Abstract
Theory predicts that speciation rates should be accelerated in organisms undergoing sexual selection. In systems involving female choice, sexual selection acts directly on traits that may be important in prezygotic reproductive isolation, potentially fostering rapid divergence of such traits among allopatric populations. Despite the appeal of this concept, it has proven difficult to document. We provide genetic, behavioral, and simulation data illustrating that the striking and possibly recent divergence in traits of male behavior and morphology among populations of the jumping spider Habronattus pugillis can be attributed to sexual selection. We have found evidence for varying degrees of lower female response and offspring viability among some between-population crosses, consistent with the early stages of speciation. We have developed a gene-tree-based method for comparing phenotypic and genetic data sets to infer selection, and have found robust statistical evidence that directional selection has acted on male traits, by confirming that their rate of fixation exceeds that of neutral mitochondrial genes. Because these traits are apparent targets of female choice, the results indicate that sexual selection is driving divergence of phenotypes potentially crucial to the speciation process.
Although theory predicts that speciation rates should be accelerated in organisms undergoing sexual selection (1–5), obtaining good empirical evidence has many difficulties (reviewed in ref. 6). Ideally, evidence would show that sexual selection has caused divergence among populations, and that the rate of divergence is greater than expected under allopatric speciation without sexual selection. However, because the process of speciation occurs throughout long periods of time, rarely are we able to document ongoing selection while simultaneously observing clade diversification.
Previous studies addressing whether sexual selection has influenced rates of diversification have primarily focused on sister clade comparisons. These studies have shown correlations between species diversity and traits believed to be indicators of sexual selection (e.g., sexual dimorphism) on phylogenetic trees (7–10). This correlative phylogenetic approach possesses the virtue of taxonomic breadth, but typically lacks direct population-level evidence that the traits in question have a selective basis. Additionally, factors other than sexual selection, such as high dispersal ability and large, fragmented geographical ranges, have also been correlated with increased species richness (11).
Likewise, numerous studies have demonstrated ongoing sexual selection within populations (e.g., refs. 12–14). However, from such results alone it has been difficult to implicate sexual selection directly in increasing the divergence rate between populations, either because of the low number of populations studied or because of the lack of an historical timeframe.
Both the comparative phylogenetic approach and the selection-experiment approach are tremendously valuable, but neither alone links the pattern of clade diversification with the process of ongoing sexual selection to demonstrate that sexual selection has spurred a diversification of lineages. Here we attempt to do so, with an approach that bridges the gap between the deep time of clade diversification and the here-and-now of selection experiments, examining the speciation process nearer to the intermediate depth in time at which it actually occurs. To do this, we analyze divergence among multiple populations and test statistically whether phenotypic evolution of sexually selected traits is outpacing neutral genetic evolution.
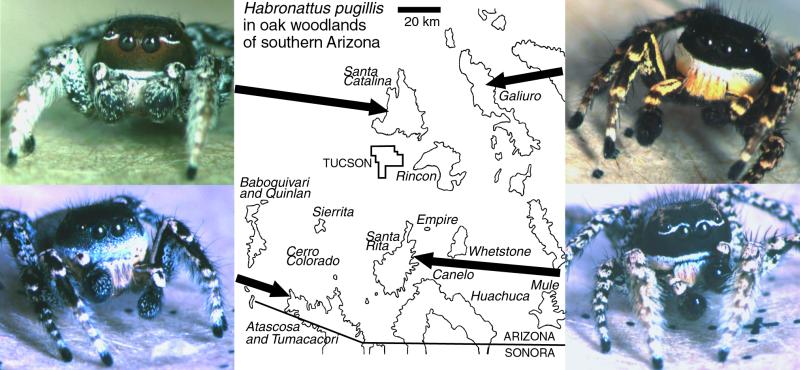
We use a system ideal for revealing the signature of sexual selection driving diversification: the many disjunct populations of the jumping spider Habronattus pugillis (Griswold), a sexually dimorphic and visually oriented spider. Allopatric populations of H. pugillis exist in woodlands on mountain ranges in southeastern Arizona (15). This region's numerous isolated mountain ranges maintain discrete patches of higher-elevation woodlands separated by low-lying areas of grassland and Sonoran Desert. Today's mountain woodlands are thought to be relicts of extensive habitats that blanketed lowland areas and provided connectivity between woodlands during the late Pleistocene or early Holocene epochs (16, 17). Despite the minimal separation of some woodland patches in space (only dozens of kilometers) and time [as recently as 10,000 to 2 million years ago (18)], populations of H. pugillis are phenotypically differentiated among the mountain ranges. The strikingly ornamented males differ among ranges in many morphological traits (Fig. 1), including shape and color of facial setae, shape of the carapace, and markings of the front appendages (15). Their courtship behaviors differ as well, with some populations including elements (palp circling, body shakes, sidling) absent from others. In contrast to males, females are relatively uniform, with simple brown, gray, and white markings in all populations. Because phenotypic differences are concentrated in male traits, particularly on body parts conspicuously displayed in courtship, and because it is difficult to explain such traits in terms of natural selection, it has been argued that these traits are sexually selected (15).
Figure 1.
Four geographic forms of H. pugillis and the mountain ranges in southern Arizona on which they occur. Mountain ranges are outlined to show the extent of oak woodland habitat according to the map of Brown and Lowe (19). H. pugillis occurs in all of the named mountain ranges. The Black Mountains (not shown) are located 30 km northwest of the Santa Catalina Mountains. Depicted here are example forms from the Santa Catalina, Galiuro, Santa Rita, and Atascosa Mountains.
In this study we develop a population genetic approach with mitochondrial gene trees to illustrate that selection, rather than genetic drift in allopatry, has acted on male traits important in promoting incipient speciation by means of sexual selection. To test whether selection has driven the divergence of these traits, we sought to determine whether patterns of phenotypic divergence in males were matched by patterns in neutrally evolving, maternally inherited genetic markers. We developed coalescent-based simulations to ask whether the rate of fixation of male traits is greater than expected, based on the rate of fixation of mitochondrial haplotypes. Sexual selection by female choice reduces the amount of time needed for fixation of male phenotypic traits, yet does not reduce the effective population size of neutral, unlinked, mitochondrial genes. Therefore, the rate of fixation of mitochondrial genes can serve as a control against which to measure the effects of sexual selection. The null expectation is that neutral nuclear genes, with their expected 4-fold larger effective population size, take longer to become fully sorted than mitochondrial genes. Here, we demonstrate statistically that in H. pugillis male phenotypic traits show a greater degree of fixation than mitochondrial haplotypes, indicating that selection has acted on the male traits. The test we have developed may be generally useful for examining selection on male phenotype when little is known about the genetic basis of the traits in question.
Finding that historical selection has accelerated the rate of divergence among populations, however, does not guarantee that it would be strong enough to maintain population discreteness, should populations come into sympatry again. Because the habitat of H. pugillis was probably fragmented and reconnected multiple times during the Pleistocene epoch (18) because of climate change, we sought to determine to what extent interbreeding might be expected should present-day populations come into secondary contact. We conducted mating trials in the laboratory with spiders from three mountain ranges, and found that varying amounts of indicators of both prezygotic and postzygotic isolation exist among certain allopatric populations, suggesting that at least some populations may continue to maintain their distinctiveness.
Materials and Methods
Relationships and Evolution of mtDNA Sequences.
We collected 78 H. pugillis from 13 mountain ranges in southeastern Arizona, as well as one Habronattus oregonensis and one Habronattus geronimoi as outgroups. We sequenced an 815-bp region of the mitochondria encoding the 5′ half of ND1 through the 3′ half of 16S as reported by Masta (18, 20). To determine whether the genes were appropriate neutral markers, we used the McDonald and Kreitman (21) test and compared the ratio of nonsynonymous-to-synonymous polymorphisms in ND1 within the 78 H. pugillis with the ratio fixed with respect to the outgroup species H. geronimoi. These two ratios should be the same if the gene is evolving in accordance with the neutral theory of Kimura (22).
To determine whether the spiders' mitochondrial genes were as distinct among ranges as male phenotype, we reconstructed gene relationships by using parsimony and maximum likelihood (ML), using PAUP* (23). Parsimonious trees were sought with heuristic searches involving 2,001 random addition sequences and tree bisection reconnection rearrangements (equal weighting, unordered states), holding a maximum of 50,000, 200, or 50 trees at each replicate. One thousand bootstrap replicates were performed. ML trees were sought by first reconstructing a single tree (assumptions: lset nst = 2, basefreq = empirical, tratio = 2, rates = equal) and with nearest-neighbor interchange swapping, by using this tree to estimate rate parameters which were then used as assumptions for five random addition sequence heuristic searches with subtree pruning regrafting rearrangements.
Statistics of Contrast Between Phenotype and Gene Tree.
We used two different approaches to compare phenotype with the gene tree. First, because male morphological characters are uniform within each mountain range, we assessed whether the sequence data reject the hypothesis that mitochondrial sequences from each range form a clade. Sequences from each mountain range were constrained to be monophyletic and the degree of support for departures from monophyly were assessed with the Templeton (24) and Kishino–Hasagawa (25) tests of tree congruence, implemented by PAUP*.
Second, we assessed whether the rate of fixation of mitochondrial sequences was different from the rate of fixation of male phenotypic characters. When two or more populations become separated and gene flow ceases, stochastic extinction of neutral parental haplotypes will occur in the daughter populations, until all but one parental type becomes extinct. The amount of time required for a neutral haplotype to become fixed in a daughter population (i.e., for lineage sorting to go to completion) depends on the effective population size (Ne). After 4Ne generations of population isolation, it is highly probable that lineage sorting of neutral nuclear haplotypes will have gone to completion and the populations will be reciprocally monophyletic (26, 27). Directional selection, however, acts to decrease the time needed for sorting, or fixation, of a gene (e.g., ref. 28). To test statistically the contrast between phenotypic and mitochondrial differentiation, we focused on samples from four ranges (Santa Rita, Huachuca, Galiuro, and Santa Catalina). We chose only four ranges because our simulation test would increase in complexity with more ranges, and we chose those particular populations because they were represented by the largest sample size (10 individuals each). They are phenotypically representative of the diversification in the sense that, like almost any other set of ranges we could have chosen, many fixed phenotypic differences exist among them (15). We used the “s” statistic (the number of parsimony steps) of Slatkin (29) calculated for these four ranges as a measure of incompleteness of sorting on the gene trees. Our test consists of three parts: (i) to determine observed s on our reconstructed mitochondrial trees, (ii) to compare this s against s values from simulated gene trees to estimate time since divergence, and (iii) to simulate nuclear gene trees in populations with the estimated divergence times to determine the probability of fixation of differences in nuclear-controlled phenotypic characters under the assumption of neutrality. We performed the test alternately by using two different assumptions about population divergence (star phylogeny vs. dichotomous branching) and two different methods to calculate s. In the first method of calculating s, incompleteness of lineage sorting among all four ranges was considered, as if a single locus had alternative alleles fixed in each of the four ranges. In the second method, incompleteness of lineage sorting in one range versus the other three was considered, as if one of the ranges, but not necessarily the other three, fixed a unique distinguishing allele.
First, to consider incompleteness of lineage sorting among all four ranges, s was calculated by assigning to each haplotype from the four focal mountain ranges a character state representing its source range. The number of parsimony steps in this character on the gene tree was calculated by using all of the trees, which yielded the observed s values. To account for errors in tree reconstruction, we conservatively chose the smallest value of s that emerged from the thousands of equally parsimonious trees (s = 6 instead of more common values up to 10).
For each set of parameters, computer simulations generated 10,000 gene trees by gene coalescence within a population tree, with Ne in each population = 500 (much lower than would be realistic, but all of our calculations are scaled by Ne). Branch lengths (generations since isolation) of the population tree were estimated, conservatively, as the greatest time that would yield at least a 5% probability of producing a gene tree with an s as high or higher than that observed (the longer the time, the lower the expected s). Then, to consider nuclear genes with four times the population size of mitochondrial genes, the lengths of branches in the population tree were divided by four (equivalent to multiplying the population size by 4), and simulations run to estimate the probability of complete sorting. Complete sorting among the four populations (i.e., every population fixed for a different allele) is equivalent to an s value of 3. Use of the upper 95% confidence limit for divergence time is conservative for our test, because the greater the time the more likely complete sorting will occur in the simulations for the nuclear genes.
All computer simulations were performed with the programming system MESQUITE (W.P.M. and D. R. Maddison, see http://mesquiteproject.org). Computer files detailing the parameters and allowing reproduction of the simulations are available at http://mesquiteproject.org/MastaMaddison/simulations.html.
Intermountain Matings.
To determine a proxy for the extent of reproductive isolation among mountain populations, we focused on populations from three mountain ranges whose males exhibit very different ornaments and courtship behaviors (Galiuro, Santa Rita, and Sierrita). For between-range mating trials, virgin female spiders were lab-reared or collected as immatures, and raised in individual vials until they molted to sexual maturity. Some inseminated adult females were also collected and maintained in the laboratory for obtaining comparative data on within-range fecundity. Males were either laboratory-reared to maturity, or collected as adults. To examine the degree of prezygotic isolation among spiders from different ranges, we studied female responses. We used latency time (time from the initiation of courtship by a male to the time of female acceptance and mating) in single male–female interactions as a measure of female response. We did not use simultaneous choice experiments with multiple caged males or video images (30) for two reasons. First, no known acceptance behavior occurs in females other than copulation (J. Withgott, W.P.M., and S.E.M., personal observation). Second, one-on-one interactions are probably more representative of natural conditions, for the spiders' low densities in the wild make it unlikely that a female would be exposed to two courting males at a time. Latency is expected to correlate with acceptance in the wild, because courtship takes place in the open, and the risk of predation may well limit the time a female can safely spend assessing a mate. As measures of postzygotic isolation we scored the number of viable offspring (from parents that produced eggs) resulting from within and between-range matings.
In all trials, one female and one male were placed together in a 15-cm arena and allowed to court until mating occurred, or were removed after 10–15 min if males refused to court. Inseminated females were maintained in 7-dram vials, each containing an oak leaf to which egg cases could be attached, and kept on a 12/12 h light/dark cycle. Spiderlings were removed from the mother's vial as soon as they emerged from egg cases. Remaining egg sacs were dissected, and spiderling molts, hatched eggs, and unhatched eggs were counted.
Because data on behavior and fecundity did not conform to parametric assumptions, nonparametric tests (Wilcoxon tests and Kruskal–Wallis one-way analyses of variance by ranks) were used to determine whether populations or crosses differed significantly from one another.
Results
Gene sequences for the 815-bp region of the 5′ half of ND1, tRNALeu(CUN), and 3′ 16S used in this study have been deposited in GenBank (accession nos. AF239933-AF239941, AF239948, AF255805-AF255844, and AF255853-AF255882). The ND1 sequences contained 5 synonymous to 64 nonsynonymous polymorphisms within H. pugillis versus 3 synonymous to 18 nonsynonymous fixed differences with H. geronimoi (not significantly different; P = 0.383 Fisher's exact test). Therefore, the McDonald and Kreitman test results are consistent with neutral evolution of the ND1 gene, and therefore presumably the adjacent, linked 16S gene.
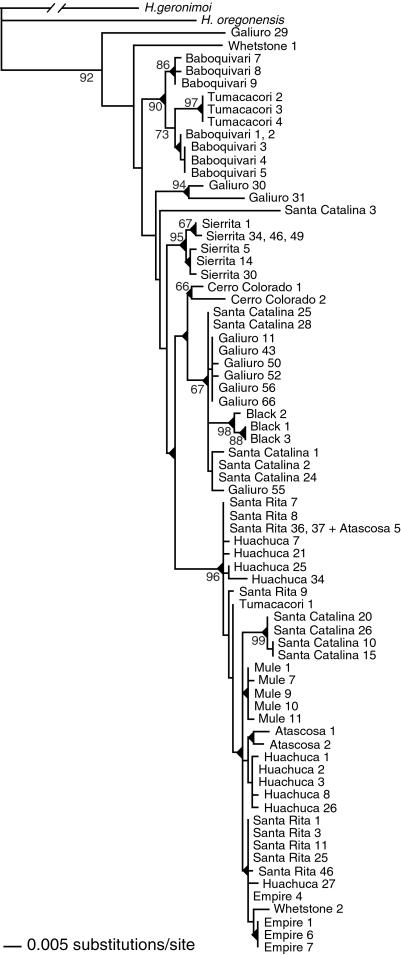
All phylogenetic analyses yielded estimates of relationships that were substantially concordant with one another. The ML tree (Fig. 2) shows some concordance with geography and therefore male phenotype: four clades each contain all and only individuals collected from a single mountain range; for three of these, bootstrap support is greater than 65%. Sequences from the remaining nine ranges do not form distinct monophyletic groups. In particular, sequences from the Santa Catalina and Galiuro Mountains show deep within-population divergences on the gene tree, whereas some sequences from the two ranges are very closely related. Both tree-congruence tests suggest that the lack of concordance of the gene tree with male phenotype likely reflects sequence relationships accurately. Trees generated under the constraint of geographic monophyly were 63 steps longer than the unconstrained tree, significantly longer by both the Templeton (ref. 24; P < 0.0001) and Kishino–Hasagawa (ref. 25; P < 0.0001) tests.
Figure 2.
Gene tree for mitochondrial haplotypes of H. pugillis from 13 mountain ranges in Arizona. Shown is one of several equally likely trees from ML analysis. Bootstrap values (from parsimony analysis) are given next to nodes; triangles at the bases of clades indicate the clades also were found in all parsimony searches.
In contrast to the sequences of the gene tree, male phenotypic traits are almost completely sorted, i.e., males within each range are generally phenotypically uniform. Maddison and McMahon (15) found that nine of these mountain populations were fixed in all of the 22 phenotypic traits scored, two ranges fixed in 21 traits, and one range fixed in 20 traits. Furthermore, differences between ranges were striking; even the genetically closely related (see Fig. 2) Santa Catalina and Galiuro individuals showed fixed differences in at least 11 of 22 phenotypic characters scored. Not only are male traits fully sorted on most ranges, but some are unique to a single mountain range, suggesting they evolved since isolation of the populations. Such autapomorphies include palp-circling during courtship in Santa Rita Mountain males, and the prominent scale-covered cheek patch in Galiuro Mountain males.
The gene-tree simulations gave very low (P < 0.001–0.02) probabilities that neutral genes would show fixed differences among populations. The minimum conceivable s value for four populations is 3, which is equivalent to every population being fixed for a different allele. The observed s contrasting four populations was 6–10, yielding an upper 95% confidence limit on branch lengths of 1.39N generations for the star phylogeny, and 0.98N for the dichotomous phylogeny. When the branch lengths were reduced to 0.3475N and 0.245N generations, respectively, to mimic nuclear genes, an s as low as 4 occurred rarely (<10 of the 10,000 replicates) whereas complete fixation (s = 3) occurred in none of simulated gene trees (yielding estimates of P < 0.001). Thus, assuming the fixed phenotypic differences among populations reflect fixed differences in underlying nuclear genes, this phenotypic differentiation is highly unlikely to have arisen under neutrality. Selection is implied.
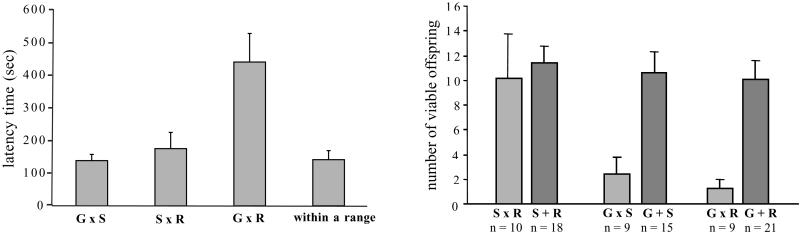
In the behavioral trials, latency to mating was significantly longer in crosses between individuals from the Galiuro and Santa Rita Mountains than in either of the other two crosses (Fig. 3 Left).
Figure 3.
(Left) Means ± SE latency times to mating for each of three between-range crosses tested in mating trials. G = Galiuro; S = Sierrita; R = Santa Rita. Galiuro × Santa Rita crosses showed significantly longer latencies to mating than did either of the other crosses (χ2 = 9.35, df = 2, P = 0.0093; Kruskal–Wallis test with Tukey–Kramer test for pair comparisons). Sample sizes are: n = 15 Galiuro × Sierrita (14 Gf, 1 Sf); n = 14 Sierrita × Santa Rita (5 Sf, 9 Rf); n = 15 Galiuro × Santa Rita (10 Rf, 5 Gf); n = 5 pooled within ranges (G = 1, S = 1, R = 3). (Right) Means ± SE number of viable offspring for each of three between-range crosses and three pooled combinations of within-range matings. Two results are significant: (i) Sierrita × Santa Rita crosses produced significantly more offspring than Galiuro × Santa Rita crosses (χ2 = 7.50, df = 2, P = 0.0235; Kruskal–Wallis test with Tukey–Kramer test for pair comparisons); and (ii) each of the two between-range crosses involving Galiuro individuals produced significantly fewer offspring than pooled means of within-range matings of spiders from the two ranges involved in each between-range cross. That is, Sierrita × Galiuro crosses were less fecund than within-range Sierrita matings plus within-range Galiuro matings (S = 62.5, z = 2.99, P = 0.0028); whereas Galiuro × Santa Rita crosses were less fecund than within-range Galiuro matings plus within-range Santa Rita matings (S = 57, z = 3.74, P = 0 0002).
Analysis of the number of viable offspring (from parents that produced eggs) indicates that postzygotic isolation exists between some but not all crosses. Crosses between Sierrita and Santa Rita individuals produced significantly more offspring than crosses between Galiuro and Santa Rita individuals (P = 0.0235; Fig. 3 Right). Fecundity differences were not due to the time pairs spent mating, because time spent mating did not differ among crosses (means = 82, 80, 77 s; P = 0.9456). In addition, crosses involving Galiuro individuals produced significantly fewer viable offspring than within-range matings (Fig. 3 Right). This reduction in number of offspring was driven by females: Galiuro females crossed with males from either the Sierrita or Santa Rita ranges showed a significant decrease in number of offspring (P = 0.0091), whereas between-range matings performed with Galiuro males showed no such decrease (P = 0.9761). Males did influence latency times, however, because females from other ranges took significantly longer to mate when presented with Galiuro males (P = 0.0136). Thus, among these ranges, male identity influences the degree of prezygotic isolation, as measured by latency time, whereas female identity influences the degree of postzygotic isolation, as measured by number of viable offspring.
Discussion
We looked back in time by using a coalescent approach to demonstrate that sexual selection drove diversification of H. pugillis populations. Such an approach may be generally useful for recently diverged populations in which one may suspect sexual selection is causing male trait evolution to outpace that of neutral genes. We found that males of H. pugillis on the mountain ranges of southeastern Arizona are phenotypically highly differentiated among populations, whereas their mitochondrial genes are not. The lack of differentiation of female-inherited mitochondrial genes is concordant with females' lack of morphological differentiation among populations. This indicates that directional selection has acted on male but not female morphology. We also found varying degrees of prezygotic and postzygotic isolation; looking forward in time, this suggests some populations will likely complete the process of speciation.
Our coalescent test assumes an ancestral population that was polymorphic for different male traits, and that polyphyly of mitochondrial sequences from a given mountain range is caused by retention of ancestral polymorphisms present before spider populations became separated. However, it is likely that many male traits arose after population isolation, because many are unique to different mountain populations. Our observation of genealogical diversity of sequences from the same population together with low divergence among some sequences from different populations is consistent with retention of ancestral haplotypes present before the populations became separated. Although polyphyly could also result from migration between mountain ranges, Masta (18) provided reasons supporting incomplete lineage sorting over migration as the major cause of polyphyly in this system. Nonetheless, Maddison and McMahon (15) found suggestions of introgression of some male traits among some mountain ranges. However, even if migration has occurred among ranges, it does not weaken our argument regarding selection. If anything, it would indicate that selection has been strong, because any migrant phenotypes must either have been overwhelmed by local selection, or quickly proceeded to fixation, to yield the observed within-range phenotypic uniformity. Male traits controlled on a Y chromosome could remain distinct in the face of female-biased migration, but H. pugillis has XXO sex chromosomes (W.P.M., unpublished data), and thus no Y chromosome.
Our statistical test can be used only on populations that are not monophyletic for their mitochondrial haplotypes, and have at least one phenotypic character fixed. These criteria may often be met by populations in the early stages of divergence. Our test may be used when little is known about the heritability of traits, and when it is therefore not possible to use a more conventional test (e.g., ref. 31) of neutral phenotypic evolution. Furthermore, our test has the added advantage of requiring no information on the absolute time since population divergence, because it uses the relative amount of divergence between mitochondrial and nuclear-encoded phenotypic traits.
Our test could be compromised if the genetic basis of the phenotypic traits is complex such that fixation of alleles is not needed to effect a fixed phenotypic difference (e.g., polygenic traits with thresholds). However, unless we have severely underestimated the probability of fixed phenotypic differences under neutrality, the fact that multiple traits show fixed differences makes a compelling argument for selection. Our statistical test analyzed the probability of only a single nuclear-encoded trait being fixed. In fact, up to 22 traits of body form, pelage, color, and behavior show fixed differences, and unless they are all pleiotropic effects of a single gene, the probabilities for fixation of their multiple underlying genes would compound. The probability of fixation of all of these genes by chance would be very small indeed.
The fact that female response and hybrid viability are lowered to varying degrees among some crosses indicates that at least some populations will remain distinct if climatic change reconnects the woodland habitats and drives spider populations into sympatry again. Mating trials between Galiuro and Santa Rita individuals showed high latency times to acceptance, and very low viability of offspring. Postzygotic isolation may have arisen as a consequence of genetic divergence in allopatry between individuals from these ranges, which resulted in subsequent genetic incompatibilities. Alternatively, the reduction in the number of viable offspring in these crosses may not be true postzygotic isolation, but rather a result of the female choosing not to use the sperm she obtained during copulation. The data hints at the latter explanation, because only when females were from the Galiuro Mountains was the number of offspring in crosses involving Galiuro individuals significantly reduced. In either case we can conclude that, at least between these populations, the process of speciation by means of sexual selection in allopatry is well underway.
However, females will mate readily with males from at least some other ranges. Thus, male trait divergence is not necessarily coupled with divergence in female response as measured by latency time. Such a lack of coupling may be expected during the early stages of sexual selection-induced differentiation either because female preferences would evolve more slowly (32) or because male ornaments in some populations could evolve to exploit preexisting female sensory biases (33, 34) that would be retained by females in other populations.
It has been suggested that divergence by sexual selection may also be driven by environmental or ecological differences among allopatric populations when the selective context of a trait is changed (32, 35). Although this possibility cannot be ruled out absolutely in our system, we feel that the geographical proximity of H. pugillis populations and the resulting similarity of habitats of these populations greatly minimize such differences. Our interpretation is consistent with other observations that sexual selection may cause phenotypic divergence without necessarily causing ecological divergence (36). Finding a marked difference in the rate of evolution of phenotypic male traits versus maternally inherited neutral genes in such a geographically focused system of allopatric populations in similar environments further underscores the power of sexual selection as a potent force for evolutionary diversification.
Acknowledgments
We thank G. Bien-Willner and J. Borgmeyer for assistance with spider maintenance and mating trials, and R. Hudson, L. Knowles, T. Markow, N. Moran, M. Nachman, and J. Withgott for discussion and support. This work was supported by a Flinn Foundation Genetics grant and a National Science Foundation Research Training Grant in Biological Diversification at the University of Arizona (to S.E.M.), National Science Foundation Doctoral Dissertation Improvement Grant DEB9520668 (to S.E.M. and W.P.M.), and a David and Lucile Packard Fellowship (to W.P.M.).
Footnotes
References
- 1.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Clarendon Press; 1958. [Google Scholar]
- 2.Kirkpatrick M. Evolution (Lawrence, Kans) 1982;36:1–12. doi: 10.1111/j.1558-5646.1982.tb05003.x. [DOI] [PubMed] [Google Scholar]
- 3.Lande R. Proc Natl Acad Sci USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lande R, Arnold S J. J Theor Biol. 1985;117:651–664. doi: 10.1016/s0022-5193(85)80245-9. [DOI] [PubMed] [Google Scholar]
- 5.West-Eberhard M J. Q Rev Biol. 1983;58:155–183. [Google Scholar]
- 6.Panhuis T M, Butlin R, Zuk M, Tregenza T. Trends Ecol Evol. 2001;16:364–371. doi: 10.1016/s0169-5347(01)02160-7. [DOI] [PubMed] [Google Scholar]
- 7.Arnqvist G, Edvardsson M, Friberg U, Nilsson T. Proc Natl Acad Sci USA. 2000;97:10460–10464. doi: 10.1073/pnas.97.19.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barraclough T G, Harvey P H, Nee S. Proc R Soc London Ser B. 1995;259:211–215. [Google Scholar]
- 9.Mitra S, Landel H, Pruett-Jones S. Auk. 1996;113:544–551. [Google Scholar]
- 10.Møller A P, Cuervo J J. Evolution (Lawrence, Kans) 1998;52:859–869. [Google Scholar]
- 11.Owens I P F, Bennett P M, Harvey P H. Proc R Soc London Ser B. 1999;266:933–939. [Google Scholar]
- 12.Endler J A, Houde A E. Evolution (Lawrence, Kans) 1995;49:456–468. doi: 10.1111/j.1558-5646.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- 13.Uy J A C, Borgia G. Evolution (Lawrence, Kans) 2000;54:273–278. doi: 10.1111/j.0014-3820.2000.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu C-I, Hollocher H, Begun D J, Aquadro C F, Xu Y, Wu M-L. Proc Natl Acad Sci USA. 1995;92:2519–2523. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddison W, McMahon M. Syst Biol. 2000;49:400–421. doi: 10.1080/10635159950127312. [DOI] [PubMed] [Google Scholar]
- 16.Betancourt J L, Devender T R V, Martin P S. Packrat Middens: The Last 40,000 Years of Biotic Change. Tucson, AZ: Univ. of Arizona Press; 1990. [DOI] [PubMed] [Google Scholar]
- 17.Van Devender T R, Spaulding W G. Science. 1979;204:701–710. doi: 10.1126/science.204.4394.701. [DOI] [PubMed] [Google Scholar]
- 18.Masta S E. Evolution (Lawrence, Kans) 2000;54:1699–1711. doi: 10.1111/j.0014-3820.2000.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 19.Brown D E, Lowe C H. Biotic Communities of the Southwest. Salt Lake City: Univ. of Utah Press; 1994. [Google Scholar]
- 20.Masta S E. Mol Biol Evol. 2000;17:1091–1100. doi: 10.1093/oxfordjournals.molbev.a026390. [DOI] [PubMed] [Google Scholar]
- 21.McDonald J H, Kreitman M. Nature (London) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 23.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1998. , Version 4.0. [Google Scholar]
- 24.Templeton A. Evolution (Lawrence, Kans) 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 25.Kishino H, Hasagawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 26.Neigel J E, Avise J C. In: Evolutionary Processes and Theory. Karlin S, Nevo E, editors. New York: Academic; 1986. pp. 515–534. [Google Scholar]
- 27.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson R R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 29.Slatkin M. Genetics. 1989;121:609–612. doi: 10.1093/genetics/121.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark D L, Uetz G W. Anim Behav. 1990;40:884–890. [Google Scholar]
- 31.Lande R. Evolution (Lawrence, Kans) 1976;30:314–334. doi: 10.1111/j.1558-5646.1976.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 32.Schluter D, Price T. Proc R Soc London Ser B. 1993;253:117–122. doi: 10.1098/rspb.1993.0089. [DOI] [PubMed] [Google Scholar]
- 33.Ryan M. Oxford Surv Evol Biol. 1990;7:156–195. [Google Scholar]
- 34.Ryan M J, Rand A S. Philos Trans R Soc London B. 1993;340:187–195. [Google Scholar]
- 35.Endler J A. In: Speciation and Its Consequences. Otte D, Endler J A, editors. Sunderland, MA: Sinauer; 1989. pp. 625–648. [Google Scholar]
- 36.Price T. Philos Trans R Soc London B. 1998;353:251–260. [Google Scholar]