Abstract
The E2F family of transcription factors are downstream effectors of the retinoblastoma protein, pRB, pathway and are essential for the timely regulation of genes necessary for cell-cycle progression. Here we describe the characterization of human and murine E2F8, a new member of the E2F family. Sequence analysis of E2F8 predicts the presence of two distinct E2F-related DNA binding domains suggesting that E2F8 and, the recently, identified E2F7 form a subgroup within the E2F family. We show that E2F transcription factors bind the E2F8 promoter in vivo and that expression of E2F8 is being induced at the G1/S transition. Purified recombinant E2F8 binds specifically to a consensus E2F-DNA-binding site indicating that E2F8, like E2F7, binds DNA without the requirement of co-factors such as DP1. E2F8 inhibits E2F-driven promoters suggesting that E2F8 is transcriptional repressor like E2F7. Ectopic expression of E2F8 in diploid human fibroblasts reduces expression of E2F-target genes and inhibits cell growth consistent with a role for repressing E2F transcriptional activity. Taken together, these data suggest that E2F8 has an important role in turning of the expression of E2F-target genes in the S-phase of the cell cycle.
INTRODUCTION
The E2F family of transcription factors are downstream effectors of the retinoblastoma protein (pRB) pathway and are essential for orchestrating the timely expression of a large number of genes required for cell-cycle progression and proliferation (1,2). In the majority of human cancers, the control of E2F transcriptional regulation is deregulated and is caused either by direct mutational inactivation of the RB1 gene or through aberrant expression of genes regulating pRB (3).
In G0/G1 of the cell cycle, the transcriptional activities of the E2Fs are restrained by members of pRB family, pRB, p107 and p130, and this suppression is dependent on the ability of these proteins to sequester and thereby repress E2F activity (4). Several lines of evidence have shown that E2Fs are required for S-phase entry (5,6). Overexpression of E2F is sufficient to induce quiescent cells to enter S-phase (6) and, more recently, it was demonstrated that E2F1-3 triple knockout cells are defective for S-phase entry (7). Furthermore, in Rb−/− mouse embryo fibroblasts the G1 phase is shorter than in wild-type cells.
Cell-cycle progression is driven by mitogenic growth signals, which result in the synthesis and accumulation of D-type cyclins. Associated cyclin-dependent kinases initiate the phosphorylation of the pRB family members, dissociating them from the E2F transcription factors. As a result, an auto-regulatory amplification loop of transcription is activated at the late G1/early S-phase of the cell-cycle resulting in increased transcriptional activity of E2F family members, which in turn activates the expression of a large number of genes involved in cell-cycle progression, proliferation and DNA replication (2). The transcriptional activity of the E2Fs oscillates through the cell cycle and to terminate and exit S-phase a decrease in E2F activity is required (8).
Mammalian cells express at least seven members of the E2F family (E2F1–7). In addition, several E2F isoforms are generated by alternative splicing for some of the E2F family members adding another layer of complexity to the functions of E2Fs (6). The E2F family can be divided into four subgroups on the basis of their structure, affinity for members of the pRB family, complex formation with members of the DP family, transcriptional function and expression pattern. E2F1–6 are characterized by the presence of two highly conserved domains necessary for sequence specific DNA binding and dimerization with DP proteins, respectively. E2F1–3a are considered to be transcriptional activators and, when overexpressed, can drive quiescent cells into S-phase (6). Their transcriptional repertoires are exclusively controlled by pRB and E2F1–3a expression is induced at the G1/S-phase of the cell cycle and directs the expression E2F-regulated genes. In contrast, E2F3b, E2F4 and E2F5 occupy E2F-regulated promoters in the G0/G1 phase of the cell cycle and interact with all three pRB family members, which repress transcription by recruitment of chromatin-remodelling complexes, histone- and DNA-modifying enzymes such HDAC and DNA methyltransferases (6). E2F4 and E2F5 lack nuclear localization signals and are excluded from the cell nucleus during S-phase, but relocalizes to the nucleus after complex formation with the pocket proteins (6). The third group is represented by E2F6, which is also a repressor of transcription that forms a structurally and functionally separate group within the E2F family. E2F6 lacks a transcriptional activation domain and sequences necessary for complex formation with members of the pRB family. Instead, E2F6 represses transcription through interactions with the Polycomb group of proteins (6). The exact role of E2F6 in transcriptional regulation is presently unclear, but recent data suggest that E2F6 specifically down-regulates E2F-target genes activated at the G1/S boundary of the cell cycle (9). E2F6 also appears to play a role in development and differentiation. Accordingly, a mild homeotic phenotype has been reported for E2F6−/− knockout mice (10). Recently, E2F7, a novel member of the E2F family was identified (11–13). E2F7 is structurally unique in that it harbours a tandem repeat of E2F DNA-binding domains. Furthermore E2F7 interacts with DNA independently of DP proteins. E2F7b expression is restricted to the S-phase of the cell cycle and has been shown to repress a subset of E2F-regulated genes.
Here we describe the identification of a novel member of the E2F family, E2F8. E2F8 shows a high degree of resemblance to E2F7 and shares the unique structure of E2F7 by having two distinct domains exhibiting a high degree of similarity to the DNA-binding domain of the E2F family. We show that E2F8 expression is cell-cycle regulated and is activated by E2Fs at G1/early S-phase of the cell cycle and that members of the E2F family occupy the E2F8 promoter in vivo. E2F8 binds consensus E2F sites in a DP-independent manner and represses transcription of E2F-regulated promoters. Ectopic expression of E2F8 inhibits cellular proliferation. Altogether, the restricted expression and repressive transcriptional function of E2F8 suggests a role for E2F8 in negative feedback loop repressing E2F-activated promoters. Our results extend and are consistent with two recent reports on the identification of mouse and human E2F8 (14,15), published while this manuscript was in preparation.
MATERIALS AND METHODS
Plasmids
The promoter reporter constructs encoding various promoters fused to the luciferase reporter gene and expression constructs used for transcriptional assays pGL3-6xE2F, pGL3-E2F1, pGL3-CDC6, pGL3-CyclinE1, pGAL-luc, pCMV-E2F1 and pCMV-E2F7, respectively, have been described previously (12,22,23).
cDNA cloning of human and murine E2F8
The putative open reading frames (ORFs) of human and murine E2F8 were amplified by PCR from HeLa cDNA and a ProQuest Mouse Embryo (day 8.5) cDNA library from Invitrogen (Carlsbad, CA), respectively, using primers: 5′–3′ and 5′–3′. PCR products were gel purified and cloned into pCR2.1-TOPO (InVitrogen). Three clones were sequenced on both strands to generate a consensus sequence. To generate expression vectors, hE2F8 was subcloned into the EcoRI site of pEntr3C, and mE2F8 was transferred into pDONR221. These are both Gateway compatible vectors (Invitrogen). Using these entry clones E2F8 was transferred into pCMV, pCMV-Ha, pCMV-myc and pBabepuro. A double mutant in the conserved DNA-binding domain of hE2F8 changing amino acids 118–119 from leucine-glycine to glutamate-phenylalanine was generated in pEntr3C using a standard mutagenesis method.
Real-time quantitative PCR (qPCR)
Total RNA was purified from U2OS and HeLaS3 cells using RNAeasy (Qiagen, West Sussex). Total RNA (2 µg) was reverse transcribed using a TaqMan reverse transcription reagents from ABI (NJ) according to the manufacturer's instructions. For RNA quantification, 150 ng of reversed transcribed total RNA were analysed by real-time PCR using SYBR Green PCR Master Mix and an ABI prism 7700 Sequence Detection system. All reactions were analysed in triplicates. Primer sequences for human E2F8, E2F7, β-actin, Cyclin E1, Cyclin A2 and Cyclin B1 are available upon request.
Tissue culture
HeLaS3 and U20S were grown at 37°C in 5% CO2. HeLaS3 cells were arrested in S-phase by a double thymidine block as has been described previously (24). After 24 h of plating cells at a density of 2 × 106cells per 15 cm dish, the cells were blocked with 2 mM thymidine for 17–18 h, released from the arrest for 9 h and arrested a second time with thymidine. After 18 h of incubation, the cells were released and collected at different time points. To obtain populations of cells in mitosis, cells were arrested in 2 mM thymidine for 17–18 h, released for 4 h and blocked in 100 ng/ml nocodazole for 12 h. Floating mitotic cells were collected, washed twice in 1× phosphate-buffered saline (PBS) and replated at a density of 4 × 106 cells in each 15 cm dish and followed for 12 h.
FACS analysis
HeLaS3 or U2OS cells were harvested, washed in PBS and fixed by addition of ice-cold ethanol to a final concentration of 75%. The cells were washed once in PBS and resuspended in PBS containing 10 µg/ml of propidium iodide, 0.25 mg/ml of RNaseA and incubated overnight at 4°C. For BrdU incorporation studies, briefly, human TIG3 cells were pulsed for 15 min with 0.033 mM of BrdU, washed in PBS, trypsinized and fixed in ethanol as described above. The cells were denatured for 20 min at room temperature in 2 M HCl. The cells were neutralized with natrium borate, washed in PBS and incubated with anti-BrdU antibody (BectonDickinson). Bound antibody was detected using a secondary FITC-conjugated anti-mouse IgG antibody. Subsequently, the cells were incubated in the presence of RNase and propidium iodide for 30 min at 37°C. The samples were analysed by flowcytometry.
Recombinant E2F8
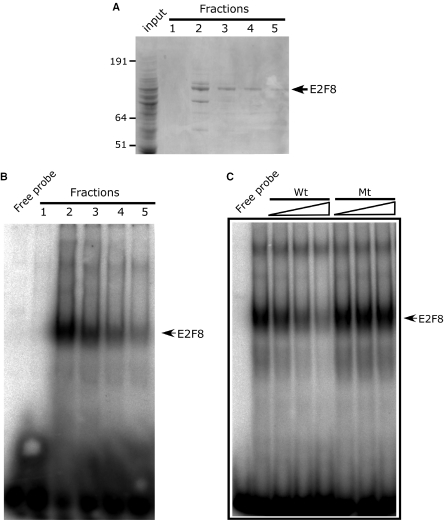
Full-length N-terminally hexahistidine-tagged human and murine E2F8 baculovirus transfer vectors were generated by excision of E2F8 cDNAs from pCR2.1topo and in frame subcloning into EcoRI digested pAcHLT-A. Recombinant baculoviruses were generated by co-transfection of baculovirus transfer vector containing the desired gene and Bsu36I linearized Bakpak6 baculovirus DNA essentially as described previously. Histidine-tagged E2F8 was expressed in Trichoplusia ni, High Five, cells by recombinant baculoviruses using a multiplicity of infection of 10. The cells were incubated at 28°C and harvested 40–44 h post infection, washed twice in PBS, resuspended in 25 mM HEPES–KOH, pH 7.6, 5 mM KCl and 1.5 mM MgCl2 and lysed by Dounce homogenization. After 20 min of incubation the lysates were adjusted to 350 mM NaCl and further incubated for 30 min and cleared by centrifugation for 30 min at 20 000 g. The supernatant was loaded on to a Cobalt–Sepharose column (Clontech) (1.5 ml of resin per 109 cells) equilibrated in buffer A (25 mM HEPES–KOH, pH 7.6, 0.5 mM MgCl2, 0.5 mM DTT and 10% glycerol) adjusted to 350 mM NaCl. The column was washed with the same buffer and eluted with buffer A adjusted to 100 mM NaCl and 100 mM imidazole. The eluted fractions were analysed by SDS–PAGE, flash frozen in liquid N2, and stored at −80°C. All procedures were performed on ice or at 4°C in the presence of complete EDTA-free protease inhibitor (Boehringer Mannheim).
Generation of antibodies to E2F8
Polyclonal antibodies were generated by immunizing rabbits with affinity-purified glutathione S-transferase fused to the C-terminal of murine E2F8 (amino acids 551–860).
Electrophoretic mobility shift assay (EMSA)
DNA-binding reactions were carried out in 25 µl of 20 mM HEPES–KOH (pH 7.8), 100 mM NaCl, 0.5 mM EDTA, 1 mM DTT and 10% glycerol supplemented with 0.5 µl of rabbit reticulocyte lysate and 50 µg of BSA/ml as non-specific protein carriers. Recombinant E2F8 were incubated for 10 min at room temperature in the presence of non-specific carrier DNA before addition of the radio-labelled probe (5 × 104 c.p.m.) and then incubated at room temperature for a further 20 min before being separated by electrophoresis in 0.25× TBE through 5% native polyacrylamide gels at 4°C for 3 h at 200 V. Gels were fixed in 10% acetic acid, dried, exposed and analysed by phosphor imaging. The sequences of the double-stranded (ds)-oligonucleotides used in the assays were: adenovirus E2 promoter E2F-site (E2Fwt), 5′-GATCAGTTTCGCGCCCTTTCTCAAGATC and corresponding mutant (E2Fmt), 5′-GATCAGTTTATATCCCTTTCTCAAGATC.
Chromatin immunoprecipitation (ChIP) assays
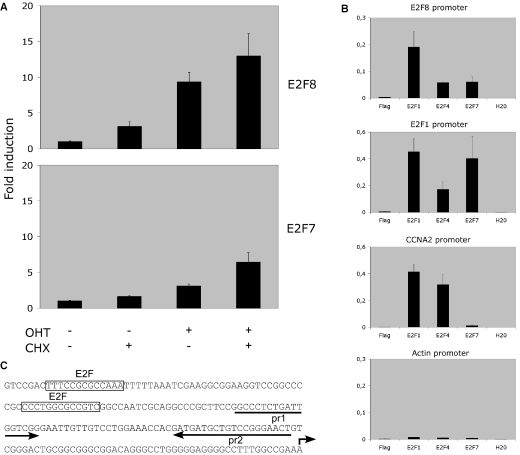
ChIP and data analysis were carried out as described previously. Briefly, human osteosarcoma U2OS cells or human TIG3 fibroblasts were cross-linked by the addition of formaldehyde to 1% final concentration, the reaction was stopped by addition of glycine, cells washed in TBS and harvested into SDS buffer. Following centrifugation, cells were resuspended in immunoprecipitation buffer and chromatin was sonicated to an average size of 250 ± 750 bp. Lysates were subsequently precleared with protein A–Sepharose beads. Precleared chromatin was incubated at 4°C overnight with antibodies specific for E2F1 (Sc-193), E2F3 (Sc-878), E2F4 (Sc-866) or E2F7 (12), or with an unrelated anti-Flag antibody (F3165; Sigma). Immunocomplexes were recovered with protein A–Sepharose beads. After extensive washes immunocomplexes were eluted from the beads, cross links reversed and material recovered by phenol/chloroform extraction and ethanol precipitation. The DNA was resuspended in 200 µl of water and 7.5 µl used per real-time qPCR using 200 nM of primers in 25 ml SYBR Green Reaction Mix. The primer sequences are for the E2F1 promoter, Cyclin A2 promoter and β-actin gene are available upon request. The primer sequences for the E2F8 promoter are indicated in Figure 5B.
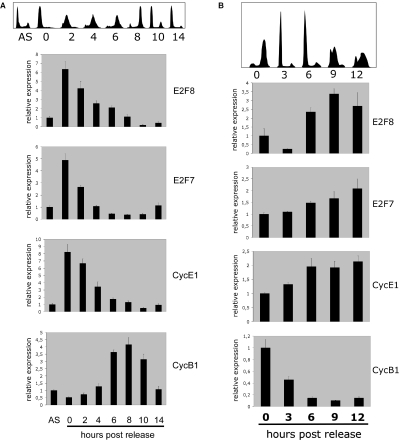
Figure 5.
E2F8 transcription is cell-cycle regulated. (A) HeLaS3 cells were synchronized to late G1/early S-phase by a double thymidine block. After release from the block, the cell-cycle profile was analysed by flow cytometry and total RNA was extracted for qPCR analysis at indicated time points. Top of the panel shows the histogram DNA profiles of propidium iodide stained cells. Asynchronous cells are labelled, AS, the time points for analysis are indicated below the figures. The E2F8, E2F7, Cyclin E1 (CycE1) and Cyclin B1 (CycB1) transcripts were quantified by qPCR (real-time PCR). The transcript levels were calculated relative to β-actin. All data were normalized to the transcript levels present in asynchronous HeLa cells. (B) U2OS cells were synchronized to late G2/early M-phase by a thymidine block followed by release into nocodazole-containing medium. After release from the block, the cell-cycle profile was analysed by flowcytometry and total RNA was extracted for qPCR analysis at indicated time points. Top of the panel shows the histogram DNA profiles of propidium iodide stained cells. The time points for analysis are indicated below the figures. After reverse transcription using random priming, the E2F8, E2F7, Cyclin E1 (CycE1) and Cyclin B1 (CycB1) transcripts were quantified by qPCR (real-time PCR). The transcript levels were calculated relative to β-actin. All data were normalized to the transcript levels present at the time of release.
Luciferase assays
To measure transcriptional activity, 4 × 104 human U2OS cells were seeded in 24-well tissue culture plates. Next day the cells were transfected with combinations of reporter plasmid and expression vectors as described in the figure legends using Lipofectamine 2000 (Invitrogen) as per the manufacturer's recommendations. The vector pCMV-LacZ expressing β-galactosidase was included as an internal control and used for normalization of the luciferase activities. Cells were harvested 24 h after transfection, and β-galactosidase and luciferase activity were measured essentially as described previously (12).
Retrovirus transduction and cell-growth experiments
To generate recombinant retroviruses expressing hE2F8 and hE2F8mt, the ORFs of hE2F8 and hE2F8mt from pEntr3C were transferred into pBabepuro by recombination generating pBabepuro-hE2F8 and pBabepuro-hE2F8mt. High titers of retroviral particles were obtained 24–48 h after transfection of the Phoenix-Eco 293 cell packing cell line. Transduction of the human TIG3-hTert-Eco was achieved by adding virus containing supernatants from the packaging cell line to the cell dishes four times within a 24 h period. Transduced cells were selected for 7 days in the presence puromycin (1 µg/µl). The puromycin resistant cells were seeded at selected densities and reseeded using a 3T3-like protocol as indicated in legends of the figures.
RESULTS
Identification and cloning of E2F8
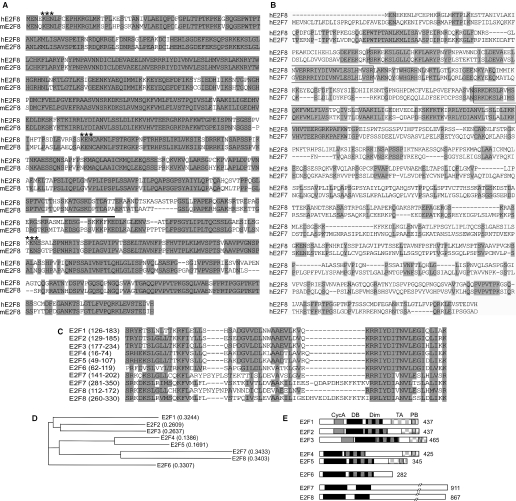
In a screen for E2F1-regulated genes, we have described recently the identification of an E2F-like gene, E2F7 (12). When searching GenBank using the E2F7 sequence as query, we noticed the existence of several human and murine expressed sequence tags and database accessions showing similarity to E2F7. Two accessions NM 024680 encoding Homo sapiens FLJ23311 protein and XM149937 encoding the Mus musculus RIKEN cDNA 4432406C08 gene, respectively, appeared to harbour full-length ORFs, since their respective start codons were preceded by in-frame stop codons. Amplification of these ORFs from HeLaS3 cDNA and a murine embryo cDNA library, respectively, resulted in single PCR products indicating no or a low abundance of alternative spliced transcripts in these tissues. Subsequent, cloning and sequencing of the PCR products resulted in consensus sequences identical to that of the FLJ23311 mRNA and RIKEN cDNA 4432406C08. The cloned transcripts, which we have designated E2F8, contained highly homologous ORFs capable of encoding proteins of 867 and 860 amino acids (identity 82.2%) with calculated molecular weights of 94 272 and 93 382 Da, respectively (Figure 1A). Alignment of human E2F7 and the human E2F8 (Figure 1B) showed an overall identity of 31.9%, with the highest conservation around the unique repeats of the two E2F-like DNA-binding domains found in E2F7. Comparison of the putative DNA-binding domains of E2F8 with other E2F members demonstrates the close relationship to E2F7 (Figure 1C). Phylogenetic analysis using a neighbour-joining (NJ) method of the human and murine E2F8 proteins based on either the full-length sequences or the sequences of the DNA-binding domains, respectively, suggest that these belong to the distinct subgroup of the E2F family presently represented by the two isoforms of E2F7 (Figure 1D). This subgroup is characterized by having two E2F-like DNA-binding domains (Figure 1E) and binds DNA independently of the DP proteins. In addition, none of the polypeptides encode the conserved sequences such as Cyclin A phosphorylation motifs, dimerization domains, pRB binding motif, marked box and transactivation domain present in the two other subgroups of the E2F family. The genomic organization of E2F8 is similar to E2F7 suggesting that they originate from a common ancestral gene. The human and murine E2F8 genes are located on chromosome 11p15.1 and short arm of chromosome 7, respectively. Both the human and murine E2F8 genes are predicted to consist of 13 exons with the putative start ATGs located in the second exons (data not shown). Together with the sequence similarity, the genomic structure strongly supports that the human and murine E2F8 genes are true orthologues.
Figure 1.
Analysis of the E2F8 ORF. (A) Alignment of the amino acid sequences predicted from the ORFs of human E2F8 and murine E2F8 using Vector NTI and a Clustal W algorithm. Identical residues are shaded in grey. A putative nuclear localization signal was identified using the ScanProsite program provided by the ExPASy World Wide Web molecular biology server of the Swiss Institute of Bioinformatics (SIB) (www.expasy.org) and is indicated by a black line. KEN boxes, which can mediate degradation through the ubiquitin-proteasome pathway, are indicated by asterisks. (B) Alignment of the amino acid sequences predicted from the ORFs of hE2F8 and hE2F7. Identical residues are shaded in grey. The highly conserved regions, which contain the two distinct DNA-binding domains aligned in (C) are boxed. (C) Alignment of the DNA-binding domains of E2F8 with the DNA-binding domains of the other E2F members. Identical and similar residues are shaded in grey. (D) Phylogenetic tree of E2F family based on NJ method of Saitou and Nei. The NJ method works on a matrix of distances between all pairs of sequence to be analysed. These distances are related to the degree of divergence between the sequences. The phylogenetic tree is calculated after the sequences are aligned. (E) Domain structure of E2F8 compared with the other members of the E2F family. Number of amino acids is indicated on the right. Shaded boxes indicate homologous regions. CycA, Cyclin A binding site; DB, DNA binding domain; Dim, dimerization domain; TA, transactivation domain; PB, pocket protein (pRB family) binding domain.
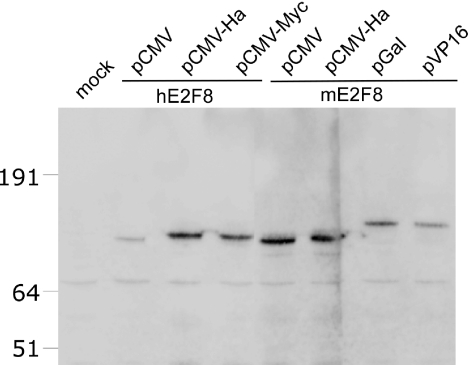
The expression of the putative ORFs of human and murine E2F8 were analysed by transient transfection of HeLa cells followed by western blotting analysis of cell lysates using a panel of expression vectors (Figure 2). Expression of E2F8 was detected using an anti-serum raised against the C-terminal of murine E2F8. In agreement with the predicted molecular weights, for both hE2F8 and mE2F8 major bands around 100 kDa were detected, which increased in size when the E2F8 was fused to the yeast GAL4 DNA-binding domain or Herpes simplex VP16. In addition, a number of bands with relatively lower molecular weights were observed, which probably represents E2F8 degradation products. In general, we found E2F8 to be very prone to degradation. Although, we observed bands in untransfected HeLa cells with similar molecular weight as the E2F8 expression products after long exposure of the blots. The non-specific reactions of the anti-sera hampered a reliable identification of endogenous expressed E2F8. Thus, the endogenous protein levels of E2F8 appeared relatively low, which is in agreement with the observed E2F8 mRNA levels as described in the following section. After transfection of U2OS cells with Ha- or myc-tagged E2F8 expression vectors and analysis by immuno-fluorescence microscopy, E2F8 exhibited localization to the nuclear compartment of the cells as judged by the overlapping staining with DAPI (data not shown).
Figure 2.
Expression of E2F8. Western blot analysis of HeLa cells transfected with different E2F8 expression constructs. HeLa cells were transiently transfected with expression vectors encoding untagged or N-terminally tagged versions of hE2F8 or mE2F8 as indicated. pCMV mediates expression of untagged hE2F8 or mE2F8, while pCMV-Ha, pCMV-Myc, pGal and pVP16 express E2F8 fused to a Ha-epitope tag, myc-epitope tag, yeast Gal4 DNA binding domain and herpes virus simplex VP16 transcriptional activation domain, respectively. Expression of E2F8 was detected using a rabbit polyclonal antibody against murine E2F8. The positions of a standard set of molecular weight markers are shown on the left.
E2F8 and E2F7 exhibit similar expression patterns
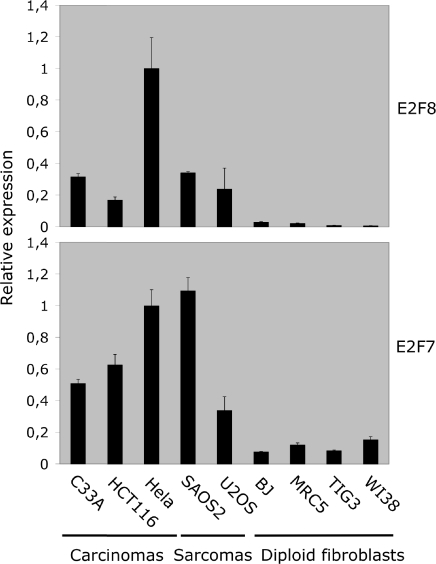
To determine the expression levels of E2F8, we screened cDNA from a panel of exponentially growing human cell lines by real-time qPCR for the expression of E2F8 and E2F7 transcripts (Figure 3). The two genes exhibited a parallel expression pattern and were more abundantly expressed in transformed cell lines of carcinoma or sarcoma origin consistent with being an E2F-target gene. The E2F8 and E2F7 primer sets used for qPCR exhibited similar amplification efficiency when compared using E2F8 and E2F7 on DNA templates indicating that the relative mRNA expression levels of E2F8 are, in general, ∼5–20% of the levels of E2F7.
Figure 3.
Expression of E2F8 and E2F7 in various cell lines. The mRNA levels of E2F8 and E2F7 were analysed by real-time qPCR. RNA was extracted from exponentially growing cell lines. Origin of cell lines: C33A, cervix carcinoma; HCT116, adenocarcinoma; HeLa, cervix carcinoma; SAOS2, osteogenic sarcoma; U2OS, osteogenic sarcoma; BJ; MRC5; TIG3; and WI38 are all diploid lung fibroblasts. The E2F8 and E2F7 transcripts were quantified by qPCR (real-time PCR). The levels of E2F8 and E2F7 transcripts were normalized relative to β-actin. All data were further normalized to the E2F8 or E2F7 levels present in HeLa cells.
E2F8 is an E2F-target gene
Several observations indicated that E2F8, in analogy to E2F7, could be an E2F-regulated gene: (i) the presence of relatively high levels of E2F8 expression in rapidly dividing cell lines; (ii) high level of expression in HeLaS3, where the pRB-E2F pathway is deregulated; and (iii) the clustering of putative E2F-binding sites around the putative transcription initiation site of the E2F8 promoter. To examine whether E2F8 is an E2F-target gene, we used U2OS cells stably expressing E2F1 fused to the estrogen receptor ligand-binding domain (ER-E2F1), which has been characterized previously (16,17). Briefly, in the presence of 4-hydroxytamoxifen (OHT) ER-E2F1 is translocated from the cytosol to nucleus of the cells and E2F-regulated genes are induced. After 4 h of OHT induction in the presence or absence of cycloheximide (CHX), total RNA was isolated and the levels of E2F8 transcripts were determined by qPCR (Figure 4A). An ∼10-fold up-regulation of E2F8 mRNA levels was observed. The magnitude of induction was comparable with that of E2F7. Since the up-regulation of transcript levels was observed in the presence of CHX, the increase in E2F8 transcript levels was independent of de novo protein synthesis indicating direct binding and activation of the promoter by ER-E2F1.
Figure 4.
E2F8 is an E2F-target gene. (A) Real-time qPCR analysis of mRNA isolated from U2OS cells stably expressing Estrogen receptor ligand-binding domain fused to E2F1. The cells were grown in the presence or absence of OHT and CHX for 4 h as indicated in the figure. The levels of E2F8 and E2F7 transcripts were normalized to β-actin. All data were further normalized to the levels E2F8 or E2F7 in non-treated cells. (B) ChIP analysis of E2F1, E2F4 and E2F7 binding to the E2F8 promoter. Asynchronously growing U2OS cells were treated with formaldehyde and enrichment of the E2F8 promoter sequences was tested by ChIP using the indicated antibodies. β-Actin, CCNA2 (Cyclin A2) and E2F1 promoters were used, respectively, as negative and positive controls. The percentage of the bound promoter versus total promoter present in the cells is indicated. (C) Sequence of the 5′ region of the putative human E2F8 promoter. A broken arrow indicates the position of the putative transcription initiation site. Boxed sequences represent consensus E2F-binding sites. The arrows labelled pr1 and pr2 show the positions of the primer set used for detection of enriched E2F8 promoter sequences in ChIP assays.
To gain further support for a role of the E2Fs in regulating E2F8 expression, ChIP experiments using specific antibodies to members of the E2F family were performed. As shown in Figure 4B and C, E2F1 and to a lesser extent E2F4 and E2F7 occupied the E2F8 promoter. In contrast to E2F1, E2F4 and E2F7 are considered to be repressors of transcription, their presence at the E2F8 promoter indicates that the promoter is regulated by both activating and repressing members of the E2F family. The CCNA2 and E2F1 promoters and β-actin gene were used as positive and negative controls, respectively. In addition, E2F3 was also found to occupy the E2F8 promoter as determined by ChIP assays in human TIG3 fibroblasts (data not shown). Taken together, these data strongly suggest that E2F8 transcription is directly regulated by members of the E2F family.
E2F8 transcription is cell-cycle regulated
The finding that E2F8 is a direct E2F-target suggests that E2F8 expression is cell-cycle regulated. To test this hypothesis, HeLaS3 cell were synchronized in early S-phase by a double thymidine block, while U2OS cells were synchronized to arrest in G2/M by a thymidine block followed by release into nocodazole-containing medium. The cells were released from the blocks and the DNA content of the cells was determined at different time points as indicated in Figure 5A and B by FACS analysis. Total RNA was purified, reverse transcribed and expression of E2F8, E2F7 and selected cyclin genes were quantitated by real-time PCR. For HeLaS3 cells, the relative level of E2F8 peaked at the time of release, early S-phase, which coincided with, two other E2F targets, E2F7 and Cyclin E1. As the cells progressed through S-phase, E2F8 transcript levels dropped and finally increased again when the cells had gone through mitosis and started re-entering S-phase. The expression of Cyclin B1 was included as a control for the synchronization of the cells. For the U2OS cells, which were arrested at G2/M, relatively low levels of E2F8 transcripts were detected at the time of release (Figure 5B). However, after mitosis and entry into the S-phase, a significant increase of E2F8 expression was observed. We also found that E2F8 expression is induced in human TIG3 fibroblasts after release from serum starvation (data not shown). Taken together, the combined cell synchronization data support the finding that E2F8 is an E2F-target gene, which is growth- and cell-cycle regulated, and is induced at the G1–S-phase transition of the cell cycle.
E2F8 binds to consensus E2F sites
The presence of two putative E2F DNA-binding domains highly homologous to domains of E2F7 suggested that E2F8 is also capable of associating with the E2F DNA-binding consensus site. To determine whether E2F8 can bind directly to E2F consensus sites, we expressed His-tagged E2F8 in insect cells using a recombinant baculovirus. Subsequently, recombinant His-tagged E2F8 was purified; fractions were analysed by SDS–PAGE (Figure 6A) and tested for site-specific DNA binding by EMSAs using a 32P-labelled ds-oligonucleotide encoding the adenovirus E2 E2F-binding site as target. A major band revealed by Coomassie staining with the predicted molecular weight of his-E2F8 was observed in fractions 2–5. This band was recognized by an E2F8 anti-serum when analysed by western blotting (data not shown) confirming the identity of the purified expression product. The purified fractions were tested by EMSA and gave rise to a slower migrating complex with a signal intensity paralleling the levels of E2F8 present in the fraction (Figure 6B). The binding was efficiently competed by addition of increasing amounts of unlabelled E2F ds-oligonucleotide (Wt), while a mutation of the core GCGC sequence of the ds-oligonucleotide (Mt) abolished its inhibitory activity (Figure 6C). Altogether, these data strongly suggest that E2F8 by itself binds directly to E2F consensus sites.
Figure 6.
Recombinant E2F8 binds the E2F DNA-binding consensus site. (A) SDS–PAGE analysis of recombinant his-tag purified E2F8 expressed in insect cells. Input is the cleared lysate used for chromatography. Fractions 1–5 denote the fractions eluted from chromatography column. An arrow at the right of the gel indicates the position of recombinant E2F8, and positions of a standard set of molecular weight markers are shown on the left. The gel was stained with Coomassie brilliant blue. (B) EMSA analysis of purified E2F8 containing fractions. One microlitre of each fraction (100–10 ng) shown in (A) were incubated with a 32P-labelled ds-oligonucleotide containing the consensus E2F-site derived from the Adenovirus E2 promoter. (C) Specificity of E2F8 binding by EMSA. Recombinant E2F8 (100 ng) was incubated in the presence of 32P-labelled ds-oligonucleotide containing the consensus E2F site and 3, 9 or 27 molar excess of unlabelled ds-oligonucleotide or a ds-oligonucleotide abolishing the core GCGC sequence of the Wt ds-oligonucleotide and analysed by an EMSA.
E2F8 is a repressor of E2F-activated transcription
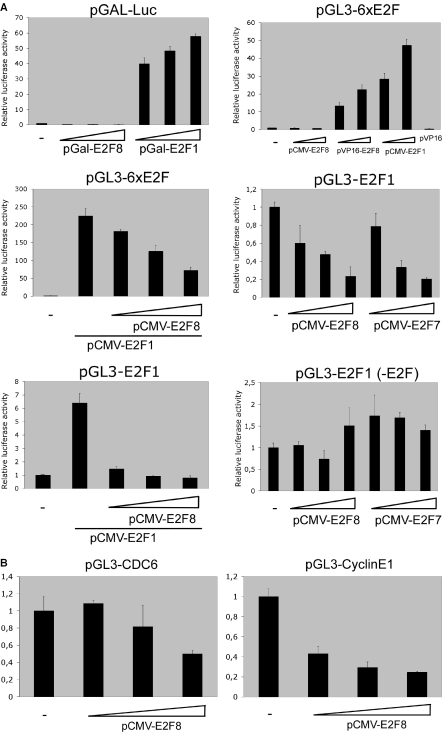
To determine a role for E2F8 in E2F transcriptional regulation, we generated a fusion protein containing E2F8 fused to the yeast GAL4-DNA binding domain. The GAL4 DNA-binding domain targets E2F8 independently of its intrinsic DNA binding activity to GAL4 binding sites. To test its activity we transfected U2OS cells with a plasmid containing five Gal4 binding-sites fused to a luciferase reporter gene with the GAL4-E2F8 expression plasmid. A β-galactosidase expression vector was co-transfected and used for normalization of the data. The GAL4 DNA-binding domain fused to E2F1 was included as a positive control (18). In addition, expression constructs encoding E2F8 or E2F8 fused to transcriptional activation domain, VP16, were tested for their ability to transactivate a reporter plasmid containing an array of six E2F sites (Figure 7A). In both assays no increase in luciferase activity was observed indicating that E2F8 lacks the capacity to function as a transcriptional activator. However, when E2F8 was fused to VP16, a concentration-dependent increase of luciferase activity comparable with E2F1 using the pGL3-6xE2F reporter was observed, demonstrating that E2F8 binds the promoter of the reporter gene. Together, these data support the idea that E2F8 is a repressor of transcription.
Figure 7.
E2F8 is a repressor of E2F-responsive promoters. (A) U2OS cells were transfected with 100 ng of the reporter constructs indicated at the top of the panel. The pGAL-Luc reporter plasmid contains the luciferase gene driven by the adenovirus E1B minimal promoter (TATA) fused to five upstream GAL4-binding sites and was co-transfected with increasing amounts of pGAL-E2F8 or pGAL-E2F1 (10, 30 and 90 ng) of expression plasmid, respectively. pGL3-6xE2F contains six consensus E2F sites upstream of a TATA box. The pGL3 E2F1 (−242) contains 242 nt upstream of the transcription initiation site of the human E2F1 promoter linked to a luciferase reporter. pGL3 E2F1 (−242-E2F) is identical to pGL3 E2F1 (−242) except that the E2F-binding sites are mutated. These reporters were co-transfected with 30, 80 or 240 ng of pCMV-E2F8 or pCMV-E2F7b in the presence or absence of constant amounts (30 ng) of pCMV-E2F1 expression plasmid as indicated below the panel. The amount of expression plasmid was kept constant in all assays by addition of empty pCMV vector. For correction of transfection efficiency, 100 ng of pCMV-lacZ was included in all assays and luciferase activity was normalized to β-galactosidase activity. All experiments were performed in triplicate and reproduced at least three times. (B) U2OS cells were transfected with 100 ng of the reporter constructs indicated at the top of the panel. pGL3-CDC6 contains −1524 to + 225 bp of the human CDC6 promoter and pGL3-CyclinE1 contains −207 to 79 bp of the human CCNE1 (Cyclin E1) promoter, respectively. These reporters were co-transfected with 30, 80 or 240 ng of pCMV-E2F8. For normalization of transfection efficiency, 100 ng of pCMV-lacZ was included in all assays as described above.
To test this hypothesis, we measured the ability of E2F8 to inhibit E2F1-dependent transactivation. As shown in Figure 7A, E2F8 represses in a concentration-dependent manner, comparable with E2F7, the transactivation of a synthetic promoter construct and the E2F1 promoter by E2F1. As a control for specificity, E2F8 and E2F7 expression constructs were unable to repress transcription from the E2F1 promoter construct having mutations in the E2F-binding sites. Also, the Cyclin E1 and CDC6 promoters, which are known targets for E2F regulation, were tested for E2F8-mediated repression (Figure 7B). Albeit the level of transcriptional down-regulation differed, all tested promoters were repressed proportional to the amount of input E2F8 expression vector. These results demonstrate that E2F8 can repress E2F1-activated transcription probably by competing or displacing activating E2Fs from promoter binding.
Ectopic expression of E2F8 inhibits cell proliferation
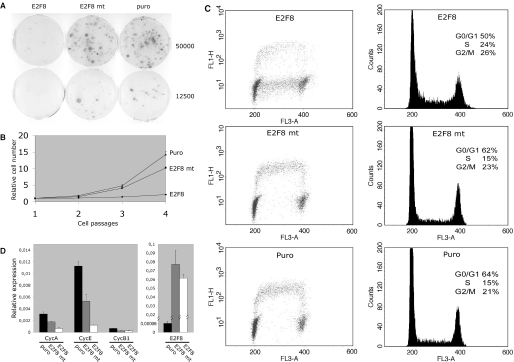
E2F1–3 are required for S-phase entry and are key players in mediating transcriptional activation of genes involved in DNA synthesis and cell-cycle progression. We have shown previously that ectopic expression of E2F7 affects cell proliferation and given the homology between E2F7 and E2F8, we would predict that ectopic expression of E2F8 could inhibit cell proliferation. To test this hypothesis, human TIG3 fibroblasts were transduced by retroviruses carrying a puromycin resistance gene and the full-length E2F8 ORF or an E2F8 (E2F8mt) mutant carrying a double mutation in the DNA-binding domain. As a control, a retrovirus only harbouring the puromycin resistance gene was included. After, 7 days of puromycin selection, the cells were plated and cell growth was evaluated in a colony forming assay or monitored by a 3T3 protocol. Only few cell colonies were observed for E2F8 transduced cells when compared with E2F8 mt and the Puro control demonstrating that ectopic expression of E2F8 inhibits cell proliferation (Figure 8A). Accordingly, only modest growth, with a tendency of slightly increased growth after each passage, was observed in a 3T3 assay ∼2.5 doublings for E2F8 over a period of four cell passages compared with ∼10- to 15-fold for E2F8 mt or Puro, respectively (Figure 8B). To analyse the inhibitory function of E2F8 in more detail, de novo DNA synthesis was determined by pulsing the transduced cells with BrdU and total DNA was stained with propidium iodide before FACS analysis. For both E2F8 mt and Puro transduced cells, a similar proportion of cells were present in the different phases of the cell cycle and all cells in S-phase also incorporated BrdU showing active DNA synthesis. In contrast, E2F8 transduced cells had an accumulation of cells in the S-phase of cell cycle and the major part of these were not incorporating BrdU indicating the absence or low level of DNA synthesis. The population of cells incorporating BrdU probably represents a subset of cells, which are expressing no or low levels of E2F8, since these become more abundant-dependent on the passage number. Also the expression of E2F8 slowly decreases after passage of the cells, probably because of the counter selection in growing cells (data not shown). The lack of DNA synthesis might be a consequence of down-regulation of E2F-target genes involved in DNA synthesis or cell-cycle control. Thus, to determine whether ectopic E2F8 expression affected the expression of E2F-target genes, the levels of Cyclin A2, Cyclin E1 and Cyclin B1 mRNA relative to β-actin were quantified by qPCR (Figure 8C). While transduction of cells with E2F8 mt resulted in a decrease compared with the Puro control, a major reduction was observed for all transcripts of E2F8-transduced cells indicating that E2F8 represses transcription of E2F-target genes. Together, the data suggests that ectopically expressed E2F8 directly binds the promoters and blocks transcription of E2F-target genes.
Figure 8.
Ectopic expression of E2F8 inhibits cell proliferation. (A) Ectopic expression of E2F8 in human TIG3 fibroblasts inhibits cell proliferation. Human TIG3-tert-ecoR fibroblasts were transduced with retroviruses encoding E2F8 (E2F8), an E2F8 mutant (E2Fmt) or virus without insert (puro). After puromycin selection, 50 000 or 12 500 of transduced cells were plated in selective medium and, after 3 weeks of incubation, the colonies were stained using crystal violet. (B) Ectopic expression of E2F8 in human TIG3 fibroblasts inhibits cell proliferation. Growth curve using a 3T3 protocol of TIG3 fibroblasts transduced with retroviruses encoding E2F8, an E2F8 mutant or a virus without insert (puro). (C) Ectopic expression of E2F8 in TIG3 fibroblasts inhibits DNA synthesis and causes S-phase accumulation. TIG3 fibroblast were transduced and selected as described above. 800 000 stably transduced cells were plated in 10 cm dishes and after 3 days of incubation the cells were pulsed with BrdU, stained with propidium iodide and analyzed by flowcytometry. The transduced gene is indicated in the top of each panel. The left panels are dot plot analysis of BrdU stained cells, while the right panels represent the corresponding histograms. (D) Ectopic expression of E2F8 represses E2F-target genes and Cyclin B1. TIG3 fibroblast were transduced and selected as described above. 800.000 stably transduced cells were plated in 10 cm dishes and, after 3 days of incubation the cells were harvested. Total RNA was extracted for qPCR analysis. After reverse transcription using random priming, the E2F8, Cyclin A2, Cyclin E1 and Cyclin B1 transcripts were quantified by qPCR (real-time PCR). The transcript levels were normalized relative to β-actin. Left panel shows relative Cyclin A2, Cyclin E1 and Cyclin B1 levels. The transduced gene is indicated below each bar in the histogram. Right panel shows the relative transcript levels of endogenous E2F8 compared with levels transduced by the retroviruses carrying E2F8 or E2Fmt, respectively.
DISCUSSION
The E2F family of transcription factors is essential for orchestrating the expression of cellular genes necessary for proliferation and cell-cycle progression (2,5). Until recently, seven members of the mammalian E2F family have been described (6). These members can be divided into four subgroups based on their phylogenetic relationship and function. In this report we describe the identification of yet another putative member of the E2F family, E2F8. Based on its unique structure, sequence homology and function, E2F8 appears to belong to the E2F subgroup of the E2F family currently represented by E2F7. A similar subfamily of transcription factors E2L1–3 has been described recently in Arabidopsis thaliana suggesting evolutionary conservation. The members of this subgroup are repressors of E2F-mediated transcription and are characterized by sharing a distinct structure different from the other E2Fs. It contains a tandem repeat of sequences showing high homology with the consensus E2F DNA-binding domain. Another feature of this subgroup is the ability to bind consensus E2F DNA-binding sites in the absence of DP co-factors (11–13).
The structure of the DNA binding domains of E2F7 and, recently E2F8, has been predicted by computational modelling to consist of three α-helices and a β-sheet and forms a homodimeric winged helix structure similar to that of the E2F-DP heterodimer (11–15,19). Here we have shown that purified recombinant E2F8 binds E2F sites suggesting that E2F8 also binds DNA as a homodimer in the absence of co-factors. The possibility that a factor associating with E2F8 could potentially be present in insect cells and contributes to binding cannot be ruled out. Although, it seems very unlikely because of the vast overexpression of E2F8 in insect cells and no detectable E2F-binding activity was observed in mock-infected cells (data not shown).
Together with pocket proteins, the E2F family controls transcription of a variety of growth and cell-cycle related genes in the different phases of the cell cycle. In the present study, using different cell synchronization protocols, we demonstrate that E2F8 is an E2F-target gene and the peak of transcription is confined to the late G1/early S-phase of the cell cycle. The transcriptional kinetics of E2F8 parallels that of Cyclin E1 and E2F7, and basal levels are reached concomitant with the peak of Cyclin A2 (data not shown) indicating that the functional role of E2F8 is restricted to S-phase. ChIP analysis demonstrated that both activating and repressing members of the E2F family occupy the E2F8 promoter in vivo suggesting that the tight control of E2F8 expression in S-phase is mediated by E2Fs. Owing to the low levels of E2F8 expression, we were unable to detect endogenously expressed E2F8 with the available antibodies we currently have. However, as mentioned previously, our observations suggest that the E2F8 protein, such as E2F7b, has a short half-life and its expression is limited to S-phase. The short half-life may be explained by the presence of two or three ‘KEN’ motifs present in E2F8, which are conserved in E2F7. Such motifs are present in substrates of the APC/C complex and target proteins for degradation upon exit of mitosis and in non-proliferating cells (20).
E2F8 represses a variety of E2F-responsive promoters in transient transfection assays. Furthermore, ectopic expression of E2F8 causes down-regulation of E2F-target genes and cell-cycle arrest. In contrast, a DNA-binding domain mutant of E2F8 exhibits no cell-cycle arrest and impaired repression of E2F-target genes, suggesting that E2F8 binds to a variety of E2F-target genes and that repression is mediated by direct binding to E2F-dependent promoters. Of note, the ectopic expression of E2F8 ablates cell-cycle progression and DNA replication in a major proportion of cells in S-phase, which probably reflects the down-regulation of E2F-target genes required for DNA synthesis. Taken together, these observations demonstrate that, when overexpressed, E2F8 acts as a general repressor of E2F-activated transcription. Previously, E2F7 was shown to be involved in the regulation of a subset of E2F-regulated genes (12). When taking into consideration that endogenous E2F8-expression levels are 5- to 20-fold lower than E2F7 in the cell lines we have analysed, it would indicate that E2F8 has a more restricted role in gene regulation. Other E2Fs have been shown to interact with promoters in a combinatorial fashion, where activator E2Fs are dependent on an adjacent CCAAT sites that is bound by the NF-Y transcription factor and binding of a repressor E2Fs are dependent on an adjacent CHR element in certain promoters. In general, it has been proposed that combinatorial interactions involving E2F proteins provide a basis for the specificity of transcription control in the pRB/E2F pathway (21). Some recent data have provided some candidate target genes potentially modulated by E2F repressors. E2F6, which modulates transcription through the recruitment of Polycomb group genes, specifically appears to bind and repress E2F-activated genes at the G1/S transition during S-phase (9). Similarly, E2F7 occupies promoters and represses genes expressed early in the S-phase such as E2F1, CDC6 and CCNE1 while it does not bind to the promoters of CDC2 and CCNA2. The common theme from these studies is that repression of early E2F targets might be necessary for the balanced and timely regulation of genes involved in S-phase traversal. Whether E2F8 regulates an overlapping or specific subset of genes in the G1/S transition or modulates a different repertoire of genes is, presently, unclear. A conclusive determination of E2F targets regulated by endogenous expressed E2F8 will require the development of ChIP grade E2F8 antibodies and a detailed analysis of E2F8 occupancy at E2F-target genes in vivo.
Several of the E2Fs appear to have overlapping functions suggesting E2F7 and E2F8 also might have redundant functions. However, they show very little conservation outside the DNA-binding domains and only short stretches of motifs in the N- and C- termini are similar indicating specific functions of the two polypeptides. The molecular mechanism of repression for E2F7 and E2F8 has not been elucidated. We have evidence that E2F7 binds a repressor through specific motifs in the C-terminal (L. D. Stefano and K. Helin, unpublished data). These motifs are not present in E2F8 suggesting a different mechanism of gene repression and a distinct role in gene regulation. Experiments are in progress directed at understanding the biological function, identifying target genes and the mechanisms of repression of E2F8.
Acknowledgments
We thank members of the Helin laboratory for fruitful discussions. E.T. was supported by an EMBO fellowship. This work was supported by grants from Lundbeckfonden, the Danish Cancer Society, the Association for International Cancer Research and the Associazione Italiana per la Ricerca sul Cancro. Funding to pay the Open Access publication charges for this article was provided by the grants mentioned above.
Conflict of interest statement: None declared.
REFERENCES
- 1.Blais A., Dynlacht B.D. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Bracken A.P., Ciro M., Cocito A., Helin K. E2F target genes: unraveling the biology. Trends Biochem. Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nature Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 4.Dimova D.K., Dyson N.J. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 5.Trimarchi J.M., Lees J.A. Sibling rivalry in the E2F family. Nature Rev. Mol. Cell. Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 6.Attwooll C., Denchi E.L., Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L., Timmers C., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., et al. The E2F1–3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 8.Krek W., Xu G., Livingston D.M. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 9.Giangrande P.H., Zhu W., Schlisio S., Sun X., Mori S., Gaubatz S., Nevins J.R. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storre J., Elsässer H.-P., Fuchs M., Ullmann D., Livingston D.M., Gaubatz S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002;3:695–700. doi: 10.1093/embo-reports/kvf141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruin A., Maiti B., Jakoi L., Timmers C., Buerki R., Leone G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2003;278:42041–42049. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- 12.Di Stefano L., Jensen M.R., Helin K. E2F7, a novel E2F featuring DP independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan N., Delavaine L., Graham A., Reilly C., Wilson J., Brummelkamp T.R., Hijmans E.M., Bernards R., La Thangue N.B. E2F-7: a distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene. 2004;23:5138–5150. doi: 10.1038/sj.onc.1207649. [DOI] [PubMed] [Google Scholar]
- 14.Logan N., Graham A., Zhao X., Fisher R., Maiti B., Leone G., Thangue N.B. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene. 2005;24:5000–5004. doi: 10.1038/sj.onc.1208703. [DOI] [PubMed] [Google Scholar]
- 15.Maiti B., Li J., de Bruin A., Gordon F., Timmers C., Opavsky R., Patil K., Tuttle J., Cleghorn W., Leone G. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2005;280:18211–18220. doi: 10.1074/jbc.M501410200. [DOI] [PubMed] [Google Scholar]
- 16.Vigo E., Muller H., Prosperini E., Hateboer G., Cartwright P., Moroni M.C., Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller H., Bracken A.P., Vernell R., Moroni M.C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J.D., Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helin K., Harlow E., Fattaey A.R. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng N., Fraenkel E., Pabo C.O., Pavletich N.P. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 1999;13:666–674. doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ang X.L., Harper J.W. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci. STKE. 2004;2004:pe31. doi: 10.1126/stke.2422004pe31. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W., Giangrande P.H., Nevins J.R. Temporal control of cell cycle gene expression mediated by E2F transcription factors. Cell Cycle. 2005;4:633–636. doi: 10.4161/cc.4.5.1650. [DOI] [PubMed] [Google Scholar]
- 22.Hateboer G., Wobst A., Petersen B.O., Le Cam L., Vigo E., Sardet C., Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helin K., Wu C.-L., Fattaey A.R., Lees J.A., Dynlacht B.D., Ngwu C., Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 24.Ballabeni A., Melixetian M., Zamponi R., Masiero L., Marinoni F., Helin K. Human Geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]