. ESI-HRMS: calcd for C21H24N6NaO3S [M + Na]+: 463.1523; found: 463.1498.
6-((2-((4-((Methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (24)
Grey solid, 142.7 mg, 90.9% yield, mp: 275 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.69 (s, 1H), 10.84 (s, 1H), 10.51 (s, 1H), 7.93 (d, J = 7.0 Hz, 1H), 7.70 (s, 1H), 7.51 (d, J = 8.1 Hz, 2H), 7.36 (d, J = 8.4 Hz, 2H), 7.24 (dd, J = 8.4, 2.0 Hz, 1H), 7.04 (d, J = 8.4 Hz, 1H), 6.41 (d, J = 7.1 Hz, 1H), 4.44 (s, 2H), 2.87 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 161.32, 155.06, 153.03, 143.71, 137.46, 132.46, 132.05, 128.04, 126.19, 122.82, 118.16, 110.09, 104.95, 59.46, 39.78. ESI-HRMS: calcd for C19H17N5NaO4S [M + Na]+: 434.0893; found: 434.0935.
N 4-(1H-Indol-5-yl)-N2-(3-((methylsulfonyl)methyl)phenyl)pyrimidine-2,4-diamine (25)
Off-white solid, 36.9 mg, 22.9% yield, mp: 110 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.00 (s, 1H), 9.13 (s, 1H), 9.06 (s, 1H), 7.90 (d, J = 5.8 Hz, 1H), 7.81 (s, 1H), 7.79–7.76 (m, 1H), 7.76–7.73 (m, 1H), 7.33 (d, J = 8.6 Hz, 1H), 7.30 (t, J = 2.7 Hz, 1H), 7.20–7.11 (m, 2H), 6.89 (dt, J = 7.6, 1.3 Hz, 1H), 6.34 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 6.11 (d, J = 5.8 Hz, 1H), 4.26 (s, 2H), 2.81 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 161.75, 160.12, 156.09, 141.80, 133.27, 131.76, 129.45, 128.82, 128.23, 126.40, 123.62, 121.55, 119.32, 117.50, 111.81, 101.48, 60.30, 39.77. ESI-HRMS: calcd for C20H20N5O2S [M + H]+: 394.1332; found: 394.1295.
1-Ethyl-3-(4-((2-((3-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)phenyl)urea (26)
Yellow solid, 104.1 mg, 68.8% yield, mp: 203 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.14 (d, J = 16.9 Hz, 2H), 8.29 (s, 1H), 7.92 (d, J = 5.8 Hz, 1H), 7.78–7.69 (m, 2H), 7.47 (d, J = 8.5 Hz, 2H), 7.33–7.27 (m, 2H), 7.20 (t, J = 7.8 Hz, 1H), 6.91 (dt, J = 7.5, 1.3 Hz, 1H), 6.12 (d, J = 5.8 Hz, 1H), 6.00 (t, J = 5.6 Hz, 1H), 4.34 (s, 2H), 3.07 (qd, J = 7.2, 5.6 Hz, 2H), 2.87 (s, 3H), 1.01 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 161.10, 160.03, 156.16, 155.83, 141.72, 136.17, 133.94, 129.47, 128.91, 123.95, 121.74, 119.53, 118.72, 99.01, 60.32, 34.50, 16.07. ESI-HRMS: calcd for C21H25N6O3S [M + H]+: 441.1703; found: 441.1732.
6-((2-((3-((Methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (27)
Off-white solid, 144.7 mg, 92.2% yield, mp: 279 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.67 (s, 1H), 10.72 (s, 1H), 10.40 (s, 1H), 7.91 (d, J = 7.0 Hz, 1H), 7.75 (s, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.44 (s, 1H), 7.36 (t, J = 7.9 Hz, 1H), 7.23–7.15 (m, 2H), 7.04 (d, J = 8.3 Hz, 1H), 6.38 (d, J = 7.0 Hz, 1H), 4.42 (s, 2H), 2.87 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 161.34, 155.07, 153.14, 143.69, 137.45, 132.48, 130.54, 129.59, 128.08, 124.98, 123.04, 118.13, 110.15, 104.94, 100.20, 59.64, 39.68. ESI-HRMS: calcd for C19H18N5O4S [M + H]+: 412.1074; found: 412.1110.
N-(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (28)
Brown solid, 91.1 mg, 56.4% yield, mp: 136 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.08 (s, 1H), 9.90 (s, 1H), 9.67 (s, 1H), 9.55 (s, 1H), 7.85 (d, J = 6.6 Hz, 2H), 7.56 (d, J = 8.4 Hz, 2H), 7.35–7.30 (m, 2H), 7.11 (dd, J = 9.3, 2.7 Hz, 3H), 6.37 (t, J = 2.8 Hz, 1H), 6.22 (d, J = 6.6 Hz, 1H), 2.90 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 161.46, 158.91, 158.61, 133.72, 130.42, 128.12, 126.74, 122.42, 121.74, 117.21, 116.20, 113.87, 111.88, 101.69, 39.39. ESI-HRMS: calcd for C19H19N6O2S [M + H]+: 395.1285; found: 395.1257.
N-(4-((4-((4-(3-Ethylureido)phenyl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (29)
Grey solid, 82.4 mg, 54.4% yield, mp: 187 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.34 (s, 1H), 9.18 (s, 1H), 9.10 (s, 1H), 8.29 (s, 1H), 7.90 (d, J = 5.9 Hz, 1H), 7.64 (d, J = 8.9 Hz, 2H), 7.46 (d, J = 8.5 Hz, 2H), 7.28 (d, J = 8.9 Hz, 2H), 7.06 (d, J = 8.9 Hz, 2H), 6.10 (d, J = 5.8 Hz, 1H), 6.00 (t, J = 5.6 Hz, 1H), 3.11–3.01 (m, 2H), 2.87 (s, 3H), 1.01 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.60, 161.07, 159.66, 155.82, 155.45, 138.33, 136.26, 133.81, 131.87, 122.55, 122.17, 121.62, 120.57, 118.67, 98.81, 34.49, 16.05. ESI-HRMS: calcd for C20H24N7O3S [M + H]+: 442.1656; found: 442.1685.
N-(4-((4-((2-Oxo-2,3-dihydrobenzo[d]oxazol-6-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (30)
Brown solid, 34.4 mg, 21.9% yield, mp: 260 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.50 (s, 1H), 9.62 (s, 1H), 9.45 (s, 1H), 9.38 (s, 1H), 7.93 (d, J = 6.0 Hz, 2H), 7.58 (d, J = 8.6 Hz, 2H), 7.15 (d, J = 2.0 Hz, 1H), 7.15–7.06 (m, 2H), 6.98 (d, J = 8.4 Hz, 1H), 6.17 (d, J = 6.0 Hz, 1H), 2.90 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.60, 160.89, 159.94, 156.31, 155.27, 143.95, 138.35, 135.47, 131.90, 130.03, 125.46, 122.50, 120.60, 115.88, 110.00, 103.37, 99.20, 39.24. ESI-HRMS: calcd for C18H17N6O4S [M + H]+: 413.1027; found: 413.1012.
N-(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (31)
Green solid, 35.4 mg, 21.9% yield, mp: 146 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.97 (s, 1H), 9.53 (s, 1H), 9.07 (s, 1H), 8.99 (s, 1H), 7.89 (d, J = 5.8 Hz, 1H), 7.84 (s, 1H), 7.63 (dd, J = 8.2, 1.2 Hz, 1H), 7.51 (s, 1H), 7.34–7.26 (m, 2H), 7.15 (dd, J = 8.8, 2.1 Hz, 1H), 7.09 (t, J = 8.1 Hz, 1H), 6.71 (ddd, J = 8.0, 2.1, 1.0 Hz, 1H), 6.35 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 6.10 (d, J = 5.8 Hz, 1H), 2.94 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.61, 161.73, 160.15, 155.97, 142.47, 138.92, 133.24, 131.92, 129.44, 128.33, 126.39, 117.26, 115.46, 113.03, 111.86, 111.36, 101.62, 39.68. ESI-HRMS: calcd for C19H19N6O2S [M + H]+: 395.1285; found: 395.1309.
N-(3-((4-((4-(3-Ethylureido)phenyl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (32)
Brown solid, 64.6 mg, 42.6% yield, mp: 213 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.72 (s, 1H), 8.52 (s, 1H), 7.89 (d, J = 6.4 Hz, 1H), 7.47 (t, J = 8.4 Hz, 3H), 7.35 (s, 1H), 7.33–7.28 (m, 2H), 7.19 (t, J = 8.1 Hz, 1H), 6.86 (d, J = 7.6 Hz, 1H), 6.24 (d, J = 6.5 Hz, 1H), 6.13 (t, J = 5.6 Hz, 1H), 3.06 (qd, J = 7.2, 5.4 Hz, 2H), 2.96 (s, 3H), 1.01 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.59, 161.03, 155.87, 139.31, 137.29, 132.58, 129.77, 122.15, 118.40, 116.87, 114.73, 112.51, 39.77, 34.42, 16.05. ESI-HRMS: calcd for C20H24N7O3S [M + H]+: 442.1656; found: 442.1664.
N-(3-((4-((2-Oxo-2,3-dihydrobenzo[d]oxazol-6-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (33)
White solid, 100 mg, 63.5% yield, mp: 264 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.60 (s, 1H), 10.15 (s, 1H), 10.01 (s, 1H), 9.76 (s, 1H), 7.93 (d, J = 6.5 Hz, 1H), 7.86 (s, 1H), 7.43 (dd, J = 8.1, 2.7 Hz, 1H), 7.32 (s, 1H), 7.25–7.17 (m, 2H), 7.01 (d, J = 8.4 Hz, 1H), 6.89 (dd, J = 8.1, 2.4 Hz, 1H), 6.28 (d, J = 6.5 Hz, 1H), 2.96 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.59, 161.17, 159.39, 159.14, 155.19, 143.85, 139.47, 133.66, 129.96, 126.99, 118.73, 117.51, 117.12, 115.31, 113.00, 110.09, 104.23, 99.99, 39.78. ESI-HRMS: calcd for C18H17N6O4S [M + H]+: 413.1026; found: 413.0000.
1-(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-3-ethylurea (34)
Yellow solid, 134.9 mg, 85.0% yield, mp: 151 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.07 (s, 1H), 9.81 (s, 1H), 9.60 (s, 1H), 8.45 (s, 1H), 7.88 (s, 1H), 7.83 (d, J = 6.5 Hz, 1H), 7.48–7.42 (m, 2H), 7.35–7.26 (m, 4H), 7.13 (dd, J = 8.7, 2.0 Hz, 1H), 6.36 (t, J = 2.4 Hz, 1H), 6.20–6.12 (m, 2H), 3.07 (qd, J = 7.2, 5.5 Hz, 2H), 1.02 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.63, 161.44, 159.46, 159.21, 156.70, 155.85, 136.78, 133.61, 132.76, 130.84, 128.21, 126.68, 122.23, 118.82, 118.59, 117.10, 116.45, 111.90, 101.76, 34.49, 16.04. ESI-HRMS: calcd for C21H22N7O [M + H]+: 388.1880; found: 388.1895.
1-(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-3-ethylurea (35)
Green solid, 43.5 mg, 27.4% yield, mp: 122 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.96 (s, 1H), 9.03 (s, 1H), 8.93 (s, 1H), 8.25 (s, 1H), 7.92–7.86 (m, 2H), 7.48 (t, J = 2.0 Hz, 1H), 7.39 (ddd, J = 8.1, 2.1, 1.1 Hz, 1H), 7.34–7.25 (m, 2H), 7.16 (dd, J = 8.7, 2.0 Hz, 1H), 7.10 (ddd, J = 8.1, 2.1, 1.1 Hz, 1H), 7.00 (t, J = 8.1 Hz, 1H), 6.33 (td, J = 2.1, 1.0 Hz, 1H), 6.10 (d, J = 5.8 Hz, 1H), 6.01 (t, J = 5.6 Hz, 1H), 3.06 (qd, J = 7.2, 5.5 Hz, 2H), 1.00 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 172.61, 161.64, 160.24, 155.78, 155.68, 141.79, 141.06, 133.11, 132.08, 128.88, 128.33, 126.30, 117.06, 113.16, 111.79, 111.44, 109.29, 101.61, 34.43, 16.04. ESI-HRMS: calcd for C21H22N7O [M + H]+: 388.1880; found: 388.1895.
5-Chloro-N4-(1H-indol-5-yl)-N2-(4-((methylsulfonyl)methyl)phenyl)pyrimidine-2,4-diamine (36)
Off-white solid, 65.5 mg, 42.5% yield, mp: 214 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.05 (s, 1H), 9.28 (s, 1H), 8.75 (s, 1H), 8.05 (s, 1H), 7.70 (d, J = 2.0 Hz, 1H), 7.60–7.54 (m, 2H), 7.38–7.30 (m, 2H), 7.18 (dd, J = 8.6, 2.1 Hz, 1H), 7.02 (d, J = 8.5 Hz, 2H), 6.39 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 4.25 (s, 2H), 2.78 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.14, 157.17, 154.43, 141.30, 134.05, 131.29, 130.36, 128.03, 126.43, 121.36, 119.68, 118.94, 116.37, 111.40, 104.30, 101.66, 59.60, 39.37. ESI-HRMS: calcd for C20H18ClN5NaO2S [M + Na]+: 450.0762; found: 450.0785.
1-(4-((5-Chloro-2-((4-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)phenyl)-3-ethylurea (37)
Off-white solid, 101.8 mg, 69.7% yield, mp: 197 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.40 (s, 1H), 8.79 (s, 1H), 8.53 (s, 1H), 8.08 (s, 1H), 7.62–7.56 (m, 2H), 7.41 (d, J = 9.0 Hz, 2H), 7.38–7.32 (m, 2H), 7.18–7.13 (m, 2H), 6.23 (t, J = 5.6 Hz, 1H), 4.30 (s, 2H), 3.08 (qd, J = 7.3, 2.9 Hz, 2H), 2.81 (s, 3H), 1.02 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 157.95, 156.73, 155.81, 154.36, 141.21, 137.77, 132.23, 131.44, 124.89, 121.83, 119.29, 118.25, 104.58, 59.70, 39.60, 34.51, 16.03. ESI-HRMS: calcd for C21H23ClN6NaO3S [M + Na]+: 497.1133; found: 497.1137.
6-((5-Chloro-2-((4-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (38)
Off-white solid, 138.5 mg, 92.0% yield, mp: 286 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.65 (s, 1H), 9.78–9.75 (m, 1H), 9.35–9.31 (m, 1H), 8.19 (s, 1H), 7.64–7.58 (m, 1H), 7.51 (d, J = 8.5 Hz, 2H), 7.30 (dd, J = 8.4, 2.0 Hz, 1H), 7.19–7.13 (m, 2H), 7.05 (d, J = 8.3 Hz, 1H), 4.33 (s, 2H), 2.81 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.59, 157.57, 155.13, 154.41, 143.54, 139.17, 132.32, 131.63, 128.36, 123.99, 120.69, 109.85, 109.69, 107.37, 104.93, 59.51, 39.48. ESI-HRMS: calcd for C19H17ClN5O4S [M + H]+: 446.0684; found: 446.0691.
5-Chloro-N4-(1H-indol-5-yl)-N2-(3-((methylsulfonyl)methyl)phenyl)pyrimidine-2,4-diamine (39)
Off-white solid, 56.1 mg, 36.5% yield, mp: 177 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.11 (s, 1H), 9.46 (s, 1H), 9.07 (s, 1H), 8.08 (s, 1H), 7.65 (d, J = 2.0 Hz, 1H), 7.53 (ddd, J = 8.3, 2.3, 1.1 Hz, 1H), 7.45 (t, J = 1.9 Hz, 1H), 7.40–7.31 (m, 2H), 7.15 (dd, J = 8.6, 2.0 Hz, 1H), 6.97 (t, J = 7.9 Hz, 1H), 6.84 (dt, J = 7.6, 1.3 Hz, 1H), 6.38 (ddd, J = 2.9, 1.9, 0.9 Hz, 1H), 4.00 (s, 2H), 2.70 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 159.42, 159.13, 157.91, 155.28, 139.85, 134.52, 129.64, 128.97, 128.04, 126.86, 125.34, 122.18, 120.23, 120.00, 117.22, 111.73, 104.62, 101.80, 59.77, 39.68. ESI-HRMS: calcd for C20H19ClN5O2S [M + H]+: 428.0942; found: 428.0985.
1-(4-((5-Chloro-2-((3-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)phenyl)-3-ethylurea (40)
Off-white solid, 33 mg, 22.8% yield, mp: 288 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.33 (s, 1H), 8.71 (s, 1H), 8.57 (s, 1H), 8.05 (s, 1H), 7.62 (ddd, J = 8.3, 2.3, 1.0 Hz, 1H), 7.54 (t, J = 1.9 Hz, 1H), 7.40 (d, J = 8.8 Hz, 2H), 7.38–7.33 (m, 2H), 7.13 (t, J = 7.9 Hz, 1H), 6.88 (dt, J = 7.7, 1.3 Hz, 1H), 6.19 (t, J = 5.6 Hz, 1H), 4.25 (s, 2H), 3.07 (qd, J = 7.2, 5.5 Hz, 2H), 2.84 (s, 3H), 1.02 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 157.82, 155.81, 139.30, 138.57, 130.44, 129.89, 129.25, 126.68, 126.00, 123.07, 121.06, 117.94, 104.88, 59.75, 39.68, 34.46, 16.02. ESI-HRMS: calcd for C21H24ClN6O3S [M + H]+: 475.1314; found: 475.1347.
6-((5-Chloro-2-((3-((methylsulfonyl)methyl)phenyl)amino)pyrimidin-4-yl)amino)benzo[d]oxazol-2(3H)-one (41)
Off-white solid, 124.3 mg, 82.5% yield, mp: 284 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.66 (s, 1H), 9.79 (s, 1H), 9.34 (s, 1H), 8.18 (s, 1H), 7.63 (s, 1H), 7.58 (dd, J = 8.6, 2.2 Hz, 1H), 7.45 (t, J = 1.9 Hz, 1H), 7.28 (dd, J = 8.4, 2.0 Hz, 1H), 7.14 (t, J = 7.9 Hz, 1H), 7.06 (d, J = 8.3 Hz, 1H), 6.98 (d, J = 7.7 Hz, 1H), 4.27 (s, 2H), 2.82 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 157.82, 155.10, 153.61, 143.50, 138.68, 132.05, 130.05, 129.14, 128.66, 126.59, 123.16, 121.13, 120.97, 109.79, 107.59, 104.96, 59.77, 39.66. ESI-HRMS: calcd for C19H17ClN5O4S [M + H]+: 446.0684; found: 446.0695.
N-(4-((4-((1H-Indol-5-yl)amino)-5-chloropyrimidin-2-yl)amino)phenyl)methanesulfonamide (42)
Grey solid, 52.5 mg, 34.0% yield, mp: 238 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.04 (s, 1H), 8.96 (s, 1H), 8.62 (s, 1H), 7.98 (s, 1H), 7.73 (d, J = 2.0 Hz, 1H), 7.39–7.28 (m, 4H), 7.18 (dd, J = 8.6, 2.0 Hz, 1H), 6.79–6.72 (m, 2H), 6.38 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 2.67 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.34, 157.01, 154.43, 136.05, 133.92, 130.48, 127.98, 126.36, 121.84, 120.21, 119.44, 116.08, 111.33, 103.56, 101.64, 40.60. ESI-HRMS: calcd for C19H17ClN6NaO2S [M + Na]+: 451.0714; found: 451.0737.
N-(4-((5-Chloro-4-((4-(3-ethylureido)phenyl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (43)
Brown solid, 40.5 mg, 27.6% yield, mp: 209 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.64 (s, 1H), 9.46 (s, 1H), 9.20 (s, 1H), 8.52 (s, 1H), 8.12 (s, 1H), 8.05 (s, 1H), 7.47 (d, J = 7.0 Hz, 1H), 7.45–7.30 (m, 5H), 7.08–6.98 (m, 2H), 3.07 (q, J = 7.2 Hz, 2H), 2.87 (s, 3H), 1.02 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 159.38, 159.09, 157.14, 155.73, 138.39, 136.35, 133.30, 131.39, 125.20, 122.19, 121.57, 118.04, 115.27, 104.52, 39.37, 34.49, 16.00. ESI-HRMS: calcd for C20H23ClN7O3S [M + H]+: 476.1266; found: 476.1291.
N-(4-((5-Chloro-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (44)
Grey solid, 81.8 mg, 54.2% yield, mp: 281 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.67 (s, 1H), 9.80 (s, 1H), 9.52 (s, 1H), 9.47 (s, 1H), 8.18 (s, 1H), 7.61 (s, 1H), 7.41 (d, J = 8.8 Hz, 2H), 7.26 (dd, J = 8.4, 2.0 Hz, 1H), 7.07–6.97 (m, 3H), 2.87 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 157.77, 155.08, 143.49, 134.64, 134.55, 131.97, 128.62, 122.45, 121.53, 120.88, 109.66, 107.59, 104.69, 39.39. ESI-HRMS: calcd for C18H16ClN6O4S [M + H]+: 447.0637; found: 447.0658.
N-(3-((4-((1H-Indol-5-yl)amino)-5-chloropyrimidin-2-yl)amino)phenyl)methanesulfonamide (45)
Yellow solid, 29.2 mg, 18.9% yield, mp: 215 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.04 (s, 1H), 9.53 (s, 1H), 9.18 (s, 1H), 8.69 (s, 1H), 8.03 (s, 1H), 7.71 (d, J = 2.0 Hz, 1H), 7.51 (ddd, J = 8.3, 2.1, 1.0 Hz, 1H), 7.39–7.29 (m, 2H), 7.24 (d, J = 2.6 Hz, 1H), 7.20 (dd, J = 8.6, 2.1 Hz, 1H), 6.86 (t, J = 8.1 Hz, 1H), 6.64 (ddd, J = 8.0, 2.2, 1.0 Hz, 1H), 6.38 (ddd, J = 2.9, 1.9, 0.9 Hz, 1H), 2.88 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.32, 157.16, 154.34, 141.94, 138.80, 134.06, 130.53, 129.37, 128.07, 126.44, 119.62, 116.23, 115.38, 113.41, 111.49, 111.27, 104.52, 101.73, 39.53. ESI-HRMS: calcd for C19H18ClN6O2S [M + H]+: 429.0895; found: 429.0871.
N-(3-((5-Chloro-4-((4-(3-ethylureido)phenyl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (46)
White solid, 36 mg, 24.6% yield, mp: 217 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.57 (s, 1H), 9.25 (s, 1H), 8.66 (s, 1H), 8.36 (s, 1H), 8.04 (s, 1H), 7.51 (dd, J = 8.3, 2.3 Hz, 1H), 7.48–7.43 (m, 2H), 7.36–7.32 (m, 1H), 7.31 (d, J = 7.0 Hz, 2H), 7.04 (t, J = 8.1 Hz, 1H), 6.70 (ddd, J = 8.0, 2.2, 1.0 Hz, 1H), 6.04 (t, J = 5.6 Hz, 1H), 3.07 (qd, J = 7.2, 5.5 Hz, 2H), 2.92 (s, 3H), 1.02 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.21, 156.53, 155.78, 154.55, 141.87, 138.90, 137.48, 132.42, 129.45, 124.45, 118.23, 115.47, 113.56, 111.34, 104.67, 39.71, 39.50, 34.51, 16.04. ESI-HRMS: calcd for C20H22ClN7NaO3S [M + Na]+: 498.1086; found: 498.1105.
N-(3-((5-Chloro-4-((2-oxo-2,3-dihydrobenzo[d]oxazol-6-yl)amino)pyrimidin-2-yl)amino)phenyl)methanesulfonamide (47)
Brown solid, 100.4 mg, 66.5% yield, mp: 283 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.56 (s, 1H), 9.60 (s, 1H), 9.41 (s, 1H), 8.94 (s, 1H), 8.10 (s, 1H), 7.72 (d, J = 2.0 Hz, 1H), 7.51–7.43 (m, 1H), 7.34 (dd, J = 8.4, 2.0 Hz, 1H), 7.32 (s, 1H), 7.10–7.00 (m, 2H), 6.73 (dd, J = 8.1, 2.1 Hz, 1H), 2.93 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 157.81, 155.12, 143.49, 139.41, 138.94, 131.86, 129.75, 128.66, 120.82, 117.12, 115.73, 112.74, 109.72, 107.43, 105.07, 39.77. ESI-HRMS: calcd for C18H16ClN6O4S [M + H]+: 447.0637; found: 447.0612.
1-(4-((4-((1H-Indol-5-yl)amino)-5-chloropyrimidin-2-yl)amino)phenyl)-3-ethylurea (48)
Brown solid, 80 mg, 52.7% yield, mp: 234 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.04 (s, 1H), 8.98 (s, 1H), 8.63 (s, 1H), 8.11 (s, 1H), 7.99 (s, 1H), 7.73 (d, J = 1.9 Hz, 1H), 7.42–7.38 (m, 2H), 7.38–7.29 (m, 2H), 7.20 (dd, J = 8.6, 2.1 Hz, 1H), 7.03 (d, J = 8.5 Hz, 2H), 6.38 (t, J = 2.6 Hz, 1H), 5.91 (t, J = 5.6 Hz, 1H), 3.04 (qd, J = 7.2, 5.5 Hz, 2H), 0.99 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.47, 157.01, 155.81, 154.47, 134.95, 134.80, 133.95, 130.62, 128.05, 126.40, 119.92, 119.45, 118.67, 115.95, 111.38, 101.70, 34.46, 16.09. ESI-HRMS: calcd for C21H21ClN7O [M + H]+: 422.1490; found: 422.1508.
1-(3-((4-((1H-Indol-5-yl)amino)-5-chloropyrimidin-2-yl)amino)phenyl)-3-ethylurea (49)
Off-white solid, 40.1 mg, 26.4% yield, mp: 226 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.01 (s, 1H), 9.06 (s, 1H), 8.64 (s, 1H), 8.19 (s, 1H), 8.02 (s, 1H), 7.76 (d, J = 2.0 Hz, 1H), 7.37–7.26 (m, 4H), 7.21 (dd, J = 8.6, 2.0 Hz, 1H), 7.00 (dt, J = 8.2, 1.3 Hz, 1H), 6.79 (t, J = 8.2 Hz, 1H), 6.37 (ddd, J = 3.0, 1.9, 0.9 Hz, 1H), 5.98 (t, J = 5.6 Hz, 1H), 3.04 (qd, J = 7.2, 5.5 Hz, 2H), 0.99 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 158.47, 157.01, 155.61, 154.30, 141.32, 140.96, 133.96, 130.62, 128.82, 128.06, 126.37, 119.43, 115.95, 113.05, 111.68, 111.44, 109.13, 104.20, 101.73, 34.41, 16.03. ESI-HRMS: calcd for C21H21ClN7O [M + H]+: 422.1490; found: 422.1508.
N-(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(pyrrolidin-1-yl)acetamide (50)
Yellow solid, 57.5 mg, 32.9% yield, mp: 88 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.97 (s, 1H), 9.49 (s, 1H), 9.03 (s, 1H), 8.94 (s, 1H), 7.91–7.81 (m, 2H), 7.69–7.61 (m, 2H), 7.48–7.40 (m, 2H), 7.34–7.25 (m, 2H), 7.14 (dd, J = 8.7, 2.0 Hz, 1H), 6.36 (ddd, J = 3.0, 2.0, 0.9 Hz, 1H), 6.08 (d, J = 5.8 Hz, 1H), 3.17 (s, 2H), 2.60–2.52 (m, 4H), 1.71 (td, J = 6.7, 3.2 Hz, 4H). 13C NMR (125 MHz, DMSO-d6) δ 168.74, 161.67, 160.29, 156.12, 137.35, 133.13, 132.62, 132.03, 128.31, 126.33, 120.38, 119.67, 111.78, 101.59, 60.08, 54.28, 23.99. ESI-HRMS: calcd for C24H26N7O [M + H]+: 428.1293; found: 428.2165.
N-(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(pyrrolidin-1-yl)acetamide (51)
Yellow solid, 66.5 mg, 37.9% yield, mp: 85 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.95 (s, 1H), 9.46 (s, 1H), 9.02 (d, J = 15.8 Hz, 2H), 7.89 (d, J = 5.7 Hz, 2H), 7.78 (t, J = 2.1 Hz, 1H), 7.51 (dd, J = 8.2, 2.1 Hz, 1H), 7.33–7.23 (m, 2H), 7.23–7.20 (m, 1H), 7.17 (dd, J = 8.7, 2.0 Hz, 1H), 7.08 (t, J = 8.1 Hz, 1H), 6.31 (t, J = 2.5 Hz, 1H), 6.11 (d, J = 5.8 Hz, 1H), 3.16 (s, 2H), 2.51 (td, J = 4.8, 2.1 Hz, 4H), 1.67 (h, J = 3.1 Hz, 4H). 13C NMR (125 MHz, DMSO-d6) δ 168.90, 160.22, 156.01, 141.83, 139.01, 133.07, 132.07, 128.93, 128.29, 126.31, 116.93, 115.41, 113.12, 111.78, 111.25, 101.59, 59.76, 54.20, 23.91. ESI-HRMS: calcd for C24H26N7O [M + H]+: 428.2193; found: 428.2171.
N-(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(4-methylpiperazin-1-yl)acetamide (52)
Off-white solid, 72.1 mg, 38.5% yield, mp: 153 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.98 (s, 1H), 9.49 (s, 1H), 9.04 (s, 1H), 8.96 (s, 1H), 7.88 (d, J = 5.7 Hz, 2H), 7.68–7.63 (m, 2H), 7.45–7.40 (m, 2H), 7.35–7.26 (m, 2H), 7.14 (dd, J = 8.5, 2.0 Hz, 1H), 6.36 (ddd, J = 2.9, 1.9, 0.8 Hz, 1H), 6.08 (d, J = 5.8 Hz, 1H), 3.05 (s, 2H), 2.53–2.47 (m, 4H), 2.41–2.26 (m, 4H), 2.14 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 168.24, 161.66, 160.28, 156.12, 137.42, 133.12, 132.47, 132.02, 128.30, 126.33, 120.54, 120.32, 119.78, 117.14, 114.31, 113.03, 111.79, 101.59, 62.28, 55.07, 53.20, 46.24. ESI-HRMS: calcd for C25H29N8O [M + H]+: 457.2458; found: 457.2438.
N-(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(4-methylpiperazin-1-yl)acetamide (53)
Yellow solid, 17.2 mg, 9.2% yield, mp: 146 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.95 (s, 1H), 9.45 (s, 1H), 9.03 (d, J = 17.1 Hz, 2H), 7.90 (t, J = 6.7 Hz, 2H), 7.76 (t, J = 2.0 Hz, 1H), 7.51 (dd, J = 8.0, 2.0 Hz, 1H), 7.30–7.26 (m, 2H), 7.24 (dd, J = 7.9, 1.9 Hz, 1H), 7.20–7.14 (m, 1H), 7.09 (t, J = 8.0 Hz, 1H), 6.30 (t, J = 2.5 Hz, 1H), 6.11 (d, J = 5.8 Hz, 1H), 3.02 (s, 2H), 2.45–2.38 (m, 4H), 2.27 (s, 4H), 2.10 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 168.55, 161.54, 160.22, 156.01, 141.90, 138.92, 133.06, 132.13, 128.98, 128.30, 126.29, 115.39, 112.92, 111.78, 111.09, 101.58, 62.14, 55.08, 53.09, 46.19. ESI-HRMS: calcd for C25H29N8O [M + H]+: 457.2458; found: 457.2431.
N-(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(4-(pyrrolidin-1-yl)piperidin-1-yl)acetamide (54)
White solid, 86.7 mg, 41.4% yield, mp: 115 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.99 (s, 1H), 9.48 (s, 1H), 9.05 (s, 1H), 8.96 (s, 1H), 7.88 (d, J = 5.7 Hz, 2H), 7.70–7.63 (m, 2H), 7.48–7.41 (m, 2H), 7.35–7.26 (m, 2H), 7.14 (dd, J = 8.6, 2.0 Hz, 1H), 6.36 (t, J = 2.5 Hz, 1H), 6.09 (d, J = 5.8 Hz, 1H), 3.02 (s, 2H), 2.80 (dt, J = 11.6, 3.8 Hz, 2H), 2.42 (d, J = 6.0 Hz, 4H), 2.12 (td, J = 11.6, 2.5 Hz, 2H), 1.91 (tt, J = 10.4, 4.0 Hz, 1H), 1.81–1.74 (m, 2H), 1.62 (h, J = 3.0 Hz, 4H), 1.52–1.41 (m, 2H). 13C NMR (125 MHz, DMSO-d6) δ 168.45, 161.66, 160.29, 156.11, 137.41, 133.13, 132.49, 132.04, 128.31, 126.32, 120.27, 119.74, 117.13, 113.01, 111.78, 101.58, 62.51, 61.23, 52.58, 51.41, 31.60, 23.49. ESI-HRMS: calcd for C29H35N8O [M + H]+: 511.2928; found: 511.2894.
N-(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)-2-(4-(pyrrolidin-1-yl)piperidin-1-yl)acetamide (55)
White solid, 83.6 mg, 40.0% yield, mp: 111 °C. 1H NMR (500 MHz, DMSO-d6) δ 10.97 (s, 1H), 9.48 (s, 1H), 9.06 (s, 1H), 9.02 (s, 1H), 7.90 (d, J = 5.7 Hz, 2H), 7.77 (t, J = 2.1 Hz, 1H), 7.55–7.48 (m, 1H), 7.33–7.22 (m, 3H), 7.18 (dd, J = 8.6, 2.1 Hz, 1H), 7.10 (t, J = 8.1 Hz, 1H), 6.30 (t, J = 2.6 Hz, 1H), 6.12 (d, J = 5.8 Hz, 1H), 3.02 (s, 2H), 2.77 (dt, J = 12.5, 3.7 Hz, 2H), 2.62–2.57 (m, 4H), 2.17–2.14 (m, 1H), 2.10 (td, J = 11.7, 2.4 Hz, 3H), 1.77 (dd, J = 13.4, 4.2 Hz, 2H), 1.72–1.61 (m, 5H). 13C NMR (125 MHz, DMSO-d6) δ 168.70, 161.56, 160.24, 156.01, 141.92, 138.97, 133.07, 132.14, 128.96, 128.31, 126.29, 116.88, 115.39, 112.91, 111.77, 111.12, 101.57, 98.60, 62.13, 61.17, 52.25, 51.36, 30.83, 23.36. ESI-HRMS: calcd for C29H35N8O [M + H]+: 511.2928; found: 511.2892.
4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)-N-(2-(dimethylamino)ethyl)benzamide (56)
White solid, 67 mg, 39.3% yield, mp: 203 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.05 (s, 1H), 9.34 (s, 1H), 9.19 (s, 1H), 8.20 (t, J = 5.7 Hz, 1H), 7.94 (d, J = 5.8 Hz, 1H), 7.84 (d, J = 8.8 Hz, 2H), 7.69 (d, J = 8.8 Hz, 2H), 7.54 (d, J = 8.6 Hz, 1H), 7.36–7.28 (m, 1H), 7.19 (d, J = 8.8 Hz, 1H), 6.54–6.47 (m, 1H), 6.37 (t, J = 2.5 Hz, 1H), 6.18 (d, J = 5.8 Hz, 1H), 3.37–3.33 (m, 2H), 3.30 (dd, J = 13.3, 7.0 Hz, 1H), 2.46–2.42 (m, 2H), 2.22 (s, 6H). 13C NMR (125 MHz, DMSO-d6) δ 166.45, 161.78, 159.95, 156.11, 144.40, 133.33, 131.75, 128.31, 128.22, 126.67, 126.46, 118.01, 117.49, 111.85, 101.56, 58.78, 45.68, 37.68. ESI-HRMS: calcd for C23H26N7O [M + H]+: 416.2193; found: 416.2209.
3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)-N-(2-(dimethylamino)ethyl)benzamide (57)
White solid, 37.5 mg, 22.0% yield, mp: 103 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.00 (s, 1H), 9.14 (d, J = 6.6 Hz, 2H), 8.24 (q, J = 5.9 Hz, 1H), 8.05–7.95 (m, 2H), 7.91 (dd, J = 5.8, 2.0 Hz, 1H), 7.33–7.26 (m, 4H), 7.23 (t, J = 7.9 Hz, 1H), 7.21–7.16 (m, 1H), 6.32 (t, J = 2.5 Hz, 1H), 6.16 (t, J = 6.2 Hz, 1H), 3.32 (q, J = 6.5 Hz, 2H), 2.46–2.43 (m, 2H), 2.21 (s, 6H). 13C NMR (125 MHz, DMSO-d6) δ 167.27, 161.68, 160.18, 156.00, 141.72, 135.49, 133.14, 132.04, 128.64, 128.29, 126.33, 122.22, 119.80, 118.82, 117.07, 111.82, 101.57, 58.13, 45.07, 37.22. ESI-HRMS: calcd for C23H26N7O [M + H]+: 416.2193; found: 416.2197.
(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(4-methylpiperazin-1-yl)methanone (58)
White solid, 26.3 mg, 15.0% yield, mp: 97 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.02 (t, J = 2.3 Hz, 1H), 9.29 (s, 1H), 9.14 (s, 1H), 7.93 (d, J = 5.8 Hz, 1H), 7.82 (dd, J = 8.7, 2.6 Hz, 3H), 7.34 (d, J = 8.6 Hz, 1H), 7.31 (t, J = 2.7 Hz, 1H), 7.24–7.17 (m, 2H), 7.14 (dd, J = 8.7, 2.0 Hz, 1H), 6.38–6.31 (m, 1H), 6.15 (d, J = 5.8 Hz, 1H), 3.50–3.40 (m, 4H), 2.32–2.21 (m, J = 4.9 Hz, 4H), 2.15 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 169.79, 161.83, 160.01, 156.15, 143.11, 133.38, 131.70, 129.70, 128.37, 128.30, 127.75, 126.46, 118.36, 117.60, 113.73, 113.19, 111.83, 101.52, 55.14, 46.18, 43.91. ESI-HRMS: calcd for C24H26N7O [M + H]+: 428.2193; found: 428.2198.
(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(4-methylpiperazin-1-yl)methanone (59)
White solid, 80.4 mg, 45.9% yield, mp: 119 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.00 (s, 1H), 9.18 (s, 1H), 9.10 (s, 1H), 7.95–7.87 (m, 2H), 7.78 (s, 1H), 7.73 (t, J = 1.9 Hz, 1H), 7.36–7.25 (m, 2H), 7.20 (t, J = 7.8 Hz, 2H), 6.83 (dt, J = 7.4, 1.3 Hz, 1H), 6.35 (t, J = 2.4 Hz, 1H), 6.13 (d, J = 5.8 Hz, 1H), 3.53 (s, 2H), 3.31–3.20 (m, 2H), 2.26–2.18 (m, 4H), 2.11 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ 169.67, 161.76, 160.11, 156.02, 141.70, 136.59, 133.29, 131.85, 128.94, 128.31, 126.39, 120.06, 119.44, 117.27, 111.86, 101.58, 54.77, 47.53, 46.14. ESI-HRMS: calcd for C24H26N7O [M + H]+: 428.2193; found: 428.2200.
(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(morpholino)methanone (62)
White solid, 53.7 mg, 31.6% yield, mp: 178 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.04 (s, 1H), 9.46 (s, 1H), 9.32 (s, 1H), 7.93 (d, J = 5.9 Hz, 1H), 7.86–7.77 (m, 3H), 7.38–7.30 (m, 2H), 7.27–7.21 (m, 2H), 7.15 (dd, J = 8.7, 2.0 Hz, 1H), 6.37 (ddd, J = 3.0, 2.0, 0.9 Hz, 1H), 6.18 (d, J = 6.0 Hz, 1H), 3.56 (t, J = 4.6 Hz, 4H), 3.47 (s, 4H). 13C NMR (125 MHz, DMSO-d6) δ 169.84, 161.81, 159.22, 142.79, 133.50, 131.43, 128.53, 128.28, 127.75, 126.55, 118.72, 117.64, 113.85, 111.86, 101.55, 66.70, 21.60. ESI-HRMS: calcd for C23H23N6O2 [M + H]+: 415.1877; found: 415.1893.
(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(morpholino)methanone (63)
Brown solid, 71 mg, 41.8% yield, mp: 263 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.20 (s, 1H), 10.87 (s, 1H), 10.68 (s, 1H), 7.91 (d, J = 7.2 Hz, 1H), 7.74 (s, 1H), 7.67 (d, J = 7.2 Hz, 1H), 7.48 (s, 1H), 7.35 (dd, J = 6.0, 3.1 Hz, 3H), 7.22 (s, 1H), 7.19–7.14 (m, 1H), 6.45 (s, 1H), 6.34 (s, 1H), 3.55–3.45 (m, 4H), 3.15 (d, J = 25.8 Hz, 4H). 13C NMR (125 MHz, DMSO-d6) δ 168.88, 152.70, 137.65, 136.91, 129.65, 128.04, 127.10, 123.79, 120.69, 101.88, 66.50, 26.02. ESI-HRMS: calcd for C23H23N6O2 [M + H]+: 415.1877; found: 415.1891.
(4-((4-((1H-indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(4-(pyrrolidin-1-yl)piperidin-1-yl)methanone (64)
White solid, 29.5 mg, 14.9% yield, mp: 181 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.01 (s, 1H), 9.27 (s, 1H), 9.13 (s, 1H), 7.92 (d, J = 5.8 Hz, 1H), 7.80 (t, J = 8.2 Hz, 3H), 7.33 (d, J = 8.6 Hz, 1H), 7.29 (t, J = 2.8 Hz, 1H), 7.18 (s, 1H), 7.17–7.10 (m, 2H), 6.35 (t, J = 2.5 Hz, 1H), 6.14 (d, J = 5.8 Hz, 1H), 2.95 (s, 1H), 2.43 (d, J = 6.2 Hz, 6H), 2.17 (tt, J = 10.2, 4.0 Hz, 1H), 1.78 (d, J = 13.2 Hz, 2H), 1.62 (q, J = 3.4 Hz, 5H), 1.36–1.25 (m, 2H). 13C NMR (125 MHz, DMSO-d6) δ 169.72, 161.82, 160.02, 156.15, 142.98, 133.36, 131.72, 129.49, 128.30, 128.18, 128.14, 126.42, 118.35, 117.58, 113.66, 113.20, 111.84, 101.53, 61.24, 55.44, 51.30, 31.69, 23.46. ESI-HRMS: calcd for C28H32N7O [M + H]+: 482.2662; found: 482.2632.
(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(4-(pyrrolidin-1-yl)piperidin-1-yl)methanone (65)
White solid, 40 mg, 20.3% yield, mp: 113 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.00 (s, 1H), 9.16 (s, 1H), 9.08 (s, 1H), 7.90 (dd, J = 8.6, 3.7 Hz, 2H), 7.77 (s, 1H), 7.68 (s, 1H), 7.33–7.28 (m, 2H), 7.21–7.17 (m, 2H), 6.81 (d, J = 7.5 Hz, 1H), 6.35 (d, J = 2.5 Hz, 1H), 6.12 (d, J = 5.8 Hz, 1H), 2.95 (s, 1H), 2.89–2.81 (m, 1H), 2.43 (d, J = 5.9 Hz, 5H), 2.16 (dq, J = 10.5, 5.2 Hz, 1H), 1.83 (d, J = 14.2 Hz, 1H), 1.71 (s, 1H), 1.61 (q, J = 3.3 Hz, 5H), 1.33–1.25 (m, 2H). 13C NMR (125 MHz, DMSO-d6) δ 169.57, 161.78, 160.12, 156.02, 141.67, 136.97, 133.29, 131.84, 128.92, 128.31, 126.40, 119.95, 119.22, 117.40, 117.08, 113.24, 111.85, 101.57, 61.10, 51.25, 49.14, 31.83, 23.44. ESI-HRMS: calcd for C28H32N7O [M + H]+: 482.2662; found: 482.2628.
N 4-(1H-Indol-5-yl)-N2-(4-((4-(pyrrolidin-1-yl)piperidin-1-yl)methyl)phenyl)pyrimidine-2,4-diamine (68)
White solid, 143 mg, 74.7% yield, mp: 76 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.08 (s, 1H), 9.57 (d, J = 57.8 Hz, 2H), 7.92 (d, J = 6.1 Hz, 1H), 7.77 (d, J = 8.1 Hz, 2H), 7.36–7.32 (m, 3H), 7.26 (d, J = 8.1 Hz, 2H), 7.13 (dd, J = 8.6, 2.1 Hz, 1H), 6.36 (t, J = 2.5 Hz, 1H), 6.19 (d, J = 6.1 Hz, 1H), 4.07 (s, 2H), 3.25–3.12 (m, 3H), 3.01 (s, 2H), 2.69 (s, 2H), 2.17 (d, J = 12.8 Hz, 2H), 2.02–1.69 (m, 8H). 13C NMR (125 MHz, DMSO-d6) δ 161.71, 159.45, 159.20, 158.95, 133.34, 131.64, 128.28, 126.51, 119.37, 118.90, 117.30, 116.52, 114.09, 111.87, 101.54, 60.13, 55.46, 51.55, 50.44, 27.16, 23.09. ESI-HRMS: calcd for C28H34N7 [M + H]+: 468.2870; found: 468.2843.
N 4-(1H-Indol-5-yl)-N2-(3-((4-(pyrrolidin-1-yl)piperidin-1-yl)methyl)phenyl)pyrimidine-2,4-diamine (69)
White solid, 60.8 mg, 31.7% yield, mp: 71 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.08 (t, J = 2.3 Hz, 1H), 9.22 (s, 1H), 9.17 (s, 1H), 7.90 (d, J = 5.8 Hz, 1H), 7.81 (s, 1H), 7.70–7.61 (m, 2H), 7.38–7.28 (m, 2H), 7.18–7.09 (m, 2H), 6.82 (d, J = 7.5 Hz, 1H), 6.34 (dd, J = 3.1, 1.9 Hz, 1H), 6.14 (d, J = 5.9 Hz, 1H), 3.35 (t, J = 6.5 Hz, 3H), 2.99–2.82 (m, 4H), 1.97–1.91 (m, 3H), 1.84 (s, 3H), 1.61–1.51 (m, 2H), 1.40–1.31 (m, 3H), 1.30–1.22 (m, 1H). 13C NMR (125 MHz, DMSO-d6) δ 161.77, 159.68, 159.53, 159.28, 159.03, 158.78, 155.13, 141.41, 133.37, 131.73, 128.82, 128.29, 126.46, 122.46, 120.48, 118.87, 117.57, 116.49, 113.59, 111.93, 101.62, 60.91, 55.41, 51.42, 51.01, 28.62, 23.04. ESI-HRMS: calcd for C28H34N7 [M + H]+: 468.2870; found: 468.2862.
General procedure to prepare compound 60, 61, 66, and 67
Intermediate 16 (100 mg, 0.41 mmol, 1 equiv.) and appropriate Boc-anilines (14a, 14b, 15a, and 15b) (1.3 equiv.), TFA (0.4 mL per 100 of intermediate) in 2-propanol (4 mL) was heated in a sealed tube at 100 °C for 12 hours. The mixture was cooled to room temperature and evaporated. The crude was immediately treated with TFA in DCM (1 : 1) and stirred for overnight and room temperature. Solvent was evaporated and redissolved 10% MeOH/DCM. Solution was stirred with Amberlyst-A21 for 1 hour. The solvent was removed and purified further with column chromatography.
(4-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(piperazin-1-yl)methanone (60)
Following procedure, compound 60 was prepared using Boc-aniline 14a. White solid, 28 mg, 16.5% yield, mp: 155 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.02 (s, 1H), 9.30 (s, 1H), 9.14 (s, 1H), 7.92 (d, J = 5.8 Hz, 1H), 7.83–7.81 (m, 2H), 7.36–7.27 (m, 2H), 7.25–7.22 (m, 2H), 7.16–7.08 (m, 2H), 6.54–6.47 (m, 1H), 6.36 (ddd, J = 3.0, 2.0, 0.9 Hz, 1H), 6.14 (d, J = 5.8 Hz, 1H), 5.53 (s, 1H), 3.56–3.53 (m, 4H), 2.92 (p, J = 2.9 Hz, 4H). 13C NMR (125 MHz, DMSO-d6) δ 169.90, 161.85, 160.01, 156.14, 143.18, 133.39, 131.72, 128.44, 128.30, 127.54, 126.46, 118.36, 117.62, 113.72, 111.84, 101.53, 45.34. ESI-HRMS: calcd for C23H24N7O [M + H]+: 414.2037; found: 414.2011.
(3-((4-((1H-Indol-5-yl)amino)pyrimidin-2-yl)amino)phenyl)(piperazin-1-yl)methanone (61)
Following procedure, compound 61 was prepared using Boc-aniline 14b. White solid, 12 mg, 7.1% yield, mp: 173 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.00 (s, 1H), 9.17 (s, 1H), 9.10 (s, 1H), 7.94–7.85 (m, 2H), 7.77 (d, J = 5.9 Hz, 1H), 7.74–7.68 (m, 1H), 7.35–7.26 (m, 2H), 7.19 (dt, J = 9.2, 7.2 Hz, 2H), 6.87–6.79 (m, 1H), 6.34 (t, J = 2.5 Hz, 1H), 6.13 (d, J = 5.8 Hz, 1H), 3.65–3.44 (m, 4H), 2.86–2.58 (m, 4H). 13C NMR (125 MHz, DMSO-d6) δ 169.85, 161.77, 160.10, 156.01, 141.75, 141.69, 136.59, 135.91, 133.28, 131.86, 128.99, 128.30, 126.42, 120.37, 119.61, 117.48, 117.28, 111.86, 101.56, 55.46, 44.37. ESI-HRMS: calcd for C23H24N7O [M + H]+: 414.2037; found: 414.2010.
N 4-(1H-Indol-5-yl)-N2-(4-(piperazin-1-ylmethyl)phenyl)pyrimidine-2,4-diamine (66)
Following procedure, compound 66 was prepared using Boc-aniline 15a. White solid, 11.2 mg, 6.9% yield, mp: 81 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.20 (s, 1H), 10.73 (s, 1H), 10.58 (s, 1H), 7.92 (d, J = 7.0 Hz, 1H), 7.88 (d, J = 9.5 Hz, 1H), 7.56 (d, J = 8.1 Hz, 2H), 7.37 (q, J = 3.9 Hz, 2H), 7.26 (t, J = 8.8 Hz, 2H), 7.13 (d, J = 8.8 Hz, 1H), 6.36 (d, J = 12.4 Hz, 2H), 3.68 (s, 2H), 3.41 (s, 2H), 3.15–3.09 (m, 4H), 2.69 (s, 2H), 2.51 (d, J = 5.1 Hz, 1H). 13C NMR (125 MHz, DMSO-d6) δ 159.72, 159.47, 159.22, 130.61, 128.10, 127.05, 121.97, 118.69, 116.32, 112.07, 101.81, 60.86, 49.09, 42.83. ESI-HRMS: calcd for C23H26N7 [M + H]+: 400.2244; found: 400.2221.
N 4-(1H-Indol-5-yl)-N2-(3-(piperazin-1-ylmethyl)phenyl)pyrimidine-2,4-diamine (67)
Following procedure, compound 67 was prepared using Boc-aniline 15b. White solid, 15.4 mg, 9.4% yield, mp: 103 °C. 1H NMR (500 MHz, DMSO-d6) δ 11.18 (s, 1H), 7.92 (dd, J = 7.1, 2.8 Hz, 1H), 7.77 (s, 1H), 7.52 (d, J = 8.1 Hz, 1H), 7.44 (s, 1H), 7.36 (td, J = 7.9, 2.9 Hz, 2H), 7.22 (td, J = 7.8, 2.9 Hz, 1H), 7.13 (d, J = 8.8 Hz, 1H), 7.01 (d, J = 7.5 Hz, 1H), 6.32 (d, J = 10.7 Hz, 2H), 3.35 (s, 2H), 2.99 (s, 4H), 2.46–2.43 (m, 4H). 13C NMR (125 MHz, DMSO-d6) δ 159.73, 159.48, 159.22, 138.68, 134.15, 129.87, 129.17, 128.14, 126.97, 124.79, 122.00, 121.13, 120.78, 118.76, 117.71, 116.38, 112.10, 101.81, 61.64, 49.37, 43.30. ESI-HRMS: calcd for C23H26N7 [M + H]+: 400.2244; found: 400.2219.
Biological testing
Plasmodium falciparum 3D7 screening
The antimalarial activity against chloroquine sensitive strain (3D7) was performed at the Drug Discovery Unit, University of Dundee, as described previously.50,51P. falciparum chloroquine sensitive reference strain, were maintained in a 5% suspension of human red blood cells (obtained from East of Scotland Blood Transfusion Service, Ninewells Hospital, Dundee, UK) cultured in RPMI 1640 medium (pH 7.3) supplemented with 0.5% Albumax II (Gibco Life Technologies, San Diego, CA, USA), 12 mM sodium bicarbonate, 0.2 mM hypoxanthine, and 20 mg L−1 gentamicin at 37 °C, in a humidified atmosphere of 1% O2, 3% CO2 with a balance of nitrogen. Growth inhibition was qualified using a fluorescence assay utilizing the binding of SYBR green to double stranded DNA, which emits a fluorescent signal at 528 nm after excitation at 458 nm. Mefloquine (potency range 30–60 nM) was used as a drug control to monitor the quality of the assay (Z′ = 0.6 to 0.8, signal to background >3, where Z′ is a measure of the discrimination between the positive and negative controls on a screen plate. Hits were confirmed by full-potency curves with dose–response curves determined from a minimum of 2 independent experiments, with key molecules tested a minimum of 4. Parasites were incubated for 72 h. with compound in a 10-point, 1 in 3 dilution series, from a top concentration of 25 μM. Compound bioactivity was expressed as IC50, the effective concentration of compound causing 50% parasite death.
HepG2 cytotoxicity
HepG2 cytotoxicity assays were conducted by the University of Dundee, HepG2 (Human Caucasian hepatocyte carcinoma cell), to estimate potency/potential toxicity in mammalian cell lines.52 Cells were provided by ECACC with certificate of analysis confirming STR profiling and were mycoplasma tested every time new stabilities were made. Cells were screened utilizing a resazurin-based fluorescence assay, (Λex = 528 nm, λem = 590 nm), in a 384 well plate format. HepG2 cells (1 × 105 cells per mL) were incubated with compound in a 10-point, 1 in 3 dilution series from a top concentration of 25 μM, for 72 h. The selected repeat compounds were incubated with a top concentration of 100 μM for confirmation. Doxorubicin was used as a drug control to monitor the quality of the assay. After the 72 hours incubation period resazurin (0.27 mM in cell culture medium) was added and the plates were incubated for further 3–4 h before the fluorescent signal was measured using a PHERAstar (BMG Labtech, GER). Compound bioactivity was expressed as IC50, the effective concentration of compound causing 50% cell death.
Human kinase screening
The compounds were tested at 1 μM concentration at the MRC Protein Phosphorylation & Ubiquitylation Unit at the University of Dundee.53 Briefly, all assays (25.5 μL) were carried out robotically at room temperature (21 °C). Assays were performed for 30 min using Multidrop Micro reagent dispensers (Thermo Electron Corporation, Waltham, MA, U.S.A.) in a 96-well format. The concentration of magnesium acetate in the assays was 10 mM and [γ-33P]ATP (800 c.p.m. pmol−1) was used at 5, 20 or 50 μM as indicated, in order to be at or below the Km for ATP for each enzyme. The assays were initiated with MgATP, stopped by the addition of 5 μL of 0.5 M orthophosphoric acid and spotted on to P81 filter plates using a unifilter harvester (PerkinElmer, Boston, MA, U.S.A.).
Physicochemical properties
Octanol/water partition coefficient
The 1-octanol/water partition coefficient at pH7.4 was determined using the shake flask technique.54,55 Approximately 1 mg of sample was dispensed into a 10 mL glass screw-cap vial to which 2 mL octanol was added. The solution was sonicated for 1 hour. Compounds that dissolved incompletely were diluted and further sonicated until completely dissolved. A final volume of 2 mL of octanol solution and 2 mL of 0.02 M phosphate buffer (PBS) pH7.4 were shaken for 6 h using as IKA HS 260 shaker. The resulting solution was centrifuged, and the octanol and the water layers were extracted and separated. The amount of sample present in the octanol and PBS solutions was determined by HPLC-UV (Hitachi HPLC with a Sunshell C8, 2.6 μm column). 100 μL injection of the aqueous sample and 10 μL of octanol solution (pre-diluted 100-fold using DMSO). The log D7.4 was calculated from the area under the curve obtained from the 2 samples, corrected for the dilution factors and injection volume.
PBS aqueous solubility
The equilibrium solubility of samples was determined in PBS at pH7.4.55,56 Approximately 1 mg of sample was placed in a 10 mL glass screw-cap vial and dissolved in 1 mL of 0.02 M PBS at pH7.4. The saturated solution and then filtered using a syringe filter (13 mm, 0.2 μm a filter, Xiboshi). A standard solution of each sample was generated by dissolving a known amount of sample in 1 mL of DMSO. The amount of sample present in the PBS and DMSO solutions were determined by HPLC-UV (Hitachi HPLC with a Sunshell C8, 2.6 μm column). 100 μL injection of the aqueous sample and 10 μL of DMSO solution (pre-diluted 100-fold using DMSO). The solubility was calculated based on the ratio of the area under the curve obtained from the 2 samples, corrected for the dilution factors and injection volume.
Intrinsic clearance
Test compound (0.5 μM) was incubated in separate experiments with male Han Wistar rat liver microsomes (Xenotech LLC TM; 0.5 mg mL−1 50 mM potassium phosphate buffer, pH 7.4) and human pooled liver microsomes (Gibco TM; 0.5 mg mL−1 50 mM potassium phosphate buffer, pH 7.4) and each reaction started with addition of excess NADPH (8 mg mL−1 50 mM potassium phosphate buffer, pH 7.4). Immediately, at time zero, then at 3, 6, 9, 15 and 30 min an aliquot (50 μL) of the incubation mixture was removed into acetonitrile (100 μL) to stop the reaction. Internal standard (50 ng mL−1 in 82 : 18 water : acetonitrile) was added to all samples, the samples centrifuged (22 °C, 3750 rpm for 10 min) to sediment precipitated protein and the plates then sealed prior to UPLC-MS/MS analysis using an Acquility UPLC coupled with Waters Xevo MicroTQS (Waters Corporation, USA). XLfit (IDBS, UK) was used to calculate the exponential decay and consequently the rate constant (k) from the ratio of peak area of test compound to internal standard at each timepoint. The rate of intrinsic clearance (CLint) of each test compound was then calculated per g of liver. Verapamil (0.5 μM) was used as a positive control to confirm acceptable assay performance. Experiments were performed using a single timecourse experiment. SimCYP microsomal scaling factors for human were set to 39.7 mg microsomal protein per g liver and 46 mg microsomal protein per g liver for rat.
MDCK passive permeability
MDCK-II cells (ECACC) were maintained in culture (DMEM, Gibco Cat: 61965-026 supplemented with 1% penicillin/streptomycin, 10% FCS) until required. For experimentation, cells were seeded onto individual transwell ‘Thincerts’ (Greiner, Cat 662610) at a density of 35 000 cells per well. Cells were grown at 37 °C, 5% CO2 for 4 days. On day 5, the media was replaced with fresh media and incubated for 1 hour. Media was removed and replaced with Dulbeccos PBS (Gibco, 14287-080), and cell inserts incubated for a further 1 hour. Dosing solutions containing 3 μM test compound, 10 μM Lucifer Yellow (1% DMSO) were prepared. 1.2 mL of PBS (1% DMSO) was added to wells of a 24-well cell culture plate (Corning, Cat 353504). 0.35 mL of dosing solution was added in duplicate to the apical side of the transwell and transwells transferred into the receiver plate solutions. Transwell plates were then incubated for 1 hour, after which inserts were removed to an empty plate to prevent any further permeation of compound. 100 μL of solution from donor, receiver wells were removed to a 96 well plate alongside 100 μL of dosing solution. 150 μL of acetonitrile containing internal standard (e.g. 100 ng mL−1 sulfadimethoxine) is then added to all samples prior to analysis by LC-MS/MS. Bupropion (positive controls) and atenolol (negative control) were run alongside test compounds. To confirm monolayer integrity, a further 100 μL from each compartment is added of the 96 well F-bottomed microtitre plate containing the Lucifer Yellow standard curve for fluorescence determination of Lucifer Yellow concentrations. Papp (apparent permeability) values were calculated using the following equation:
Molecular docking
Crystal structures were used for PfCDPK2 (PDB ID: 4MVF) and PfPK6 (PDB ID: 9CMZ), while 3D structure models for PfCDPK1 and PfPKB were generated using AlphaFold.20 Briefly, water molecules and cofactors were removed for protein preparation. Compound 68 was docked using GOLD 5.3 (ref. 57) with H-bonds constraint corresponding to the hinge H-bond donor. In addition, the binding mode interactions were visualized using PyMOL 3.1.1.58
Cheminformatics analysis
Reported antimalarial molecules were extracted from ChEMBL database. Molecules (≤900 Da) with reported IC50/EC50 was selected for further analysis. The average value was considered for multiple IC50/EC50. Molecules that displayed huge differences in various values (standard deviation >1) were excluded. Ionization classifications were determined based on acid and base pKa values, identifying molecules as either neutral, basic, acidic, or zwitterionic. Data were subjected to an appropriate statistical test of either Student t-test or one-way ANOVA with Dunnet's multiple-comparison test.
The relationship between Pf pIC50 and molecular descriptors computed using Chemaxon JChem59 and DataWarrior program.60 Our previous result was used as a training set. Compounds 22–69 were subjected to a test set. A partial least square regression model in SIMCA P14.61 Two cross-validated components were optimally fitted, explaining 65% of the total variance in the training set (r2 = 0.65. q2 = 0.50). Y-Randomization trials (26 iterations) confirmed that a random model could not be generated by chance with the same performance in fit and cross-validation.
Author contributions
GG performed synthesis, cheminformatics analysis and wrote the draft. MPG instigated the project, performed analysis and wrote the draft. MCA performed Pf assay, NM performed HepG2 and permeability assay. DG, LW and KDR analysed data.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
GG would like to acknowledge the support provided by King Mongkut's Institute of Technology Ladkrabang (KDS 2021/023). MPG would like to acknowledge funding from the National Research Council of Thailand and King Mongkut's Institute of Technology Ladkrabang (NRCT-N42A650386).
Data availability
Supplementary information is available. See DOI: https://doi.org/10.1039/D5MD00353A.
The supporting information contains additional Figures and Tables describing SAR and model statistics. Also provided are 1H-NMR, 13C-NMR, HPLC-UV chromatograms and high-resolution mass spectroscopy data.
References
- WHO, World Malaria Report 2023, Geneva, 2023 [Google Scholar]
- Björkman A. Gil P. Alifrangis M. Lancet Infect. Dis. 2024;24:e540–e541. doi: 10.1016/S1473-3099(24)00427-4. [DOI] [PubMed] [Google Scholar]
- Arendse L. B. Wyllie S. Chibale K. Gilbert I. H. ACS Infect. Dis. 2021;7:518–534. doi: 10.1021/acsinfecdis.0c00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski Jr. R. Pharmacol. Res. 2024;200:107059. doi: 10.1016/j.phrs.2024.107059. [DOI] [PubMed] [Google Scholar]
- Attwood M. M. Fabbro D. Sokolov A. V. Knapp S. Schiöth H. B. Nat. Rev. Drug Discovery. 2021;20:839–861. doi: 10.1038/s41573-021-00252-y. [DOI] [PubMed] [Google Scholar]
- Nishal S. Jhawat V. Gupta S. Phaugat P. Oncol. Res. 2022;30:221–230. doi: 10.32604/or.2022.027549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. Nielsen T. E. Clausen M. H. Trends Pharmacol. Sci. 2015;36:422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Tewari R. Straschil U. Bateman A. Böhme U. Cherevach I. Gong P. Pain A. Billker O. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogwera K. S. P. Chibale K. Arendse L. B. Trends Parasitol. 2023;39:720–731. doi: 10.1016/j.pt.2023.06.001. [DOI] [PubMed] [Google Scholar]
- Arendse L. B. Wyllie S. Chibale K. Gilbert I. H. ACS Infect. Dis. 2021;7:518–534. doi: 10.1021/acsinfecdis.0c00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesely K. Noomuna P. Vieth M. Hipskind P. Haldar K. Pantaleo A. Turrini F. Low P. S. PLoS One. 2020;15:e0242372. doi: 10.1371/journal.pone.0242372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustiere R. Vanelle P. Primas N. Molecules. 2020;25:1–35. doi: 10.3390/molecules25245949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A. Molina-Cruz A. Brzostowski J. Liu P. Luo Y. Gunalan K. Li Y. Ribeiro J. M. C. Miller L. H. Proc. Natl. Acad. Sci. U. S. A. 2018;115:774–779. doi: 10.1073/pnas.1715443115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghartey-Kwansah G. Yin Q. Li Z. Gumpper K. Sun Y. Yang R. Wang D. Jones O. Zhou X. Wang L. Bryant J. Ma J. Boampong J. N. Xu X. Cell Transplant. 2020;29:963689719884888. doi: 10.1177/0963689719884888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther G. J. Hillesland H. K. Keyloun K. R. Reid M. C. Lafuente-Monasterio M. J. Ghidelli-Disse S. Leonard S. E. He P. Jones J. C. Krahn M. M. Mo J. S. Dasari K. S. Fox A. M. Boesche M. El Bakkouri M. Rivas K. L. Leroy D. Hui R. Drewes G. Maly D. J. Van Voorhis W. C. Ojo K. K. PLoS One. 2016;11:e0149996. doi: 10.1371/journal.pone.0149996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmer M. J. Wang J. Istvan E. S. Luth M. R. Collins J. E. Huttlin E. L. Wang L. Mittal N. Hao M. Kwiatkowski N. P. Gygi S. P. Chakrabarti R. Deng X. Goldberg D. E. Winzeler E. A. Gray N. S. Chakrabarti D. ACS Infect. Dis. 2023;9:1004–1021. doi: 10.1021/acsinfecdis.3c00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galal K. A. Truong A. Kwarcinski F. de Silva C. Avalani K. Havener T. M. Chirgwin M. E. Merten E. Ong H. W. Willis C. Abdelwaly A. Helal M. A. Derbyshire E. R. Zutshi R. Drewry D. H. J. Med. Chem. 2022;65:13172–13197. doi: 10.1021/acs.jmedchem.2c00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo F. Sanz L. Vidal J. de Cozar C. Alvarez E. Lavandera J. Vanderwall D. Green D. Kumar V. Hasan S. Brown J. Peishoff C. Cardon L. Garcia-Bustos J. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Ong H. W. de Silva C. Avalani K. Kwarcinski F. Mansfield C. R. Chirgwin M. Truong A. Derbyshire E. R. Zutshi R. Drewry D. H. ACS Med. Chem. Lett. 2023;14:1774–1784. doi: 10.1021/acsmedchemlett.3c00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M. Bertoni D. Magana P. Paramval U. Pidruchna I. Radhakrishnan M. Tsenkov M. Nair S. Mirdita M. Yeo J. Kovalevskiy O. Tunyasuvunakool K. Laydon A. Zidek A. Tomlinson H. Hariharan D. Abrahamson J. Green T. Jumper J. Birney E. Steinegger M. Hassabis D. Velankar S. Nucleic Acids Res. 2024;52:D368–D375. doi: 10.1093/nar/gkad1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toviwek B. Riley J. Mutter N. Anderson M. Webster L. Hallyburton I. Gleeson D. Read K. D. Gleeson M. P. RSC Med. Chem. 2022;13:1587–1604. doi: 10.1039/d2md00218c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toviwek B. Phuangsawai O. Konsue A. Hannongbua S. Riley J. Mutter N. Anderson M. Webster L. Hallyburton I. Read K. D. Gleeson M. P. Bioorg. Med. Chem. 2021;46:116348. doi: 10.1016/j.bmc.2021.116348. [DOI] [PubMed] [Google Scholar]
- Phuangsawai O. Beswick P. Ratanabunyong S. Tabtimmai L. Suphakun P. Obounchoey P. Srisook P. Horata N. Chuckowree I. Hannongbua S. Ward S. E. Choowongkomon K. Gleeson M. P. Eur. J. Med. Chem. 2016;124:896–905. doi: 10.1016/j.ejmech.2016.08.055. [DOI] [PubMed] [Google Scholar]
- Martin M. P. Zhu J.-Y. Lawrence H. R. Pireddu R. Luo Y. Alam R. Ozcan S. Sebti S. M. Lawrence N. J. Schönbrunn E. ACS Chem. Biol. 2012;7:698–706. doi: 10.1021/cb200508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson M. P. J. Med. Chem. 2008;51:817–834. doi: 10.1021/jm701122q. [DOI] [PubMed] [Google Scholar]
- Gleeson P. Bravi G. Modi S. Lowe D. Bioorg. Med. Chem. 2009;17:5906–5919. doi: 10.1016/j.bmc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Valluri H. Bhanot A. Shah S. Bhandaru N. Sundriyal S. J. Med. Chem. 2023;66:8382–8406. doi: 10.1021/acs.jmedchem.3c00206. [DOI] [PubMed] [Google Scholar]
- Ishikawa M. Hashimoto Y. J. Med. Chem. 2011;54:1539–1554. doi: 10.1021/jm101356p. [DOI] [PubMed] [Google Scholar]
- Toviwek B. Phuangsawai O. Konsue A. Hannongbua S. Riley J. Mutter N. Anderson M. Webster L. Hallyburton I. Read K. D. Gleeson M. P. Bioorg. Med. Chem. 2021;46:116348. doi: 10.1016/j.bmc.2021.116348. [DOI] [PubMed] [Google Scholar]
- Liao J. J.-L. J. Med. Chem. 2007;50:409–424. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- Yu B. Tang L.-d. Li Y.-l. Song S.-h. Ji X.-l. Lin M.-s. Wu C.-F. Bioorg. Med. Chem. Lett. 2012;22:110–114. doi: 10.1016/j.bmcl.2011.11.061. [DOI] [PubMed] [Google Scholar]
- Tala S. D. Ou T. H. Lin Y. W. Tala K. S. Chao S. H. Wu M. H. Tsai T. H. Kakadiya R. Suman S. Chen C. H. Lee T. C. Su T. L. Eur. J. Med. Chem. 2014;76:155–169. doi: 10.1016/j.ejmech.2014.02.018. [DOI] [PubMed] [Google Scholar]
- Hernandez-Nunez E. Tlahuext H. Moo-Puc R. Torres-Gomez H. Reyes-Martinez R. Cedillo-Rivera R. Nava-Zuazo C. Navarrete-Vazquez G. Eur. J. Med. Chem. 2009;44:2975–2984. doi: 10.1016/j.ejmech.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Dios A. D., Li T., Cabrejas L. M. M., Pobanz M. A., Shih C., Wang Y., Zhong B., Blas J. A. and Uralde-Garmendia B. L. D., WO2006/009741Al, 2006
- Ding X. Stasi L. P. Ho M.-H. Zhao B. Wang H. Long K. Xu Q. Sang Y. Sun C. Hu H. Yu H. Wan Z. Wang L. Edge C. Liu Q. Li Y. Dong K. Guan X. Tattersall F. D. Reith A. D. Ren F. Bioorg. Med. Chem. Lett. 2018;28:1615–1620. doi: 10.1016/j.bmcl.2018.03.045. [DOI] [PubMed] [Google Scholar]
- Ong H. W. Truong A. Kwarcinski F. de Silva C. Avalani K. Havener T. M. Chirgwin M. Galal K. A. Willis C. Kramer A. Liu S. Knapp S. Derbyshire E. R. Zutshi R. Drewry D. H. Eur. J. Med. Chem. 2023;249:115043. doi: 10.1016/j.ejmech.2022.115043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D. B. Nguyen W. Looker O. Razook Z. McCann K. Barry A. E. Scheurer C. Wittlin S. Famodimu M. T. Delves M. J. Bullen H. E. Crabb B. S. Sleebs B. E. Gilson P. R. ACS Infect. Dis. 2023;9:1695–1710. doi: 10.1021/acsinfecdis.3c00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleran J. A. Ashraf K. Delvillar M. Eck T. Fondekar R. Miller E. B. Hutchinson A. Dong A. Seitova A. De Souza M. L. Augeri D. Halabelian L. Siekierka J. Rotella D. P. Gordon J. Childers W. E. Grier M. C. Staker B. L. Roberge J. Y. Bhanot P. J. Med. Chem. 2024;67:12459–12459. doi: 10.1021/acs.jmedchem.4c01411. [DOI] [PubMed] [Google Scholar]
- Davis A. M. Keeling D. J. Steele J. Tomkinson N. P. Tinker A. C. Curr. Top. Med. Chem. 2005;5:421–439. doi: 10.2174/1568026053828411. [DOI] [PubMed] [Google Scholar]
- Milner E. McCalmont W. Bhonsle J. Caridha D. Cobar J. Gardner S. Gerena L. Goodine D. Lanteri C. Melendez V. Roncal N. Sousa J. Wipf P. Dow G. S. Malar. J. 2010;9:51. doi: 10.1186/1475-2875-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X. Luong T. L. Reese N. Gaona H. Collazo-Velez V. Vuong C. Potter B. Sousa J. C. Olmeda R. Li Q. Xie L. Zhang J. Zhang P. Reichard G. Melendez V. Marcsisin S. R. Pybus B. S. J. Pharmacol. Toxicol. Methods. 2014;70:188–194. doi: 10.1016/j.vascn.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Thomas M. G. McGonagle K. Rowland P. Robinson D. A. Dodd P. G. Camino-Diaz I. Campbell L. Cantizani J. Castaneda P. Conn D. Craggs P. D. Edwards D. Ferguson L. Fosberry A. Frame L. Goswami P. Hu X. Korczynska J. MacLean L. Martin J. Mutter N. Osuna-Cabello M. Paterson C. Pena I. Pinto E. G. Pont C. Riley J. Shishikura Y. Simeons F. R. C. Stojanovski L. Thomas J. Wrobel K. Young R. J. Zmuda F. Zuccotto F. Read K. D. Gilbert I. H. Marco M. Miles T. J. Manzano P. De Rycker M. J. Med. Chem. 2023;66:10413–10431. doi: 10.1021/acs.jmedchem.3c00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleghorn L. A. T. Wall R. J. Albrecht S. MacGowan S. A. Norval S. De Rycker M. Woodland A. Spinks D. Thompson S. Patterson S. Corpas Lopez V. Dey G. Collie I. T. Hallyburton I. Kime R. Simeons F. R. C. Stojanovski L. Frearson J. A. Wyatt P. G. Read K. D. Gilbert I. H. Wyllie S. J. Med. Chem. 2023;66:8896–8916. doi: 10.1021/acs.jmedchem.3c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arang N. Kain H. S. Glennon E. K. Bello T. Dudgeon D. R. Walter E. N. F. Gujral T. S. Kaushansky A. Nat. Commun. 2017;8:1232. doi: 10.1038/s41467-017-01345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E. Tobin A. B. Kannan N. Doerig C. Philos. Trans. R. Soc., B. 2012;367:2607–2618. doi: 10.1098/rstb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T. G. Morahan B. John von Freyend S. Boeuf P. Grau G. Garcia-Bustos J. Doerig C. Mol. Biochem. Parasitol. 2016;208:2–15. doi: 10.1016/j.molbiopara.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Guiguemde W. Shelat A. Bouck D. Duffy S. Crowther G. Davis P. Smithson D. Connelly M. Clark J. Zhu F. Jimenez-Diaz M. Martinez M. Wilson E. Tripathi A. Gut J. Sharlow E. Bathurst I. El Mazouni F. Fowble J. Forquer I. McGinley P. Castro S. Angulo-Barturen I. Ferrer S. Rosenthal P. Derisi J. Sullivan D. Lazo J. Roos D. Riscoe M. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo F. J. Sanz L. M. Vidal J. de Cozar C. Alvarez E. Lavandera J. L. Vanderwall D. E. Green D. V. Kumar V. Hasan S. Brown J. R. Peishoff C. E. Cardon L. R. Garcia-Bustos J. F. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Seanego T. D. Chavalala H. E. Henning H. H. de Koning C. B. Hoppe H. C. Ojo K. K. Rousseau A. L. ChemMedChem. 2022;17:e202200421. doi: 10.1002/cmdc.202200421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallyburton I. Grimaldi R. Woodland A. Baragana B. Luksch T. Spinks D. James D. Leroy D. Waterson D. Fairlamb A. H. Wyatt P. G. Gilbert I. H. Frearson J. A. Malar. J. 2017;16:446. doi: 10.1186/s12936-017-2085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norcross N. R. Baragaña B. Wilson C. Hallyburton I. Osuna-Cabello M. Norval S. Riley J. Stojanovski L. Simeons F. R. C. Porzelle A. Grimaldi R. Wittlin S. Duffy S. Avery V. M. Meister S. Sanz L. Jiménez-Díaz B. Angulo-Barturen I. Ferrer S. Martínez M. S. Gamo F. J. Frearson J. A. Gray D. W. Fairlamb A. H. Winzeler E. A. Waterson D. Campbell S. F. Willis P. Read K. D. Gilbert I. H. J. Med. Chem. 2016;59:6101–6120. doi: 10.1021/acs.jmedchem.6b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragana B. Hallyburton I. Lee M. C. Norcross N. R. Grimaldi R. Otto T. D. Proto W. R. Blagborough A. M. Meister S. Wirjanata G. Ruecker A. Upton L. M. Abraham T. S. Almeida M. J. Pradhan A. Porzelle A. Luksch T. Martinez M. S. Luksch T. Bolscher J. M. Woodland A. Norval S. Zuccotto F. Thomas J. Simeons F. Stojanovski L. Osuna-Cabello M. Brock P. M. Churcher T. S. Sala K. A. Zakutansky S. E. Jimenez-Diaz M. B. Sanz L. M. Riley J. Basak R. Campbell M. Avery V. M. Sauerwein R. W. Dechering K. J. Noviyanti R. Campo B. Frearson J. A. Angulo-Barturen I. Ferrer-Bazaga S. Gamo F. J. Wyatt P. G. Leroy D. Siegl P. Delves M. J. Kyle D. E. Wittlin S. Marfurt J. Price R. N. Sinden R. E. Winzeler E. A. Charman S. A. Bebrevska L. Gray D. W. Campbell S. Fairlamb A. H. Willis P. A. Rayner J. C. Fidock D. A. Read K. D. Gilbert I. H. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J. Plater L. Elliott M. Shpiro N. Hastie C. J. Mclauchlan H. Klevernic I. Simon J. Arthur C. Alessi D. R. Cohen P. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenlock M. C. Potter T. Barton P. Austin R. P. J. Biomol. Screening. 2011;16:348–355. doi: 10.1177/1087057110396372. [DOI] [PubMed] [Google Scholar]
- Somnarin T. Pobsuk N. Chantakul R. Panklai T. Temkitthawon P. Hannongbua S. Chootip K. Ingkaninan K. Boonyarattanakalin K. Gleeson D. Paul Gleeson M. Bioorg. Med. Chem. 2022;76:117092. doi: 10.1016/j.bmc.2022.117092. [DOI] [PubMed] [Google Scholar]
- Wenlock M. C. Austin R. P. Potter T. Barton P. J. Lab. Autom. 2011;16:276–284. doi: 10.1016/j.jala.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Verdonk M. L. Cole J. C. Hartshorn M. J. Murray C. W. Taylor R. D. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- PyMOL, Molecular Graphics System, Version 3.1.1, Schrödinger, LLC [Google Scholar]
- ChemAxon, https://chemaxon.com/products/jchem-for-office
- Sander T. Freyss J. von Korff M. Rufener C. J. Chem. Inf. Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- S. Simca P14, https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary information is available. See DOI: https://doi.org/10.1039/D5MD00353A.
The supporting information contains additional Figures and Tables describing SAR and model statistics. Also provided are 1H-NMR, 13C-NMR, HPLC-UV chromatograms and high-resolution mass spectroscopy data.










