Abstract
Buchnera is a mutualistic intracellular symbiont of aphids. Their association began about 200 million years ago, with host and symbiont lineages evolving in parallel since that time. During this coevolutionary process, Buchnera has experienced a dramatic decrease of genome size, retaining only essential genes for its specialized lifestyle. Previous studies reported that genome size in Buchnera spp. is very uniform, suggesting that genome shrinkage occurred early in evolution, and that modern lineages retain the genome size of a common ancestor. Our physical mapping of Buchnera genomes obtained from five aphid lineages shows that the genome size is not conserved among them, but has been reduced down to 450 kb in some species. Here we show evidence of six species with a genome size smaller than Mycoplasma genitalium, the smallest bacterial genome reported thus far (580 kb). Our findings strongly suggest that the Buchnera genome is still experiencing a reductive process toward a minimum set of genes necessary for its symbiotic lifestyle.
Molecular characterization of various microbial genomes has revealed that many pathogenic and mutualistic intracellular bacterial species have smaller genomes than their free-living relatives (1, 2). The reduction of genome size is associated with the loss of a great number of genes, as an adaptation to their life conditions, in which many molecules can be obtained from the host. The smallest bacterial genome reported thus far corresponds to Mycoplasma genitalium, an intracellular parasite of epithelial cells, and comprises a circular chromosome of 580 kb with only 470 coding genes (3). The recently sequenced genome of Buchnera sp. APS, primary (P) endosymbiont of the aphid Acyrthosiphon pisum, is also extremely reduced (one circular chromosome of 641 kb, with only 564 coding genes, plus two small plasmids) (4). The concept of the “minimal genome” as the minimum number of genes necessary to support cellular life is appealing, and several attempts have been made to define it by comparing the genomes of intracellular bacterial pathogens (5). Intracellular symbionts may be of great interest in this kind of study because they do not need to maintain genes that pathogens require for survival and to evade host detection. In fact, in our revision of the sequence of Buchnera sp. APS (4) we found that it has only five (0.9%) genes without an homologue in Escherichia coli.
Buchnera is a γ proteobacterium that maintains a mutualistic endosymbiotic association with aphids (6, 7). The association is obligate for both partners: Buchnera cannot be cultured outside the aphid host, whereas aphids need the bacteria for normal growth and reproduction because they provide the host with the nutrients (mainly essential amino acids) that are in short supply in its strict phloem diet (8, 9). Buchnera is confined within specialized cells called bacteriocytes and is maternally transmitted. Besides Buchnera, aphids often harbor additional bacteria that are commonly referred as secondary (S) endosymbionts. These bacteria can be found in the aphid guts, in tissues surrounding the P bacteriocytes (or even invading the P bacteriocytes themselves), and in specialized S bacteriocytes (10–12). These endosymbionts are also subject to vertical transmission (13), but their patchy distribution among aphid populations implies that they, unlike Buchnera, also undergo horizontal transmission (10). A recent study has demonstrated they may have positive effects in host fitness (14). It is conceivable that S-endosymbionts may interact and modify the established mutualism between the aphid and Buchnera.
Phylogenetic studies have proven that the symbiosis between Buchnera and its host resulted from a single bacterial infection of the common ancestor to all extant aphids about 200 million years ago (15), leading to the cospeciation of the host and their symbionts. During this coevolutionary process, Buchnera suffered considerable genomic changes (i.e., a great reduction in genome size, an increased A+T bias, great accumulation of deleterious mutations, and the amplification of genes involved in amino acid biosynthesis) (16–18). Because pathogenic intracellular bacteria with small genomes display a wide variation in chromosome length, it would also be expected that Buchnera of different aphid lineages differ in genome size and gene content.
Here we present the physical mapping of nine Buchnera genomes obtained from five aphid subfamilies. We show that the genome size is not conserved in the different Buchnera analyzed, but rather it is still undergoing a reductive process. We present six species containing the smallest known bacterial genomes.
Materials and Methods
Aphid Material and Buchnera.
Buchnera spp. were obtained from five of the main aphid subfamilies (Aphidinae, Pemphiginae, Thelaxinae, Chaitophorinae, and Lachninae) (19) (Table 1). Because Buchnera cannot be cultured in the laboratory, aphids were collected from natural populations at their maximum expansion time. Only limited amounts of Tetraneura caerulescens, Thelaxes suberi, and Chaitophorus populeti could be collected. Acyrthosiphum pisum and Cinara tujafilina are being kept at 25°C with controlled humidity and photoperiod, on young broad bean and tuja plants, respectively.
Table 1.
Species of aphids (family Aphididae) collected for this study
| Subfamily | Tribe | Genus | Species | Code* | Location and date |
|---|---|---|---|---|---|
| Aphidinae | Macrosiphini | Acyrthosiphum | A. pisum | APS | Lyon, France, September 1998† |
| Macrosiphum | M. rosae | MRO | Godella, Spain, April 2001 | ||
| Pemphiginae | Fordini | Baizongia | B. pistaciae | BPI | Tuéjar, Spain, September 1999 |
| Eriosomatini | Tetraneura | T. caerulescens | TCA | Bugarra, Spain, April 2000 | |
| Thelaxinae | Thelaxes | Th. suberi | THS | Teruel, Spain, June 2001 | |
| Chaitophorinae | Chaitophorini | Chaitophorus | Ch. populeti | CHP | Benifaió, Spain, April 2000 |
| Lachninae | Cinarini | Cinara | C. (cinara) cedri | CCE | Llíria, Spain, May 2001 |
| C. (cupressobium) cupressi | CCU | Godella, Spain, November 2000 | |||
| C. (cupressobium) tujafilina | CTU | Almussafes, Spain, April 2001† |
Abbreviation used to identify the Buchnera from the different aphids.
Aphid populations that are been maintained in the laboratory.
Isolation of Buchnera from Aphids.
The bacteriomes containing the endosymbiont bacteria were purified from different species of aphid by an adaptation of the procedure described by Harrison et al. (20). The aphids were lightly crushed on 50 ml of isolation buffer A (21), and the homogenate was successively filtered through two layers of muslin and nylon filters with a pore size from 80 to 11 μm, to remove insect debris. The filtrate was centrifuged at 1,500 × g (or 4,000 × g for Cinara spp. and T. suberi) for 15 min at 4°C. The pellet was resuspended on 1 ml of buffer A and treated with DNase I (1 mg/ml) at 4°C for 1 h to eliminate the remaining aphid DNA. Finally, the bacteriomes were washed, collected by centrifugation as mentioned above, and resuspended in 1 ml of buffer A per 1 g of aphids used in the extraction.
Determination of Buchnera spp. Genome Size by Pulse-Field Gel Electrophoresis (PFGE).
Agarose plugs containing Buchnera genomic DNA were obtained as described (22, 23). To eliminate the remaining contaminants, the plugs were subjected to an initial PFGE with a gradient of 4.5 V/cm and pulse-time ramping from 60–120 s over 24 h in a CHEF-DRII device (BioRad). Subsequently, the plugs were removed from the wells, and the intact chromosomal DNA was digested with 40 units of the selected restriction enzymes (KspI, ApaI, and RsrII) during 16 h, following the provider's recommendations. To facilitate the penetration of the enzyme in the agarose plugs, they were first equilibrated with the corresponding restriction buffer without Mg2+, then the enzyme was added, and the plugs were kept at 4°C for 1 h before adding the Mg2+ (H. Ochman, personal communication). The enzyme-digested DNA was separated by PFGE with a gradient of 4.5 V/cm and various ramping pulse times depending on the size of fragments to be resolved (pulse-time ramping from 10–90 s during 24 h for fragments of 700–50 kb, and from 1–20 s during 15 h for fragments of 200–2 kb). Phage λ multimeric DNA and Saccharomyces cerevisiae chromosomes (BioRad, Boehringer Mannheim, and New England Biolabs) were used as molecular weight markers.
Phylogenetic Analysis of Buchnera spp.
The following 16S rDNA nucleotide sequences used for the phylogenetic analysis were obtained from the GenBank/EMBL database (accession nos. in brackets): Buchnera sp. APS (M27039), Buchnera sp. BPI (AJ296752), Buchnera sp. TCA (AJ296749), Buchnera sp. THS (AJ296757), Buchnera sp. CHV (M63252), Vibrio cholerae (X74695), E. coli (AE000460), Salmonella enterica (X80681), and Wigglesworthia glossinidia (AF022879). The 16S rDNA sequence of Buchnera CTU was cloned for this study, as described (24), and subsequently sequenced.
A phylogenetic reconstruction by maximum likelihood was performed with the program treepuzzle Version 5.0. The quartet puzzling method (25), implemented in this program, was used to obtain the support for each internal branch. Values smaller than 70 were removed.
Results and Discussion
To determine the genome size of the Buchnera genus, we analyzed the chromosomes of Buchnera obtained from five of the main aphid subfamilies (Table 1) by PFGE. The phylogenetic relationship among these lineages was previously determined, based on the analysis of the 16S rDNA of Buchnera from at least one aphid species of each subfamily under study. Some other γ proteobacteria were also included as outgroup species in the analysis: the free-living bacteria V. cholerae, E. coli, and S. enterica, and the primary endosymbiont of tsetse flies, W. glossinidia (Fig. 1).
Figure 1.
Phylogenetic relationships of Buchnera species based on their rDNA 16S nucleotide sequences. Numbers on the tree indicate the support values for each internal branch. Values on the right, after the species names, indicate the genome sizes reported elsewhere. The genome size estimated in this work by PFGE appears in brackets. CHV, Chaitophorus viminalis. *, Size estimated for the genome of Buchnera CHP.
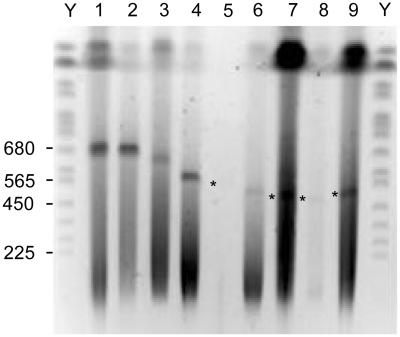
Agarose plugs containing bacterial DNA were obtained as described (see Materials and Methods). Initially, the undigested plugs were run on a PFGE gel to eliminate all of the remaining reagent impurities, contaminant aphid DNA, and broken bacterial chromosomes, which enter the gel while the intact circular bacterial chromosomal DNA remains in the plugs (26). When these gels were stained with ethidium bromide, we detected a consistent band that is characteristic of each analyzed Buchnera (Fig. 2). We confirmed by Southern blotting, using purified Buchnera sp. MRO DNA as a probe, that the detected bands correspond to Buchnera DNA (data not shown). These bands are likely to be nicked circular chromosomal DNA that has entered the gel (27). This DNA runs close to the estimated linear size, as has been proven for the known genome of Buchnera sp. APS (4).
Figure 2.
PFGE gel of undigested agarose plugs containing Buchnera genomic DNA. Lane 1, APS; lane 2, MRO; lane 3, BPI; lane 4, TCA; lane 5, THS; lane 6, CPH; lane 7, CCE; lane 8, CCU; lane 9, CTU; lane Y, yeast chromosome molecular weight marker. Some selected DNA sizes of the standards (kb) are indicated. Faint bands over the background are indicated by an asterisk.
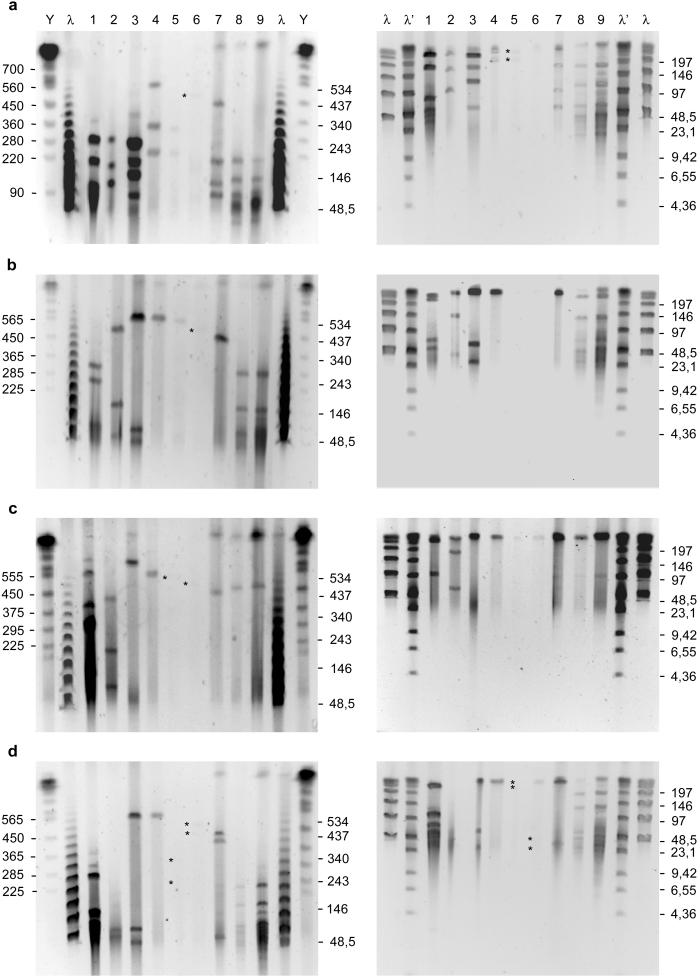
The agarose-included Buchnera DNA was then screened for cleavage with different restriction enzymes. Because of the low G+C content (30%) of the Buchnera DNA, we selected the restriction enzymes KspI, ApaI, and RsrII, which have recognition sequences rich in G and C, for physical mapping of the genomes. The restriction fragments obtained were analyzed by PFGE at different pulse times to achieve better resolution of the desired size ranges (Fig. 3). The average chromosome size was determined based on the sum of the size of the restriction fragments. We verified that the sum of the fragments corresponds with the size of the consistent band that appears on the PFGE gels without restriction digestion. The results obtained are summarized in Table 2.
Figure 3.
PFGE separation of digested genomic DNA of Buchnera under two different conditions to resolve fragments from 700–50 kb (Left) and from 200–2 kb (Right). (a) KspI digestion. (b) ApaI digestion. (c) RsrII digestion. (d) ApaI plus RsrII digestion. Lane 1, APS; lane 2, MRO; lane 3, BPI; lane 4, TCA; lane 5, THS; lane 6, CPH; lane 7, CCE; lane 8, CCU; lane 9, CTU; lane Y, yeast chromosome molecular weight markers; (λ, λ′) λ molecular weight markers. Some selected DNA sizes of the standards (kb) are indicated. Faint bands over the background are indicated by an asterisk.
Table 2.
Sizes of DNA fragments produced by digestion of Buchnera genomes with selected restriction enzymes
| Buchnera spp. | Size of DNA fragments produced by digestion with restriction enzyme(s), kb
|
Total estimated DNA length, kb
|
||||
|---|---|---|---|---|---|---|
| KspI | ApaI | RsrII | ApaI + RsrII | Sum of restriction fragments, mean ± SD | Undigested gel | |
| APS* | 252, 176, 79, 52, 48, 34 | 286, 226, 73, 52, 3.4 | 277, 264, 99 | 240, 104, 99, 73, 52, 45, 24, 3.4 | ||
| APS | 255, 176, 85, 60, 48, 40 | 290, 235, 75, 52, 3.6 | 280, 280, 100 | 240, 108, 108, 75, 52, 45, 23, 3.6 | 659 ± 4 | 670 |
| MRO | 275, 170, 110, 110 | 475, 150, 46 | 420, 185, 65 | 669 ± 3 | 670 | |
| BPI | 560, 62, 30 | 550, 64, 30 | 640 | 550, 64, 30 | 645 ± 5 | 640 |
| TCA | 335, 225 | 565 | 570 | 565 | 565 ± 4 | 565 |
| THS | 315, 215 | 545 | 545 | 320, 234 | 544 ± 10 | 550 |
| CHP | 490 | 495, 25 | 515 | 435, 48, 25 | 508 ± 13 | 520 |
| CCE | 195, 115, 70, 70 | 440 | 450 | 405, 46 | 448 ± 5 | 450 |
| CCU | 185, 120, 70, 50, 24, 24 | 265, 135, 50, 25 | 475 | 200, 136, 64, 50, 30 | 476 ± 3 | 475 |
| CTU | 185, 120, 70, 50, 24, 24 | 265, 135, 50, 25 | 480 | 200, 136, 64, 50, 30 | 477 ± 4 | 475 |
Results deduced from the sequenced Buchnera sp. APS genome (4).
In addition to the main chromosome, several lineages of Buchnera also contain small plasmids that are lost during the preliminary treatment of the agarose plugs. Leucine plasmids are present in all of the Buchnera analyzed in this study, except for Buchnera sp. CHP (18, 24, 28), and they have a size ranging from 6.3–8.2 kb. Some species also contain a cryptic leucine plasmid of 1.7–2.4 kb (24, 28). Buchnera from the Aphidinae and some tribes of the Pemphiginae subfamily also contain tryptophan plasmids of variable size, from 3.0–12.8 kb (17, 29, 30). Thus, the overall size of the Buchnera genome will not be significantly affected by the presence of such plasmids.
Great variation was found in Buchnera genome size (from 670–450 kb of average estimated size), which contrasts with what has been reported from the analysis of three species from aphids of the subfamily Aphidinae and one from the subfamily Pemphiginae (21). In that previous study, a slightly smaller size for the latter was also reported. This observation is consistent with our results for these two subfamilies. However, the analysis of species from different clades shows that the genome reduction is more dramatic for the Buchnera present in aphids from the subfamilies Chaitophorinae, Thelaxinae, and Lachninae. Although it is reasonable to assume that the vast majority of genome shrinkage may have occurred in the common Buchnera ancestor, the differences found in genome size suggest that Buchnera from different aphid lineages are still undergoing a reductive process to the minimal genome required for their survival in the aphid host. This fact is supported by the recent reduction in 25 kb of the genome size of Buchnera sp. CCE, revealed by the comparison with its close relatives from Cinara cupressi and C. tujafilina.
Buchnera species from aphids of the genus Cinara (subfamily Lachninae) show the smallest genomes. Actually, they have the smallest known genomes. The gel lines corresponding to these species consistently showed an intense smear background on the undigested gels, as well as some diffuse bands on the digested gels. The presence of abundant secondary endosymbionts that we were unable to completely eliminate from the bacteriome preparations was confirmed by a diagnostic PCR based on the amplification of 16S and 23S rRNA-encoding DNA (rDNA), and subsequent diagnostic restriction digestion (31). Almost 50% of the amplified 16S rDNA from bacteriome DNA isolated from C. tujafilina corresponded to R-type secondary endosymbiont (data not shown), and it is probably this secondary endosymbiont DNA that appears on the gels. Nevertheless, the bands corresponding to Buchnera DNA could easily be identified by their greater intensity over the background banding.
The number of genes that are present in the Buchnera spp. choromosomes can be estimated based on the fact that the known sequence of the Buchnera sp. APS chromosome contains 564 protein coding genes in 641 kb. This finding means that the genome of Buchnera sp. CCE, the smallest genome found in this study, should contain about 396 protein coding genes, which is still higher than the minimal genome estimated both from computational analysis (327–256 genes or even less, refs. 32–34) and transposon mutagenesis (350–265 genes, ref. 35). However, it must be considered that the minimal genome is a hypothetical situation that is not easily attainable in a natural environment. Nevertheless, the decrease in the genome size indicates that 395 genes can sustain independent cellular life, which is the minimum set reported so far.
In conclusion, the analysis of Buchnera from five aphid subfamilies shows that the bacterial genome is still undergoing a reductive process (16). Six of the analyzed species contain the smallest bacterial genomes ever described. However, the abundance of the secondary endosymbiont may reflect the inability of Buchnera with such small genomes to compliment properly the functions needed for their symbiotic lifestyle (14). The S symbiont could be either taking over the role of Buchnera in supplying some of the functions needed for aphid fitness, or supporting Buchnera's own cellular functions. The sequencing of these smaller genomes will give new clues about the lost genes that are essential for bacterial growth, in an attempt to define a minimal genome.
Acknowledgments
We are grateful to H. Ochman for communicating the procedure for enzyme digestion of the agarose-included chromosomal DNA. We also thank A. M. Powell for critical reading of the manuscript. This work was supported by Grant BMF2000-1383 from the Ministerio de Ciencia y Tecnología, Spain.
Abbreviation
- PFGE
pulse-field gel electrophoresis
Footnotes
Data deposition: The sequence of 16S rDNA from Buchnera sp. CTU has been deposited in the GenBank database (accesion no. AJ417833).
References
- 1.Moran A N, Wernegreen J J. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S G E, Kurland C G. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 4.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature (London) 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 5.Peterson S N, Fraser C M. Genome Biol. 2001;2:2002.1–2002.8. doi: 10.1186/gb-2001-2-2-comment2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munson M A, Baumann P, Kinsey M G. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 7.Baumann P, Moran N A, Baumann L. In: The Prokaryotes, A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. Dworkin M, editor. New York: Springer; 2000. pp. 2–68. [Google Scholar]
- 8.Douglas A E. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 9.Sandström J, Moran N. Emtomol Exp Appl. 1999;91:203–210. [Google Scholar]
- 10.Moran N A, Telang A. Bioscience. 1998;48:295–304. [Google Scholar]
- 11.Harada H, Oyaizu H, Ishikawa H. J Gen Appl Microbiol. 1996;42:17–26. doi: 10.2323/jgam.43.349. [DOI] [PubMed] [Google Scholar]
- 12.Fukatsu T, Nikoh N, Kawai R, Koga R. Appl Environ Microbiol. 2000;66:2748–2758. doi: 10.1128/aem.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D Q, Purcell A H. Curr Microbiol. 1997;34:220–225. doi: 10.1007/s002849900172. [DOI] [PubMed] [Google Scholar]
- 14.Chen D Q, Montllor C B, Purcell A H. Entomol Exp Appl. 2000;95:315–323. [Google Scholar]
- 15.Moran N A, Munson M A, Baumann P, Ishikawa H A. Proc R Soc London B. 1993;253:167–171. [Google Scholar]
- 16.Silva F J, Latorre A, Moya A. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 17.Lai C Y, Baumann L, Baumann P. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracho A M, Martínez-Torres D, Moya A, Latorre A. J Mol Evol. 1995;41:67–73. doi: 10.1007/BF00174042. [DOI] [PubMed] [Google Scholar]
- 19.Remaudière G, Remaudirère M. Catalogue des Aphididae du Monde: Homoptera Aphidoidea. Paris: Institut National de la Recherche Agronomique; 1997. [Google Scholar]
- 20.Harrison C P, Douglas A E, Dixon A F G. J Invertebr Pathol. 1989;53:427–428. [Google Scholar]
- 21.Wernegreen J J, Ochman H, Jones I B, Moran N A. J Bacteriol. 2000;182:3867–3869. doi: 10.1128/jb.182.13.3867-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riethman H, Birren B, Gnirke A. In: Analyzing DNA. Birren B, Green E D, Klapholz S, Myers R M, Roskam J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 83–248. [Google Scholar]
- 23.Charles H, Ishikawa H. J Mol Evol. 1999;48:142–150. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- 24.van Ham R C H J, Moya A, Latorre A. J Bacteriol. 1997;179:4768–4777. doi: 10.1128/jb.179.15.4768-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strimmer K, von Haesseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 26.Levene S D, Zimm B H. Proc Natl Acad Sci USA. 1987;84:4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L V, Foster J M, Tzertzinis G, Ono M, Bandi C, Slatzo B E, O'Neill S L. J Bacteriol. 2001;183:2219–2225. doi: 10.1128/JB.183.7.2219-2225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Ham R C H J, González-Candelas F, Silva F J, Sabater B, Moya A, Latorre A. Proc Natl Acad Sci USA. 2000;97:10855–10860. doi: 10.1073/pnas.180310197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Ham R C H J, Martínez-Torres D, Moya A, Latorre A. Appl Environ Microbiol. 1999;65:117–125. doi: 10.1128/aem.65.1.117-125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai C Y, Baumann P, Moran N A. Appl Environ Microbiol. 1996;62:332–339. doi: 10.1128/aem.62.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandström J P, Rusell J A, White J P, Moran N A. Mol Ecol. 2001;10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 32.Mushegian A R, Koonin E V. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mushegian A. Curr Opin Genet Dev. 1999;9:709–714. doi: 10.1016/s0959-437x(99)00023-4. [DOI] [PubMed] [Google Scholar]
- 34.Koonin E V. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchison C A, Peterson S N, Gill S R, Cline R T, White O, Fraser C M, Smith H O, Venter J C. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]