Abstract
Background:
Epigenetics in cancer has been a focus of intense research in the recent years. This phenomenon has attracted great interest because it offers potential targets for cancer therapy. To capture the intellectual landscape of this field, this article conducted a bibliometric analysis to assess the current status, frontiers and future trends of epigenetic research in cancers.
Methods:
A bibliometric analysis was performed using data from the Web of Science Core Collection up to 2023. Analytical tools such as CiteSpace, VOSviewer, and the R package “bibliometrix” were employed for comprehensive data analysis and visualization. This process identified the publication of the articles, countries, authors, institutions, journals and keywords.
Results:
The results showed that there were 51,742 articles from the WoSCC database during 1985 to 2023 for cancer epigenetics. The number of epigenetic cancer-related publications has increased annually. The most contributed country is the United States, with 850,726 citations and 55 average article citations. China ranks second, with 413,386 citations and an average of 44.7. The most productive institutions were from the USA. Plos One (1020), International Journal of Molecular Sciences (957), and Cancers (945) were the top 3 contributing journals. The top 3 most common keywords were “DNA methylation,” “expression” and “cancer.” Research hotspots within these themes highlight intensive areas of study and the potential for significant breakthroughs.
Conclusions:
This study presents an in-depth overview of the current state of epigenetics in cancers research. And the purpose of this review will be to understand the characteristics of bibliometric analyses of epigenetic cancers and anticipate the progress in understanding this field.
Keywords: bibliometrics, cancer, citespace, epigenetic, VOSviewer
1. Introduction
Cancer is a major global social, public health and economic problem, causing huge social and economic losses. There were close nearly 20 million new cancer cases and 9.7 million deaths from cancer in the year 2022, and according to the prediction, the number of new cancer cases worldwide may reach 35 million or more by 2050.[1] The exact mechanisms underlying all of the cancers still is a mysterious world. Moreover, due to the diverse and atypical clinical manifestations of cancer, the diagnosis, treatment and prognosis are affected. Therefore, it is very important to find the pathogenesis of cancer to prevent and treat cancer.
“Epigenetics” is a concept proposed by developmental biologist Conrad H. Waddington (1905–1975) in 1942, referring to the complex of developmental processes between genotype and phenotype.[2] Epigenetic regulation, which involves reversible and heritable changes in gene-expression that occur in response to external stimuli without altering the DNA sequence, is fundamental to understanding the pathogenesis of various diseases, including cancer.[3,4] This regulation includes DNA methylation, chromatin remodeling, histone modifications, and the expression of diverse noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs).[5,6] Although the exact mechanism behind cancer is still unknown, it is believed that there are genetic factors involved.[7] And there are many studies have found that epigenetics is essential for elucidating the intricate mechanisms underlying the development and progression of cancer.[4]
The growing interest in the role of epigenetics in cancer is evident from the rising number of publications. However, a comprehensive bibliometric analysis of this body of work is lacking. Bibliometric analysis is an information visualization method to identify and summarize the frontiers or hot spots in a certain area.[8] Moreover, the researcher can compare the research status among various institutions, authors, or journals, evaluate the latest cutting-edge research, understand the scientific articles, and visualize their trends through this method.
This study presents a detailed bibliometric analysis of scientific literature on epigenetics in cancer from 1985 to 2023, utilizing data from the Web of Science Core Collection (WOSCC). The research aims to show the developments and future trends in the research field of epigenetics in cancer and offer valuable insights for future investigations.
2. Materials and methods
2.1. Data source
Search for the data in the WoSCC, which is regarded as one of the most widely used and comprehensive databases for citation analysis. A single researcher performed the search independently, restricting the publication date to the period between January 1, 1985, and December 31, 2023. The keywords for searching the articles were: TS = (“epigenetic”) AND TS = (“cancer” or “tumor” or “malignancy” or “neoplasm” or “sarcoma” or “carcinoma” or “tumor”). The search was performed on October 28, 2023, and 67,270 articles were obtained in total. The following were excluded from the final selection: book chapters, editorial materials, conference papers, conference abstracts, online publications, revisions, letters, book reviews, aiming to exclude unsuitable topics and research purposes. The language was not limited. Thus, a total of 51,742 papers and reviews were ultimately selected. The data was exported in plain text format and named as “download.” See Figure 1 for details.
Figure 1.
Flow diagram for searches of databases.
2.2. Visualization analysis
For the visualization analysis of the data, CiteSpace (version 6.1.6R), VOSviewer (version 1.6.18), R-Bibliometric and Adobe Illustrator2020 are utilized.[9] CiteSpace, VOSviewer and Adobe Illustrator are used to demonstrate cluster analysis, burst detection, timeline analysis, and journal co-occurrence analysis of references. VOSviewer (1.6.11) was used to create institutions network visualizations, co-authorship, co-citation authors, and keywords and explore collaboration networks between authors/institutes/countries/journals.[10] In VOSviewer, nodes were used to represent countries, institutions, journals, and authors, and their size was determined by their co-occurrence frequency in titles and abstracts.[11] Besides, Adobe Illustrator software was used to generate geographical distribution maps of publication outputs based on year and country, among others.
3. Results
3.1. Overall analysis of publications and citations
The number of research papers published in different periods demonstrates the popularity and development tendency of research in a particular field. There were 51,742 articles from the WoSCC database during 1985 to 2023 for cancer epigenetics. The quantity of publications, citation ranks and average citation ranks per year are shown in Table 1. And it also shows the mean total number of citations (TC) per article. Table 1 shows that articles had the highest average citation ranks were published in 2023, with 61.62. The peak of average citation rank per year is 2021, with 8973. The quantitative analysis of published articles about epigenetics in cancers is shown in Figure 2. An upward trend in annual publications is observed in Figure 2A, reaching a peak in 2021 before a slight decline, yet maintaining high overall numbers. Furthermore, the quantity of different countries/regions articles were statistically analyzed. The total publication ranks of the top 20 countries/regions is displayed in Figure 2B. And Figure 2C presents total publication ranks from the 10 leading countries/regions. The result shows that the United States leads with 15,479 publications (29.92%), followed by China (9248 publications, 17.87%). These results reflect that cancer-related epigenetic research has attracted significant attention and scholarly recognition.
Table 1.
Global publication output and citation trend of epigenetic in cancers.
| Year | Mean TC per article | Publications | Mean TC per year | Total citations |
|---|---|---|---|---|
| 1998 | 7.38 | 16 | 118 | 118 |
| 1999 | 47.43 | 7 | 166 | 332 |
| 2000 | 3.78 | 138 | 174 | 521 |
| 2001 | 4.62 | 162 | 187 | 748 |
| 2002 | 4.05 | 279 | 226 | 1130 |
| 2003 | 6.60 | 385 | 424 | 2541 |
| 2004 | 8.42 | 453 | 545 | 3814 |
| 2005 | 11.21 | 591 | 828 | 6623 |
| 2006 | 13.48 | 718 | 1076 | 9681 |
| 2007 | 15.15 | 943 | 1429 | 14,288 |
| 2008 | 18.17 | 1027 | 1696 | 18,656 |
| 2009 | 21.72 | 1162 | 2103 | 25,238 |
| 2010 | 26.27 | 1416 | 2861 | 37,199 |
| 2011 | 28.63 | 1568 | 3207 | 44,898 |
| 2012 | 32.98 | 1763 | 3876 | 58,141 |
| 2013 | 31.66 | 2141 | 4237 | 67,792 |
| 2014 | 35.12 | 2303 | 4758 | 80,892 |
| 2015 | 38.12 | 2487 | 5266 | 94,794 |
| 2016 | 39.87 | 2667 | 5597 | 106,336 |
| 2017 | 41.82 | 2836 | 5930 | 118,591 |
| 2018 | 45.13 | 2771 | 5955 | 125,051 |
| 2019 | 51.63 | 3027 | 7104 | 156,282 |
| 2020 | 51.66 | 3422 | 7686 | 176,774 |
| 2021 | 56.58 | 3806 | 8973 | 215,347 |
| 2022 | 60.80 | 3598 | 8751 | 218,765 |
| 2023 | 61.62 | 2473 | 5861 | 152,393 |
TC = total number of citations.
Figure 2.
(A) Trend of publications and citations of epigenetic research in cancers from 1998 to 2023. (B) Bar graph of the top 20 productive countries. (C) Total publications radar map of the top 10 countries.
3.2. Contributions of countries/regions
During the period from 1985 to 2023, 450 countries and regions published research publications related to epigenetic cancers. Table 2 presents the 10 countries with the highest citation counts. Similarly, Figure 3A and B shows the total and average number of citations for the top 20 countries, respectively. The most contributed country is the United States, with 850,726 citations and 55 average article citations. China ranks second, with 413,386 citations and an average of 44.7. The international cooperation analysis is shown in Figure 3C. The United States has the most cooperation with many countries, China ranks second. However, the level of cooperation among other countries remains low relatively. The research analyzed the network of global collaborative with a minimum threshold of 450 papers by using VOSviewer. As shown in Figure 3D, the largest total link strength (TLS) of the United States ranks first (TLS = 136,525), followed by China (TLS = 63,904) and Germany (TLS = 23,005). In addition, the United States having the highest proportion of international cooperation (0.45), followed by China and Germany.
Table 2.
Productive countries/regions related to epigenetic in cancers.
| Country | Articles | SCP | MCP | MCP-ratio | Citations | Average article citations | TLS |
|---|---|---|---|---|---|---|---|
| USA | 15,479 | 8505 | 6974 | 0.45 | 850,726 | 55 | 136,525 |
| China | 9248 | 5166 | 4082 | 0.44 | 413,386 | 44.7 | 63,904 |
| Germany | 3334 | 2179 | 1155 | 0.35 | 141,528 | 42.5 | 23,005 |
| Italy | 2866 | 2062 | 804 | 0.28 | 125,961 | 44 | 24,676 |
| Japan | 2667 | 1852 | 815 | 0.31 | 91,745 | 34.4 | 19,869 |
| England | 2569 | 1809 | 760 | 0.3 | 104,918 | 40.8 | 22,376 |
| France | 1824 | 1361 | 463 | 0.25 | 60,356 | 33.1 | 15,540 |
| Spain | 1806 | 1290 | 516 | 0.29 | 72,999 | 40.4 | 16,236 |
| Canada | 1684 | 1146 | 538 | 0.32 | 88,191 | 52.4 | 10,424 |
| India | 1417 | 984 | 433 | 0.31 | 43,148 | 30.5 | 12,059 |
MCP = number of publications co-authored by authors from different countries, SCP = number of publications co-authored by authors from the same country, TLS = total link strength.
Figure 3.
(A) Map of cumulative citations across the 20 leading countries from 1985 to 2023. (B) The country-region-specific citation average numbers from 1985 to 2023. (C) The countries/regions cooperation networks visualization map generated by using VOS viewer. (D) The countries/regions citation network visualization map generated by using VOSviewer.
3.3. Analysis of main publishing institutions and authors
There were 450 institutions participated in research on epigenetic in cancers. The most productive institutions were from the USA. As shown in Table 3, among the top 10 productive institutions, 7 come from the USA, 2 are from France and 1 from Germany. Harvard University (2055) was the most productive institution, followed by the University of California system (1934) and the University of Texas system (1818).
Table 3.
The top 10 most productive institutions related to epigenetic in cancers.
| Affiliation | Articles | Country | TLS |
|---|---|---|---|
| Harvard University | 2055 | USA | 30,825 |
| University of California System | 1934 | USA | 40,614 |
| University of Texas System | 1818 | USA | 25,452 |
| Udice French Research Universities | 1575 | France | 17,325 |
| National Institutes of Health NIH USA | 1560 | USA | 23,400 |
| Institut National de la Sante et de la Recherche Medicale Inserm | 1356 | France | 28,476 |
| Harvard Medical School | 1344 | USA | 20,160 |
| Helmholtz Association | 1237 | Germany | 24,740 |
| Johns Hopkins University | 1168 | USA | 15,184 |
| Utmd Anderson Cancer Center | 1145 | USA | 16,030 |
TLS = total link strength.
Additionally, the research used VOS viewer to analyze the cooperation among institutions. Harvard University has the strongest collaboration capabilities, as shown in Figure 4A. The world’s top universities and institutions have made outstanding contributions to the development of the field.
Figure 4.
(A) Collaborative visualization network of institutions constructed using VOSviewer software. (B) Map of the leading 20 authors with the highest H-index. (C) The visualization network of author cooperation. (D) Visualization map of co-cited authors generated by VOSviewer.
In total, 352 authors and 221,835 co-cited authors were identified. Tables 4 and 5 summarize the top 10 most generative authors and the top 10 co-cited authors, respectively. Esteller, Manel, Herman, James G and Baylin, Stephen B ranked in the top 3 with 264, 141 and 122 papers respectively. While Yang, Ying, Pfister, Stefan M and Jones, Peter A. published fewer papers, their total citations were as high as 63,402, 62,089 and 49,740, respectively. The H-index is a measure of a scholar’s overall influence, representing the number of publications cited at least N times (N). Table 4A and Figure 4B show Baylin, Stephen B has the highest H index, which is 158, which having great influence in this field. In addition, an author’s contribution to a particular field is presented by the “article classification,” representing the author’s publications in that field as a percentage of all publications. The 2 biggest contributors in the field were Esteller, Manel and Herman, James G., with 62.26 and 33.98 respectively. Figure 4C is a visualization map of author co-authorship analysis generated by VOSviewer.
Table 4.
The top 10 most productive authors related to epigenetic in cancers.
| Author | Publications | H-index | G-index | M-index | Citations | Articles fractionalized |
|---|---|---|---|---|---|---|
| Esteller, Manel | 264 | 81 | 207 | 13.45 | 28,576 | 62.26 |
| Herman, James G. | 141 | 128 | 193 | 10.58 | 92,294 | 33.98 |
| Baylin, Stephen B. | 122 | 158 | 178 | 9.97 | 149,455 | 32.53 |
| Issa, Jean-Pierre J. | 120 | 112 | 158 | 8.62 | 51,659 | 28.99 |
| Huang, T. H. | 101 | 68 | 139 | 8.19 | 15,147 | 28.45 |
| Fraga, Mario F | 95 | 81 | 136 | 8.06 | 28,576 | 23.00 |
| Jerónimo, Carmen | 94 | 54 | 124 | 7.26 | 10,579 | 21.61 |
| Henrique, Rui Manuel Ferreira | 83 | 52 | 118 | 6.74 | 10,283 | 22.02 |
| Tao, Qian | 79 | 57 | 110 | 6.04 | 8750 | 20.68 |
| Yang, Ying | 79 | 86 | 103 | 5.77 | 63,402 | 18.59 |
Table 5.
The top 10 most productive co-cite author related to epigenetics in cancers.
| Co-cite author | Citation | TLS | Centrality |
|---|---|---|---|
| Esteller, Manel | 6346 | 24,950 | 0.18 |
| Baylin, Stephen B. | 4433 | 22,894 | 0.15 |
| Henrique, Rui Manuel Ferreira | 1574 | 20,440 | 0.12 |
| Suzuki, Hiromu | 2949 | 18,039 | 0.09 |
| Jones, Peter A. | 4466 | 17,980 | 0.09 |
| Herceg, Zdenko | 3167 | 17,853 | 0.09 |
| Jerónimo, Carmen | 1019 | 14,486 | 0.06 |
| Herman, James G. | 310 | 14,164 | 0.04 |
| Issa, Jean-Pierre J. | 902 | 13,509 | 0.03 |
| Fraga, Mario F | 4818 | 12,758 | 0.03 |
TLS = total link strength.
The top author with the highest number of citations is Baylin, Stephen B, with 149,455. In addition, Figure 4D shows the co-cited author network visualized using VOSviewer, and the highest TLS author is Esteller, Manel (TLS = 24959), followed by Baylin, Stephen B (TLS = 22,894) and Henrique, Rui Manerl Ferreira.
3.4. Analysis of the top publishing journals
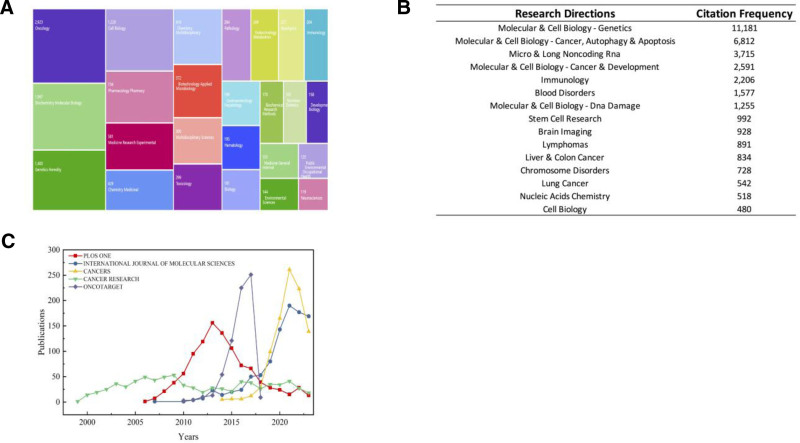
Highly cited papers are those that have been referenced frequently by other publications, which are pivotal contributions within the academic literature that have significantly influenced the landscape of a particular field. The top 10 publishing journals are shown in Table 6. Plos One (1020), International Journal of Molecular Sciences (957), and Cancers (945) were the top 3 contributing journals. Among the top 10 journals, their impact factors were relatively high, with 60% being higher than 5.0. Besides this, Plos One had both the highest citations and the highest number of H-index. R software was used to conduct the top 25 most popular research directions visualization analysis, as Figure 5A performed. Top 15 citation of research directions are shown in Figure 5B. Molecular & Cell Biology-Genetics are the top research directions in this field. Figure 5C shows the publication trends annually of the top 5 most generative journals. Cancers exhibits the highest growth rate, Oncotarget ranks second, while they all show a downward trend in recent years. Besides, Table 7 presents the top 10 most cited articles with the realm of epigenetics in cancers. The highest citation count of article titled “Molecular origins of cancer: Epigenetics in cancer” by Esteller et al has (2710) and was released in the New England Journal of Medicine (IF = 158.5). The article published in Boinformatics (IF = 5.8), titled “Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays” ranks second, which has been cited 2389 times.
Table 6.
Top 10 journals related to the research of epigenetics in cancers.
| Rank | Journal | Publications | H-index | Total citations | IF (2023) |
|---|---|---|---|---|---|
| 1 | PLoS One | 1020 | 64 | 34,336 | 3.700 |
| 2 | International Journal of Molecular Sciences | 957 | 31 | 32,782 | 6.208 |
| 3 | Cancers | 945 | 56 | 32,042 | 6.575 |
| 4 | Cancer Research | 774 | 54 | 28,120 | 13.312 |
| 5 | Oncotarget | 690 | 49 | 24,753 | 6.000 |
| 6 | Oncogene | 688 | 47 | 24,568 | 4.419 |
| 7 | Scientific Reports | 632 | 43 | 18,500 | 4.600 |
| 8 | Nature Communications | 625 | 41 | 18,426 | 16.600 |
| 9 | Epigenetics | 575 | 26 | 18,204 | 2.149 |
| 10 | Clinical Epigenetics | 560 | 24 | 17,649 | 7.259 |
IF = impact factor.
Figure 5.
(A) The top 25 most dominant directions of the research in this study. (B) The top 15 most cited research directions in this study. (C) The chart of the leading 5 journals with the most publications in terms of annual publications changes from 1998 to 2023.
Table 7.
Top 10 articles with the most citations concerning the research of Epigenetic in cancers.
| Paper | Journal | Citations | Corresponding author | IF (2023) |
|---|---|---|---|---|
| Molecular origins of cancer: Epigenetics in cancer | New England Journal of Medicine | 2710 | Esteller, M | 158.5 |
| Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays | Boinformatics | 2389 | Aryee, MJ | 5.8 |
| Cancer Epigenetics: From Mechanism to Therapy | Cell | 2300 | Dawson, MA | 64.5 |
| Epigenetic modifications and human disease | Nature Biotechnology | 2298 | Portela, A | 46.9 |
| Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers | Cell | 2101 | Lovén, J | 64.5 |
| Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates | Molecular Cell | 1914 | Hannum, G | 16.0 |
| Epigenetics in cancer | Carcinogenesis | 1911 | Sharma, S | 4.7 |
| Environmental epigenomics and disease susceptibility | Nature Reviews Genetics | 1904 | Jirtle, RL | 42.7 |
| PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations | Science Translational Medicine | 1771 | Zou, WP | 17.1 |
| Cancer epigenomics: DNA methylomes and histone-modification maps | Nature Reviews Genetics | 1702 | Esteller, M | 42.7 |
IF = impact factor.
3.5. Reference analysis
The study cited a total of 116,521 studies related to cancer epigenetics. The leading 25 strongest Citation bursts references are summarized in Figure 6A. The reference citation burst in this study began in 2000 due to the paper published by Baylin SB in 1998. The latest reference citation burst was detected in 2021 and last until now. Among them, a review article entitled “The Epigenomics of Cancer” by Jones PA published in Cell had the intensity with highest burst in 2007, attaining 17.58. Currently, most important articles are still cited frequently and it can be speculated that epigenetic of cancers research will still be a research hotspot in the next few years. Furthermore, the co-cited collaboration network is shown in Figure 6B, which reveals a diverse network of topics in cancer research, covering therapeutic strategy innovation (immunotherapy, combination therapy), environmental and inflammatory drivers, tumor heterogeneity challenges, and the focus on specific cancer types (pediatric, gastric cancer), providing researchers with a panoramic view of the field’s development.
Figure 6.
(A) Top 25 references with the strongest citation bursts. (B) Evolution path map of major themes generated by CiteSpace software.
3.6. Analysis of keywords and subject terms
According to the keyword analysis, future research directions and hotspots can be predicted to support scientific research decision-making. A total of 85,855 keywords were included in this study. Table 8 summarizes the top 20 keywords with the highest frequency. The top 3 most common keywords were “DNA methylation,” “expression” and “cancer.” Figure 7A shows the heat map of the 50 keywords with the highest occurrence rates. Figure 7B illustrates the categorization of keywords into 4 distinct color-coded clusters. DNA methylation, expression, and cancer are among the frequently used terms. Figure 7C shows the top 8 frequency keyword timeline perspective. “DNA methylation” exhibits the highest magnitude and statistically significant sustained visibility. The current hot spots are “gene expression,” “tumor microenvironment” and “prostate cancer.” Figure 7D presents the top 25 keywords with the strongest citation bursts. “Comparative genomic hybridization,” the earliest keyword burst, was detected in 2000. Later, researches related to genome and epigenetic mechanism such as “CpG island” or “promoter hypermethylation” became hot topics. The strength of “tumor microenvironment” hit 17.92 (2020–2023), which indicates the research in this field has increased rapidly in recent years. Recent popular topics such as “resistance” (2020–2023), “epigenetic modification” (2020–2023), “immunotherapy” (2021–2023), “cell” (2021–2023) and “t cell” (2021–2023) have bursts that extend until 2023, suggesting that these research topics might become new research foci in the next few years. Figure 7E shows the trend of subject evolution from 1998 to 2023. “Histone deacetylase inhibitors,” “methylation” evolved into “immunotherapy,” “cell-free DNA,” “tumor microenvironment.” To sum up, Figure 7 shows the analysis of the keywords, which depicts the evolution of research interests over time, and reflects research frontiers, hot topics, emerging trends.
Table 8.
Top 20 keywords with the most frequencies concerning the research of epigenetics in cancers.
| Rank | Keywords | Frequency | TLS | Rank | Keywords | Frequency | TLS |
|---|---|---|---|---|---|---|---|
| 1 | DNA methylation | 12,146 | 31,628 | 11 | Hypermethylation | 2583 | 8758 |
| 2 | Expression | 11,414 | 31,127 | 12 | Differentiation | 2530 | 6679 |
| 3 | Cancer | 9964 | 25,611 | 13 | Activation | 2477 | 7070 |
| 4 | Epigenetics | 6762 | 18,308 | 14 | Identification | 2460 | 7048 |
| 5 | Methylation | 6652 | 18,845 | 15 | Transcription | 2406 | 6475 |
| 6 | Gene-expression | 5781 | 12,995 | 16 | Apoptosis | 2373 | 7084 |
| 7 | Gene | 4083 | 11,932 | 17 | Mutations | 2359 | 5852 |
| 8 | Cells | 3192 | 9081 | 18 | Epigenetic regulation | 2283 | 5766 |
| 9 | Breast-cancer | 2781 | 7525 | 19 | Protein | 2216 | 5969 |
| 10 | Chromatin | 2648 | 6597 | 20 | Growth | 2122 | 6594 |
TLS = total link strength.
Figure 7.
(A) The top 50 keywords with the highest frequency. (B) Keyword cluster map constructed via VOSviewer. (C) Temporal evolution of keywords was analyzed through CiteSpace software. (D) Top 25 keywords with the strongest citation burst. (E) The trend of subject evolution from 1998 to 2023.
4. Discussion
4.1. General information discussion
This article conducted a bibliometric analysis to assess the current status, the frontier hotspots and trends of epigenetic research in cancers. The results of this study showed that the number of publications each year has been steadily increasing (Fig. 2). This reflects the continued interest. Although the circulation in 2023 decreased slightly, it still exceeded 2000, which shows the ongoing interest in cancer epigenetics. The United States, China and Germany rank in the top 3 in terms of publication and citations. The United States ranks first in the number of papers published, cited frequency and average cited frequency, and dominates the field. Because of its very strong economic strength and beneficial policy and scientific support, the United States is the largest country in this field of research. At the same time, although China is a developing country, it has an important position in the field of epigenetics research.
According to the results of countries/regions distribution, the most contributed country is the United States, with 850,726 citations and 55 average article citations (Fig. 3). And the United States has the most cooperation with many countries, China ranks second. However, the level of cooperation among other countries remains low relatively. The United States leads in cross-national impact metrics and also the highest MCP-Ratio (Table 2), which indicates that American researchers work closely with researchers in other countries.
According to research institutions, 7 of the top 10 most published institutions are from the United States (Table 3), which interprets why the United States makes contributions to a huge number of epigenetic and cancer-related articles. In addition, France accounts for 20 percent of the top 10 universities. Unfortunately, there is no any single Chinese university in the top 10.
In author and co-cited author analysis, Esteller, Manel, Herman, James G and Baylin, Stephen B ranked in the top 3 with 264, 141 and 122 papers respectively. While Yang, Ying, Pfister, Stefan M and Jones, Peter A. published fewer papers, their total citations were as high as 63,402, 62,089 and 49,740, respectively. From the visualization map of co-cited author, Esteller, Manel has the highest TLS (Fig. 4). More important articles may be published by the above team members, strengthening cooperation with these top teams is a good choice for research.
Identification of important journals analysis, Plos One, International Journal of Molecular Sciences, Cancers, Cancer Research and Oncotarget are probably the main publishing places for cancer-related articles (Table 6). It is recommended to submit more cancer-related articles to these journals. Among the most prolific publications, there are 2 journals had an IF of more than 10: Cancer Research (IF 2023, 13.312) and Nature Communications (IF 2023, 16.6). Cancers exhibits the highest growth rate, Oncotarget ranks second, while they all show a downward trend in recent years. Of particular note is that are it is an amazing choice for researchers to publish epigenetics and cancer research in these top 10 journals in the future. The most cited references are often considered the basis of research in a particular field. The most cited article is “Molecular Origins of Cancer: The Epigenetics of Cancer” (Table 7). This example illustrates how to improve cancer diagnosis and treatment from a molecular perspective.
4.2. Hotspots and frontiers of epigenetic in cancers
Bibliometrics analysis can reflect the hot spots and frontiers of this research field. Through a multiple analysis of references and keywords, these results showed that molecular and cell biology-genetics is the most popular research direction in epigenetics of cancer (Fig. 5). Through the citation analysis of reference, the result showed that the most of the topics focus on the relationship between cancer, epigenetics, etiology, diagnosis, and treatment (Table 7). According to the keyword analysis, the top 3 most common keywords were “DNA methylation,” “expression” and “cancer” (Table 8). “DNA methylation” has the largest and consistently attracting high attention (Fig. 7C). “Comparative genomic hybridization,” the earliest keyword burst, was detected in 2000. Later, researches related to genome and epigenetic mechanism such as “CpG island” or “promoter hypermethylation” became hot topics (Fig. 7D). As DNA-methylation research advances, it shows the increasingly importance of epigenetics in DNA-methylation function and cancer development.[12,13] Multiple studies have found that the inactivation of certain tumor suppressor genes is the result of hypermethylation of promoter regions, and that DNA methylation leads to cancer by causing widespread gene silencing.[14,15] In addition, DNA methylation is one of the most intensively studied epigenetic modifications in mammals, which fully demonstrates that it is a current research hotspot.[16] For example, histone modifications have been shown to be closely related to DNA methylation, regulating genome function by altering chromatin structure.[17] Global hypomethylation, which induces genomic instability, also contributes to cell transformation. Noncoding RNAs, such as microRNAs, have been found to play a key role in DNA methylation.[18] Covalent addition of methyl groups usually occurs in cytosine within CpG dinucleotides, which are concentrated in large clusters called CpG islands. It suggests that CpG can be used as a potential target for cancer treatment.[19] In conclusion, a comprehensive grasp of the epigenetic mechanism of cancer affords potential targets for the engineering of new treatments. Ancer is characterized by abnormal expression in many aspects. Through analysis, the study has identified several keywords with the strongest citation bursts. “Comparative genomic hybridization,” the earliest keyword burst, was detected in 2000. Later, researches related to genome and epigenetic mechanism such as “CPG island” or “promoter hypermethylation” became hot topics. One of the recent popular topics is “cell” (2021–2023) (Fig. 7D), which plays a very important role in the occurrence and development of cancer. For example, stem cells, often defined as cloned cells capable of self-renewal and multi-lineage differentiation, are biological tissue units responsible for the development and regeneration of tissue and organ systems. There is evidence that tissue-specific stem cells are the cells of origin for many types of cancer.[20,21] For example, cell subpopulations with CD34- and CD38- phenotypes similar to normal hematopoietic stem cells (HSCs) were found to be able to initiate human acute myeloid leukemia suggesting that normal HSCs are targets for leukemia transformation.[22,23] In addition, a number of mutations in leukemia cells were found in normal HSCs suggesting that stem cells are common targets for leukemia transformation.[24] In addition, several studies have shown that under standard culture conditions, postnatal stem cells can transform into malignant cells without any genetic manipulation.[25] For example, mesenchymal stem cells enter a malignant transformation state after long-term in vitro culture and form tumors in vivo. Taken together, abnormal epigenetics are closely linked to the pathogenesis of cancer, some of which have shown promise as markers of diagnosis and disease activity. The best-studied epigenetic changes in cancer are the methylation changes that occur within the CpG island, and the next generation sequencing platform has now provided a genome-wide map of CpG methylation.[26] And, recent cancer genome sequencing has identified recurrent mutations in DNA methyltransferase 3A (DNMT3A) in up to 25% of patients with acute myeloid leukemia. Moreover, Ten-eleven-translocation-2 (TET2) mutations also confer enhanced self-renewal properties on malignant clones. Histone acetylation: n ε-acetylation of lysine residues is a major histone modification involved in transcription, chromatin structure, and DNA repair. Acetylation neutralizes the positive charge of lysine and thus may weaken the electrostatic interaction between histones and negatively charged DNA. Enzymes that modify histones have been found, such as Lysine acetyltransferases (KATs), the first to be shown.[27] Taken together, current research focuses on DNA methylation, cancer, epigenetics, methylation, gene-expression, gene, cells, etc, through which researchers try to explore the pathogenesis of cancer.
4.3. Research frontiers and future trends of epigenetics in cancer
Bibliometrics analysis can reflect research frontiers and future trends of this research field. Based on multiple analyses of keywords and subject terms, the top 2 frequently cited keywords are “DNA methylation” and “expression,” showing that it remains a key focus of cancer and epigenetics research (Fig. 7A). Abnormal gene-expression regulation is a key element in tumorigenesis and development, and epigenetic mechanism are influential in the regulation of gene-expression.[28,29] In the genetic susceptibility to cancer, maternal inherited mitochondrial DNA (mtDNA) is believed to be involved in cancer development and prognosis, and is considered to be a therapeutic target for cancer treatment.[30] This suggests that researchers should focus more on exploring epigenetic networks and the specific role of epigenetic inheritance in different types of cancer in the future, and provide a theoretical basis for developing personalized treatment strategies.
However, the hot spot evolved from “DNA methylation” to “gene expression” and “tumor microenvironment” (Fig. 7C). DNA methylation regulates gene-expression by recruiting proteins involved in gene suppression or inhibiting the binding of transcription factors to DNA. At present, there is compelling evidence that long noncoding RNAs (lncRNAs) mediate DNA methylation under both physiological and pathological conditions. Moreover, DNA-methylation canyons are preferentially hypermethylated in cancer. Most of the DNA-methylation canyons are labeled by H3K27me3 and are associated with Polycomb proteins such as EED, EZH2, RING1B. DNA methylation in cancer cells shifts the pattern of gene-expression inhibition from a plastic polycomb-dependent H3K27Me3-mediated pattern to a more stable DNA-methylation-dependent pattern of events. Therefore, the control of DNA hypermethylation of certain oncogenes and the control of DNA damage-dependent methylation of CpG islands can help to mitigate the development of cancer. Moreover, DNA-methylation inhibitors have become a mainstay in the treatment of some hematological malignancies.[31] In addition to their ability to reactivate genes that acquire DNA methylation during cancer development, including tumor suppressors, they induce the expression of thousands of transposable elements, including endogenous retroviruses and latent cancer testicular antigens that are normally silenced by DNA methylation in most somatic cells.
From the visualization map of top 25 keywords with the strongest citation bursts in epigenetic research of cancers, the strength of “tumor microenvironment” hit 17.92 (2020–2023) (Fig. 7D), which indicates the research in this field has increased rapidly in recent years. The trend of subject evolution from 1998 to 2023 showed that “histone deacetylase inhibitors,” “methylation” evolved into “immunotherapy,” “cell-free DNA,” “tumor microenvironment” (Fig. 7E). The tumor microenvironment not only contains cancer cells and already altered cellular structures, but is also capable to recruit normal cells and release cytokines to sustain the function of tumor microenvironment. Tumor microenvironment targets the local immune response in solid tumors to help immune cells entering the tumor and function efficiently within the tumor.[32] Modulation of the tumor microenvironment is of special significance in cancer immunotherapy. Epigenetic alterations are known contributors to cancer development and aggressiveness. Additional to alterations in cancer cells, aberrant epigenetic marks are present in cells of the tumor microenvironment, including lymphocytes and tumor-associated macrophages.[33,34] Tumor microenvironment is of great interest because of the potential roles it plays in supporting or moderating tumor growth. In certain cancers with extensive stroma (e.g., PDAC and breast-cancer), stroma-targeted therapies are being considered and tested.[35–37]
And recent popular topics such as “resistance” (2020–2023), “epigenetic modification” (2020–2023), “immunotherapy” (2021–2023), “cell” (2021–2023) and “t cell” (2021–2023) have bursts that extend until 2023 (Fig. 7D), suggesting that these research topics might become new research foci in the next few years. Epigenetic modifications involve changes in chromatin structure, the methylation status of DNA fragments, and chemical transformations of histone chromosomal proteins (acetylation, methylation, ADP-ribosylation, ubiquitination, and phosphorylation), and the regulation of noncoding RNAs (ncRNAs). Several studies have demonstrated that epigenetic modifications are one of the main mechanisms underlying many diseases, including cancer. These diseases experience significant deregulation of all epigenetic components, which both affect protein expression regulation and the modification of their functions. This deregulation determines the epigenetic signature of cancer.[38] Modulating epigenetics in combination with immunotherapy might be a promising therapeutic option to improve the success of this therapy. Further studies are necessary to understand in depth the impact of the epigenetic machinery in the tumor microenvironment.
In summary, aberrant gene function and altered patterns of gene-expression are key features of cancer. And growing evidence shows that epigenetic abnormalities participate with genetic alterations to cause this dysregulation. The latest research focus on DNA methylation, epigenetic modification, tumor microenvironment and so on. The in-depth research in these fields is expected to reveal new mechanisms and provide new breakthroughs in the diagnosis and treatment of cancer.
5. Limitations
There were some limitations in this study. First, only publications from WoSCC were included in this study, which may have excluded some valuable information. Second, important non-English papers may be overlooked, leading to research bias and decreased credibility. Third, recently published articles have not had enough time to be cited.
6. Conclusion
Our bibliometric analysis provides a comprehensive overview of the current state of research in cancer epigenetics, identifying key themes, hotspots, and directions for future research. It will offer suggestions for researchers in selecting new research directions, such as epigenetic regulation and DNA methylation. This timely review scrutinizes research trends and hotspots on epigenetic cancers, which could progress the field and form the basis for forthcoming studies.
Acknowledgments
We would like to thank Web of Science Core Collection for providing the raw data for this study.
Author contributions
Data curation: Xinyi Zhong, Du Chen.
Funding acquisition: Du Chen.
Visualization: Xinyi Zhong.
Writing – original draft: Xinyi Zhong.
Writing – review & editing: Du Chen.
Abbreviations:
- AML
- acute myeloid leukemia
- DNMT3A
- DNA methyltransferase 3A
- HSCs
- hematopoietic stem cells
- KATs
- lysine acetyltransferases
- lncRNAs
- long noncoding RNAs
- MSCs
- mesenchymal stem cells
- MtDNA
- mitochondrial DNA
- NGS
- next generation sequencing
- TET2
- ten-eleven-translocation-2
This study did not require ethical approval because the information we used was all publicly sourced and did not involve interactions with human subjects.
The authors have no funding and conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
How to cite this article: Zhong X, Chen D. Global research trends on epigenetics and cancer: A bibliometric analysis. Medicine 2025;104:33(e43197).
References
- [1].Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. [DOI] [PubMed] [Google Scholar]
- [2].Li Y. Modern epigenetics methods in biological research. Methods. 2021;187:104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378:1323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–99. [DOI] [PubMed] [Google Scholar]
- [5].Bednarczyk M, Dunislawska A, Stadnicka K, Grochowska E. Chicken embryo as a model in epigenetic research. Poult Sci. 2021;100:101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. [DOI] [PubMed] [Google Scholar]
- [7].Li R, Ding Z, Jin P, et al. Development and validation of a novel prognostic model for acute myeloid leukemia based on immune-related genes. Front Immunol. 2021;12:639634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barbara G, Stanghellini V, De Giorgio R, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. [DOI] [PubMed] [Google Scholar]
- [9].van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84:523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu H, Tong L, Wang Y, Yan H, Sun Z. Bibliometric analysis of global research trends on ultrasound microbubble: a quickly developing field. Front Pharmacol. 2021;12:646626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xie L, Chen Z, Wang H, Zheng C, Jiang J. Bibliometric and visualized analysis of scientific publications on atlantoaxial spine surgery based on web of science and VOSviewer. World Neurosurg. 2020;137:435–42.e4. [DOI] [PubMed] [Google Scholar]
- [12].Zarzour A, Kim HW, Weintraub NL. Epigenetic regulation of vascular diseases. Arterioscler Thromb Vasc Biol. 2019;39:984–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cyr AR, Hitchler MJ, Domann FE. Regulation of SOD2 in cancer by histone modifications and CpG methylation: closing the loop between redox biology and epigenetics. Antioxid Redox Signal. 2013;18:1946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang TC, Wang Y-F, Vazquez-Ferrer E, et al. Sex-specific chromatin remodelling safeguards transcription in germ cells. Nature. 2021;600:737–42. [DOI] [PubMed] [Google Scholar]
- [15].Meunier L, Hirsch TZ, Caruso S, et al. DNA methylation signatures reveal the diversity of processes remodeling hepatocellular carcinoma methylomes. Hepatology. 2021;74:816–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- [17].Konishi K, Issa JP. Targeting aberrant chromatin structure in colorectal carcinomas. Cancer J. 2007;13:49–55. [DOI] [PubMed] [Google Scholar]
- [18].Aalaei-Andabili SH, Rezaei N. MicroRNAs (MiRs) precisely regulate immune system development and function in immunosenescence process. Int Rev Immunol. 2016;35:57–66. [DOI] [PubMed] [Google Scholar]
- [19].Neja SA. Site-specific DNA demethylation as a potential target for cancer epigenetic therapy. Epigenet Insights. 2020;13:2516865720964808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Reavis HD, Chen HI, Drapkin R. Tumor innervation: cancer has some nerve. Trends Cancer. 2020;6:1059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pathak S, Multani AS. Aneuploidy, stem cells and cancer. EXS. 2006;96:49–64. [DOI] [PubMed] [Google Scholar]
- [22].Yamashita M, Dellorusso PV, Olson OC, Passegué E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat Rev Cancer. 2020;20:365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. 2020;20:158–73. [DOI] [PubMed] [Google Scholar]
- [24].González-Herrero I, Rodríguez-Hernández G, Luengas-Martínez A, et al. The making of leukemia. Int J Mol Sci. 2018;19:1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Todaro F, Campolo F, Barrios F, et al. Regulation of kit expression in early mouse embryos and ES cells. Stem Cells. 2019;37:332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van Otterdijk SD, Mathers JC, Strathdee G. Do age-related changes in DNA methylation play a role in the development of age-related diseases? Biochem Soc Trans. 2013;41:803–7. [DOI] [PubMed] [Google Scholar]
- [27].Srivastava S, Kumar S, Bhatt R, Ramachandran R, Trivedi AK, Kundu TK. Lysine acetyltransferases (KATs) in disguise: diseases implications. J Biochem. 2023;173:417–33. [DOI] [PubMed] [Google Scholar]
- [28].Lu C, Wei Y, Wang X, et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer. 2020;19:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–79. [DOI] [PubMed] [Google Scholar]
- [30].Mohamed Yusoff AA. Role of mitochondrial DNA mutations in brain tumors: A mini-review. J Cancer Res Ther. 2015;11:535–44. [DOI] [PubMed] [Google Scholar]
- [31].Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat Rev Cancer. 2019;19:151–61. [DOI] [PubMed] [Google Scholar]
- [32].Song W, Musetti SN, Huang L. Nanomaterials for cancer immunotherapy. Biomaterials. 2017;148:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Song H, Liu D, Dong S, et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther. 2020;5:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Banerjee S, Modi S, McGinn O, et al. Impaired synthesis of stromal components in response to minnelide improves vascular function, drug delivery, and survival in pancreatic cancer. Clin Cancer Res. 2016;22:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Murakami M, Ernsting MJ, Undzys E, Holwell N, Foltz WD, Li S-D. Docetaxel conjugate nanoparticles that target α-smooth muscle actin-expressing stromal cells suppress breast cancer metastasis. Cancer Res. 2013;73:4862–71. [DOI] [PubMed] [Google Scholar]
- [37].Sumida T, Kitadai Y, Shinagawa K, et al. Anti-stromal therapy with imatinib inhibits growth and metastasis of gastric carcinoma in an orthotopic nude mouse model. Int J Cancer. 2011;128:2050–62. [DOI] [PubMed] [Google Scholar]
- [38].Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59:168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]