Abstract
DNA polymerase γ, Pol γ, is the key replicative enzyme in animal mitochondria. The Drosophila enzyme is a heterodimer comprising catalytic and accessory subunits of 125 kDa and 35 kDa, respectively. Both subunits have been cloned and characterized in a variety of model systems, and genetic mutants of the catalytic subunit were first identified in Drosophila, as chemically induced mutations that disrupt larval behavior (tamas). Mutations in the gene encoding the accessory subunit have not yet been described in any organism. Here, we report the consequences of null mutations upon mitochondrial DNA (mtDNA) replication and morphology, cell proliferation, and organismal viability. Mutations in the accessory subunit cause lethality during early pupation, concomitant with loss of mtDNA and mitochondrial mass, and reduced cell proliferation in the central nervous system. Surprisingly, the function of the central nervous system and muscle, as assessed in a locomotion assay, are only marginally affected. This finding is in contrast to our previous findings that disruption in the function of the catalytic subunit causes severe reduction in larval locomotion. We discuss our results in the context of current hypotheses for the function of the accessory subunit in mtDNA replication.
The cellular requirement for energy varies during development and as a consequence of different physiological signals. The differential demand is met by changes in mitochondrial proliferation and function (1). In turn, modulation of mitochondrial biogenesis depends upon gene products encoded by the mitochondrial and nuclear genomes. One important aspect of mitochondrial proliferation is appropriate mitochondrial DNA (mtDNA) replication and expression, carried out by nuclear-encoded gene products. Therefore, mitochondrial biogenesis and maintenance requires the integrated expression of two distinct genomes as well as the coordinated replication of the mitochondrial genome. A link between the cell cycle and mitochondrial biogenesis has been demonstrated recently in Drosophila. Expression of the genes encoding the mitochondrial single-stranded DNA-binding protein (lopo), mitochondrial transcription factor A (D-mtTFA), and the accessory subunit of the mitochondrial DNA polymerase (pol γ-β) depend upon the presence of binding sites for the transcription factor DREF (2–4). This factor has been shown to play a role in the orchestrated expression of genes involved in cell cycle control and nuclear DNA replication (5, 6).
Recently, mitochondrial function has emerged as an essential component of different apoptotic pathways. Several signals that lead to cell death are integrated by the mitochondria and lead to the release of cytochrome c (7). Toxic insults of different origins including hypoxia may damage the mitochondria, triggering changes in mitochondrial permeability and, thereby, activating programmed cell death. Consistent with these findings, mitochondrial dysfunction has been reported as causing or contributing to a wide variety of neural degenerative diseases and encephalomyopathies (8). For example, expression of mutant superoxide dismutase in transgenic mice, an animal model for amyotrophic lateral sclerosis, causes massive mitochondrial degeneration and subsequent motoneuron death (9). Similarly, dopamine oxidation products have been shown to open the mitochondrial permeability transition pore, thereby implicating mitochondrial dysfunction in the neuronal cell death observed in Parkinson's disease (10).
Of particular interest among the various diseases caused by inherited mitochondrial dysfunctions are the encephalomyopathies characterized by mtDNA depletion that are caused by mutations in nuclear genes. The importance of mtDNA metabolism in this subgroup of mitochondrial disorders is underscored by the recent finding (11) that recessive and dominant forms of progressive external opthalmoplegia are associated with mutations in the catalytic subunit of mitochondrial DNA polymerase, Pol γ. Similar mutations were first reported in Drosophila melanogaster. These were recovered as chemically induced mutations that disrupt larval behavior (tamas) and were shown to have an overall cell proliferation, cell growth defect, and a marked reduction in the mitochondrial mass in the central nervous system (CNS; ref. 12). Amongst the lesions described in both humans and Drosophila were amino acid substitutions in the polymerase domain and in regions of the enzyme not yet associated with a specific biochemical function.
Another essential component of the mtDNA replication machinery that functions in helix destabilization is the mitochondrial single-stranded DNA-binding protein, mtSSB. Mutations in the mtSSB gene (low power, or lopo) were identified in Drosophila as a P element insertion that disrupts the development of the adult visual system. lopo mutants show a defect in cell proliferation concomitant with a drastic decrease in mitochondrial DNA content and loss of respiration (13). These studies substantiate the usefulness of Drosophila as a model for the study of mtDNA replication, in the context of the organismal requirement for mitochondrial DNA maintenance and biogenesis.
In animal mitochondria, Pol γ is the sole polymerase responsible for mtDNA replication. It is a highly accurate and processive enzyme, comprising a catalytic subunit (α) and an accessory subunit (β). Both subunits have been cloned and characterized in a variety of systems, including Drosophila (14, 15). The catalytic subunit contains both 5′→3′ DNA polymerase and 3′→5′ exonuclease activities. Biochemical studies have demonstrated that the accessory subunit is essential for the catalytic efficiency and the high processivity of the holoenzyme (16–20). Structural and functional studies support the hypothesis that the accessory subunit has an additional role as a primer recognition factor (21–23). A cellular requirement for the accessory subunit has not yet been demonstrated, because mutations in the gene encoding it have not been described in any organism.
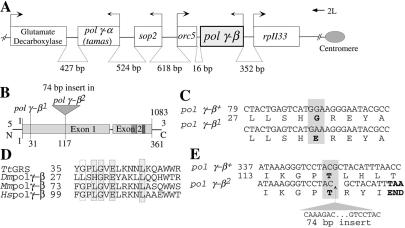
The gene encoding the accessory subunit of Pol γ in Drosophila (pol γ-β) is located in the 34D subdivision of the left arm of the second chromosome, 3.8 kb distal from the gene encoding the catalytic subunit (pol γ-α/tamas; refs. 12 and 15). This chromosomal microregion has been mutagenized extensively and studied by Ashburner and coworkers (24) and represents the first sequenced 3-Mb segment of the Drosophila genome. It is characterized by a gene density considerably above the average for the whole genome (refs. 4 and 25; Fig. 1A).
Figure 1.
Molecular characterization of mutations in the accessory subunit gene of Drosophila Pol γ. (A) Schematic representation of the ≈12-kb micro region that contains the tightly packed ORFs of six genes, including the mitochondrial DNA polymerase catalytic subunit pol γ-α and its accessory subunit pol γ-β. Arrows indicate the direction of transcription; distances between the ORFs are indicated below the schematic. (B) Key features in the pol γ-β ORF. The gene contains two exons; the positions of the sequenced mutations are indicated. (C) Nucleotide and amino acid sequence alignments of the parental strain with the pol γ-β1 mutant allele. The base substitution at nucleotide 92 changes amino acid 31 from glycine, a highly conserved amino acid, to glutamic acid. (D) Amino acid sequence alignment within motif 1 of Thermus thermophilus Glycyl-tRNA synthetase with D. melanogaster, mouse, and human Pol γ-β. The asterisk denotes the residue that is mutated in the pol γ-β1 allele. The unshaded box represents a similar amino acid, and the shaded boxes indicate conserved amino acids among all four proteins. (E) Nucleotide and amino acid sequence alignments of wild-type strain with the pol γ-β2 mutant allele. The pol γ-β2 allele contains a 74-bp insertion after nucleotide position 350, within the codon for threonine 117 within exon 1. This insertion results in premature termination of the translational product.
To evaluate the role of the accessory subunit in mtDNA synthesis and in mitochondrial function in vivo, we sought to identify mutations in the gene encoding it. The superimposition of the physical map (represented by the DNA sequence) with the genetic map (represented by the ordered complementation groups as determined by classical genetic analysis) allowed the identification of candidate mutations in the pol γ-β gene. Here, we report the molecular and phenotypic characterization of these mutations that serve to demonstrate for the first time the essential function of the accessory subunit of DNA polymerase γ in animals.
Materials and Methods
Fly Stocks and Culture.
Alleles of the 34De complementation group and a deficiency stock Df(2L)b80c1 were obtained from the Bloomington Drosophila Stock Center at Indiana University. The 34De1 parental stock was kindly provided by John Roote (Univ. of Cambridge, U.K.); the tamas (pol γ-α) alleles used in this study have been described earlier (12). The following are the genotypes of stocks that were used in this study: (i) l(2)34De1 AdhUF rds pr1 cn1/CyO(y+); (ii) In(2LR)Gla, wgGla-1 l(2)34De2/CyO(y+); (iii) yw; tam9/CyO(y+); (iv) b tam2 Adhn4/CyO(y+); (v) Canton-S; (vi) yw;Df(2L)b80c1/CyO(y+); (vii) yw;+/+;Hs-34De+; (viii) yw;S/CyO(y+); (ix) +;Sp/Sm5;Ly/Tm3Sb.
Culture medium was made of inactivated yeast, sucrose, 10% tegosept made in ethanol and 1 ml acid mix (10 parts of propionic acid:1 part of phosphoric acid per 100 ml of fly culture medium). For behavioral assays, the culture medium was supplemented with 1.25 g of β-carotene per liter of culture medium. Crosses were set with 5–10 pairs of male and virgin female flies in 100 × 25 mm vials; parents were allowed to lay eggs for 4–5 days.
Complementation Analysis.
The genetic map of the 34D region (12) (John Roote, personal communication) suggested that alleles of 34De lethal complementation group were likely candidates for mutations in the pol γ-β subunit gene. The deficiency stock Df(2L)b80c1 with breakpoints (34D3:34E2) removes genes l(2)34Db-Ance (24). The chromosomes containing the lethal alleles and the deficiency stocks were crossed into a yw;CyO(y+) background by using a yw;S/CyO(y+) stock followed by selection against Star phenotype.
To rescue the lethality associated with the mutation in pol γ-β1 allele, we crossed a transgenic line containing HS-pol γ-β+ (insert in the third chromosome) to the 34De1(pol γ-β1) fly stock. This transgene contained a miniwhite gene that confers orange eye color in a mutant white eye color background. Thus, in the genetic cross yw/yw; pol γ-β1/CyO; HS-pol γ-β+ × yw/Y; Df(2L)b80c1/CyO(y+), we observed that of 115 flies that were scored, all straight-winged flies (n = 31) exhibited the orange eye color. A heat shock stimulus was not necessary.
Larval Behavior Assay.
Eggs were collected overnight and larvae hatching in a 3-h window were picked after a 24-h incubation at 25°C. Larvae were allowed to grow in β-carotene-supplemented medium for 80–90 h in a 12 h light/12 h dark cycle. Locomotory tests were conducted on third instar larvae under safelight (20 watt lamp with Kodak GBX-2 filter). The larva was picked with a fine brush from the culture medium and was rinsed with distilled water. It was then placed on a 1% agar surface for 1 min to allow acclimation and to dislodge water. The locomotory behavior was assayed by measuring the distance traveled by the larva in 30 s. The distance traveled by the larva was measured in pixel units with a semiautomatic tracking system that was driven through a macro program written in NIH IMAGE (available on request).
Fluorescence Staining for Mitochondria and Nucleus in the Larval Brain.
Staining was performed as described in (12) with the following modifications. After dissection in PBS, individual brains were immediately transferred to a glass tube containing Mitotracker CMX-Ros (M-7512, Molecular Probes) at 1 μg/ml concentration in PBS. After 10 min, the brains were transferred to a glass tube containing 100 ng/ml of 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 3–4 min. The mitochondrial and nuclear profiles were visualized separately by using the Texas red and DAPI filters, respectively. The images were captured by using a SPOT charge-coupled device (CCD) camera, and the images were merged by using the SPOT imaging software (Diagnostic Instruments, Sterling Heights, MI).
5′-Bromo-2′-Deoxyuridine (BrdUrd) Staining.
BrdUrd was dissolved in an injection buffer (1 mg/ml in 0.1 mM NaPO4/5 mM KCl). Injection was carried out on late third instar larvae anesthetized on a CO2 pad for a few minutes by using a micromanipulator setup and pulled glass needle. BrdUrd solution (≈1 μl) was injected into the thoracic region. The larva was then placed in a Petri dish with moist filter paper for 4 h to allow incorporation. The CNS was dissected and further processed as described (26). The preparations were viewed with Nomarski optics in a Zeiss Axioskop microscope. Images were captured by using a SPOT CCD camera.
DNA Sequencing.
Genomic DNA from appropriate genotypes were extracted from larvae as described in ref. 12. Templates for DNA sequencing were produced by PCR amplification by using the primer set 5′-GAG AAT GTT GCG TCT GTT CA-3′ and 5′-GAA GGT TCC GCT AGG CTG-3′. This primer set amplified a 1.638-kb product that included a 447-bp upstream sequence of the translational start site and 120 bp after the translational stop site. To sequence the whole 1.638-kb product, we used the above mentioned primers and 5′-GGA AGA CAA TAA ACT GGA GC-3′. This primer was designed from within the ORF. Automated sequencing was done by using the cycle-sequencing protocol with Taq-FS enzyme and BigDye terminator chemistry in a Perkin-Elmer ABI 373 stretch machine. Mutations were confirmed by sequencing two independent samples. The pol γ-β1 mutation was further confirmed by sequencing the parental strain of this stock. Sequence alignments were performed by using CLUSTALX software (27).
Larval Preparation for Southern and Immunoblot Analyses.
Embryos of wild-type and homozygous or heterozygous pol γ-β flies were collected for 0–6 h on fly food plates. The embryos were incubated at 25°C for 24 h. All larvae that had already hatched at 24 h were discarded, and larvae that emerged in the next 3 h were collected and transferred to a fresh food plate. The heterozygous CyO(y+) larvae were separated from the homozygous yellow mouth-hook larvae before the transfer. Larvae were allowed to grow for 6–7 days at 25°C; they were harvested by washing several times with 0.01% Triton X-100/0.7% NaCl, frozen in liquid nitrogen, and stored at −80°C.
To prepare larval extracts for protein analysis, 150 larvae (≈0.25 g) were suspended in 0.5 ml of HB-0.28 (15 mM Hepes, pH 8.0/0.28 M sucrose/5 mM KCl/2 mM CaCl2/0.5 mM EDTA/1 mM phenylmethylsulfonyl fluoride/10 mM sodium metabisulfite/2 μg per ml leupeptin/2 mM DTT). The larvae were homogenized by grinding 10 times with a microfuge pestle on ice. The homogenate was centrifuged at 1000 × g for 30 s at 3°C in a microcentrifuge, and the supernatant fraction was collected and saved. The pellet was reground in 0.1 ml of the same buffer and centrifuged as above, twice. The resulting supernatant fractions were combined and centrifuged at 8000 × g for 3 min to pellet mitochondria. The mitochondrial pellet was washed three times by resuspension in 0.5 ml HB-0.28 followed by recentrifugation and then frozen in liquid nitrogen and stored at −80°C. In the preparation of mitochondrial extracts, the mitochondrial pellets were thawed on ice for 10 min and resuspended in 0.5 ml/gm EB-mitos [25 mM Hepes, pH 8.0/10% (vol/vol) glycerol/2 mM EDTA/0.3 M NaCl/1 mM phenylmethylsulfonyl fluoride/10 mM sodium metabisulfite/2 mg/ml leupeptin/2 mM DTT]. Sodium cholate was added to 2% final concentration, and the mixture was incubated for 30 min on ice, with gentle flicking every 5 min. The extract was centrifuged at 13,000 × g for 30 min at 3°C in a microcentrifuge, and the supernatant was removed carefully and used as the soluble mitochondrial fraction.
Results
Identification and Molecular Characterization of Mutations in the Accessory-Subunit Gene of DNA Polymerase γ.
The ≈12-kb microregion containing the gene for the accessory subunit of DNA polymerase γ is depicted in Fig. 1. Two mutant alleles isolated previously within the complementation group l(2)34De or l(2)br16 were identified as candidate mutations in pol γ-β (24). Sequence analysis demonstrated that these mutant strains indeed carried disruptions in the pol γ-β gene (Fig. 1B). The mutant allele pol γ-β1 is an EMS-induced mutation resulting in a glutamic acid substitution of a highly conserved glycine residue in the N-terminal domain of the accessory subunit (Fig. 1 C and D). The pol γ-β2 allele is a spontaneous mutation caused by an in-frame 74-bp insertion in the N-terminal domain that creates a premature stop (Fig. 1E). The lethal complementation group l(2)34De is thus demonstrated as encoding the accessory subunit of DNA polymerase γ. This conclusion was further substantiated by the finding that one copy of the pol γ-β gene under the control of the heat shock promoter hsp70 partially complements the lethality caused by the pol γ-β1 mutation (see Materials and Methods).
Loss of mtDNA Caused by Mutations in the Accessory Subunit Gene Disrupts Mitochondrial Morphology.
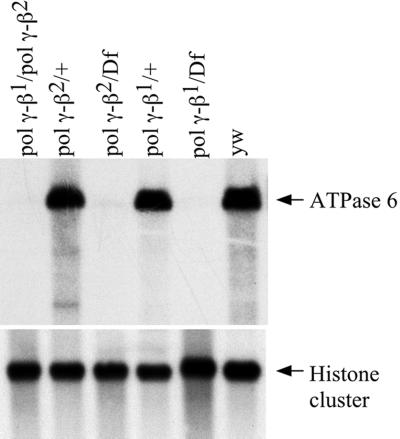
Biochemical studies have demonstrated that the accessory subunit of Drosophila Pol γ is essential to maintaining the catalytic efficiency of the holoenzyme (17, 23). Additionally, the accessory subunit is likely involved in maintaining the structural integrity of the holoenzyme through contacts between its M and C regions, with multiple sites in the exonuclease (exo) region, and part of the spacer between the exo and DNA polymerase (pol) regions of the catalytic subunit (23). Furthermore, structural modeling predicted roles for the accessory subunit in both primer recognition and in processivity (21). Thus, it was important to evaluate mtDNA content and integrity in the pol γ-β mutants. Fig. 2 shows a quantitative Southern blot of wild-type and pol γ-β mutant DNA hybridized with a mtDNA probe and with a multiple-copy genomic probe as a control. Multiple analyses of mtDNA content of moribund larvae late in the 3rd instar reproducibly failed to detect any mtDNA, demonstrating severe mtDNA depletion in pol γ-β mutants. The mechanism by which this depletion occurs is an important subject for further study, in comparison with other developmentally lethal mutants in the catalytic subunit gene, and in that for mitochondrial single-stranded DNA-binding protein.
Figure 2.
Mitochondrial DNA is absent in mutants of the accessory subunit of Drosophila Pol γ. Three micrograms of total DNA that was isolated from third instar larvae was digested with XhoI, which cleaves mtDNA once; duplicate samples of the DNAs (1 μg) were electrophoresed in a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled DNA probes from the ATPase 6 gene of mtDNA and a repeated histone gene cluster as a nuclear DNA control.
We attempted to determine the level of Pol γ-β protein in larvae by immunoblot analysis. Protein extracts were prepared from wild-type, heterozygous, and homozygous pol γ-β mutant larvae at the end of the third instar stage and subjected to SDS/PAGE followed by immunoblot analysis with high-titer affinity-purified rabbit antiserum against native Pol γ from Drosophila embryos and recombinant Pol γ-β protein. Unfortunately, the relative abundance of endogenous Pol γ-β is below the level of detection, even in an enhanced chemiluminescence analysis, so we were unable to show directly the level of the Pol γ-β protein in the pol γ-β mutants (data not shown). We do know, however, that recombinant proteins lacking the M and C region, such as what would be produced in the pol γ-β2 mutant, fail to assemble into holoenzyme, and that the free Pol γ-β protein is largely insoluble (ref. 23 and unpublished data).
To analyze the consequences of mtDNA depletion on mitochondrial physiology and morphology, we labeled brains dissected from pol γ-β mutant larvae with the selective dye Mitotracker red concomitantly with the nuclear stain DAPI. The uptake and fluorescence of Mitotracker red depends upon the presence of a membrane potential and oxidative conditions within the mitochondria and, thus, can be used to assess mitochondrial integrity as well as morphology and abundance.
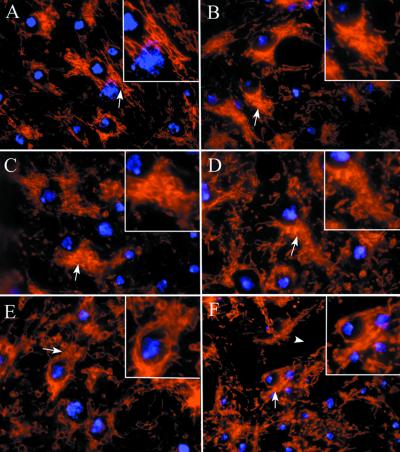
The organization of mitochondria in wild-type Drosophila neurons is similar to that described for live mammalian cells stained with the fluorescent dye JC-1 (28). Typically, mitochondria are found in a reticulum surrounding the nucleus (Fig. 3A). Reduction in pol γ-β gene function results in an overall disorganization of this reticular pattern. There is an expansion of areas showing no mitochondrial staining, together with the appearance of nuclei lacking a surrounding mitochondrial reticulum (Fig. 3 B and C). In a manner similar to that observed in mtDNA-depleted mammalian cells, the mitochondrial reticulum appears fragmented, as evidenced by the increased presence of vesicular profiles. These results also suggest, at least at this level of resolution, that mitochondrial number is not reduced drastically. Likewise, mutations that disrupt the function of the catalytic subunit of Pol γ (Pol γ-α) show a similar phenotype (Fig. 3 E and F).
Figure 3.
Mitochondrial morphology and localization are altered in mutants of Pol γ. All panels depict photomicrographs of third instar larval brain hemispheres labeled with mitochondrial probe Mitotracker (red) and the nuclear stain DAPI (blue) photographed at the same magnification. The arrows point to the mitochondrial reticulum in all panels that are magnified in Insets. In wild-type cells, mitochondria are found in a reticulum tightly arranged in the periphery of the nucleus (arrow in A). Disruption of Pol γ function either by mutations in genes encoding the accessory subunit (B, C, and D) or the catalytic subunit (E and F) disrupt this reticulum similarly. Abnormal mitochondrial morphology is characterized by a vesicular staining pattern. (A) Wild-type CNS (pol γ-β1/+). (B) pol γ-β1 mutant CNS [pol γ-β1/Df(2L)b80c1]. (C) Heteroallelic combination of two-accessory subunit mutant alleles (pol γ-β2/pol γ-β1). (D) pol γ-β2 mutant CNS [pol γ-β2/Df(2L)b80c1]. Catalytic subunit mutant CNS tam2/Df(2L)b80c1 (E) and tam9/Df(2L)b80c1 (F). The arrowhead indicates empty regions devoid of any staining.
Mutations in the Accessory Subunit Gene Are Lethal to the Organism and Impair Cell Proliferation in the CNS.
Mutations in the pol γ-β gene are strict organismal lethals in transheterozygotes with a deficiency of the region and fail to complement each other (Table 1). In all cases, lethality occurs during the early pupal period. The pol γ-β mutants begin pupariation on average 2–3 days later than wild-type controls and die soon after. These results indicate that the two mutant alleles of the pol γ-β gene represent complete lack of gene function with regard to organismal lethality and suggest the presence of a maternal contribution sufficient for embryonic and larval development.
Table 1.
Complementation analysis of accessor γ-subunit mutations
| Subunit | pol γ-β1 | Df(2l)b80c1 | tam2 |
|---|---|---|---|
| pol γ-β1 | − (0/200) | − (0/147) | + (36/112) |
| pol γ-β2 | − (0/183) | − (0/100) | + (43/120) |
Lethal mutations in the accessory subunit gene are complemented by a lethal mutation in the catalytic subunit gene of Drosophila Pol γ. Minus and plus signs indicate noncomplementation and complementation of lethality, respectively. Numerators indicate the number of viable adult flies that were heterozygous for the alleles of the pol γ-β (pol γ-β1 and pol γ-β2) and a deficiency chromosome (Df(2L)b80cl) or a mutation in the catalytic subunit gene (tam allele). Complete complementation is marked by heterozygous progeny constituting approximately 33% of the total.
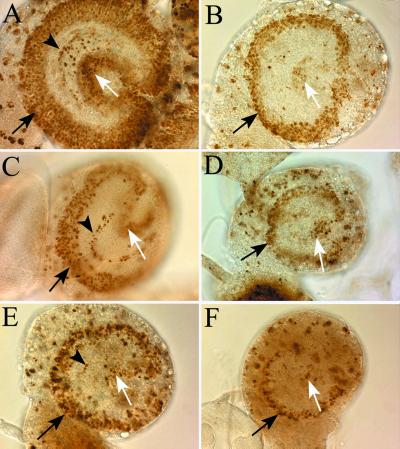
The two pol γ-β mutations complement completely null alleles of the tamas gene encoding the catalytic subunit (Table 1). Thus, a single copy of each of the genes encoding the two subunits of the Pol γ holoenzyme provides sufficient gene product for organismal viability. The external appearance of larvae lacking the accessory subunit of Pol γ is indistinguishable from wild type, whereas larvae mutant for the catalytic subunit gene are noticeably smaller (data not shown; ref. 12). Fig. 4 depicts a lateral view of brain hemispheres of these mutants seen under Nomarski optics. The CNS of larvae carrying mutations in the accessory subunit gene is smaller than that of wild-type controls but, in general, are not as small as that from catalytic-subunit gene mutants (compare Fig. 4 B to C and F; ref. 12). To assess cell proliferation, we labeled third instar larvae with BrdUrd. An overall reduction in BrdU incorporation in the CNS was observed in both mutant alleles of the pol γ-β gene (Fig. 4 B, C, and D). This finding is well illustrated by the aberrant BrdUrd labeling observed in the proliferation center of the optic lobes. Both the outer and inner proliferation centers appear to be equally affected. Mutations that disrupt the function of the catalytic subunit of Pol γ show a largely similar phenotype (Fig. 4 E and F).
Figure 4.
DNA replication and cell proliferation are impaired in mutants of Pol γ. All panels depict photomicrographs of a lateral view of third instar larval brain hemispheres dissected from larvae that had incorporated BrdU to visualize mitotically active cells found in the developing adult optic lobes. All specimens were photographed at the same magnification. In all panels, the black arrow indicates the outer proliferation center, the arrowhead indicates the lamina precursor cells, and the white arrow indicates the inner proliferation center. Disruption of Pol γ function either by mutations in genes encoding the accessory subunit (B, C, and D) or the catalytic subunit (E and F) disrupts these proliferation centers to the same extent. (A) Wild-type CNS (pol γ-β1/+). (B) pol γ-β1 mutant CNS (pol γ-β1/Df(2L)b80c1). (C) Heteroallelic combination of two accessory-subunit mutant alleles (pol γ-β2/pol γ-β1). (D) pol γ-β2 mutant CNS (pol γ-β2/Df(2L)b80c1). Catalytic-subunit mutant CNS. (E) tam2/Df(2L)b80c1. (F) tam9/Df(2L)b80c1.
Larval Behavior Is Affected Moderately by Reduction in Accessory Subunit Function.
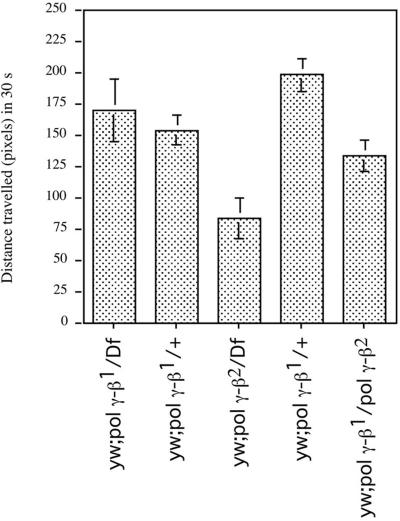
To evaluate the consequences of lack of accessory-subunit gene function for overall nervous system and muscle function, we measured larval locomotion in the absence of any light stimulus. The heteroallelic combination of the two alleles and the pol γ-β1 allele heterozygous over a deficiency of the region, did not impair significantly larval locomotion; locomotion was affected only when the larvae carried the pol γ-β2 allele over a deficiency of the region (Fig. 5). In contrast, null mutations in the catalytic-subunit gene reduced locomotion more severely and prevented the larval response to light (ref. 12 and data not shown).
Figure 5.
Genetic analysis of larval locomotion. Mean distance traveled in 30 s on an agar surface under safelight conditions. The following statistical comparisons were made with Mann–Whitney U tests at a 95% confidence interval: pol γ-β1/Df(2L)b80c1 (n = 14) vs. pol γ-β2/Df(2L)b80c1 (n = 19), test statistic (W) = 315.0, P = 0.0053; pol γ-β2/+ (n = 26) vs. pol γ-β2/Df(2L)b80c1 (n = 19), test statistic (W) = 254.0, P < 0.001; pol γ-β2/Df(2L)b80c1 (n = 19) vs. pol γ-β1/pol γ-β2 (n = 14), test statistic (W) = 261.5, P = 0.0263.
Discussion
Mutations that disrupt the function of the pol γ-β gene cause lethality in the early pupal stage. On the molecular and cellular levels, these mutations cause mtDNA depletion, aberrant mitochondrial morphology, and diminished cell proliferation in the CNS. We have not yet determined whether all tissues are affected equally. The quantitative Southern analysis was performed with DNA extracted from a total larval homogenate. Perdurance of mtDNA in the CNS, or in any other tissue that represents a small fraction of the total sample, would not likely have been detected. Moreover, different cell types have different energy requirements and, therefore, the maternal contribution of mitochondria, and/or of the pol γ-β transcript or polypeptide, will likely differ, as will their relative turnover rates. Thus, it will be of substantial interest to evaluate the temporal and spatial requirements for pol γ-β expression during zygotic development.
The two available pol γ-β mutations both map in its N-terminal region, an insertion that creates a premature stop codon and an amino acid substitution in a highly conserved glycine residue within the motif GPLGVEL in glycyl-tRNA synthetase and cause similar disruptions in Drosophila development. Both alleles, in combination with a chromosomal deficiency as well as the heteroallelic combination, cause organismal lethality at the same stage during metamorphosis. Such is also the case for nearly all other mutant phenotypes. It is reasonable to conclude that the truncated polypeptide resulting from the insertion in the N terminus will be largely or completely devoid of function and effectively represents a null mutation. That they exhibit similar phenotypes suggests that the pol γ-β1 base-substitution allele also represents a nearly complete lack-of-function mutation. Thus, we demonstrate not only that the accessory subunit of Pol γ is essential in vivo, consistent with earlier biochemical studies, but also that Gly-31 is critical for its function in mtDNA synthesis.
The biochemical consequence of the G31E substitution in the accessory subunit has not yet been determined. In vivo reconstitution of recombinant forms of Drosophila Pol γ have demonstrated that much of the N-terminal region is not required for holoenzyme assembly, yet deletion mutants exhibit a substantial loss in DNA polymerase catalytic efficiency and reduced DNA binding (23). In contrast, large deletions in human Pol γ-β that include the conserved motif GPLGVEL lack activity and apparently do not associate with the catalytic subunit on template-primer DNA in vitro (19). A recent crystallographic study suggests that mouse Pol γ-β exists as a homodimer and that the M1 domain, as defined by sequence similarity with GlyRS and including the conserved GPLGVEL motif (19, 22), is part of the interface that participates in dimerization. Notably, the M1 domain as defined by sequence similarity outside the GPLGVEL motif is almost entirely absent in Drosophila Pol γ-β, highlighting important differences in the structure of two proteins that share highly similar biochemical properties. We anticipate that Drosophila will prove a valuable model to carry out in vivo structure–function analysis by means of genetic and phenotypic characterization of additional chemically induced mutations in the accessory subunit gene.
The accessory subunit of Pol γ was initially identified in Drosophila as a 35-kDa polypeptide of unknown function copurifying with the 125-kDa catalytic subunit (29, 30). Subsequent studies demonstrated that the accessory subunit contributes to both the catalytic efficiency and the structural integrity of Drosophila Pol γ (14, 17, 18, 21). Current data support a role for the accessory subunit as a processivity clamp and primer recognition factor (16, 18–23), supporting the expectation that it would be essential for animal mtDNA replication, and that lack-of-function mutations in either the catalytic or accessory-subunit genes would disrupt development and organismal homeostasis similarly. Surprisingly, mutations in the catalytic-subunit gene appear to have more drastic consequences for the organism than mutations in the accessory-subunit gene. tamas mutant larvae are considerably smaller, with a smaller CNS and smaller, malformed imaginal discs than similarly aged larvae carrying mutations in the accessory-subunit gene. A similar disparity is found for the extent of the locomotory deficit. The difference in the stage of lethality may reflect a difference in the requirement for these gene products during development. tamas mutant larvae exhibit a significantly longer larval stage and die as third-instar foraging larvae while on the food substrate (12). These mutants rarely undergo pupariation or exhibit the behavioral and developmental changes that signal the end of larval stage and the beginning of pupal stage. In contrast, accessory-subunit mutants are able to wander and enter pupariation (2–3 days later than a wild type), dying soon after. This difference may be attributable to differences in cell growth and cell proliferation. In Drosophila, cells that will form the adult fly or imago, the so-called imaginal discs, are diploid and divide actively during the larval stages, whereas cells that form the larva never divide and are polytene (31, 32). Starvation or adverse nutritional conditions can delay larval development and the onset of pupariation, a phenocopy of mutations in the catalytic subunit gene (33). These starved larvae are, like the tamas mutant, noticeably reduced in size. Similarly, partial but not complete ablation of imaginal discs either using γ-irradiation or cell-lethal mutations delays the onset of pupariation (34, 35). These observations suggest that the developmental delay observed in larvae mutant for the catalytic-subunit gene may be caused largely by disruption of both cell growth and proliferation resulting from mitochondrial dysfunction. Recently, hypoxic conditions have been shown to induce cell-cycle arrest in Drosophila embryos, implicating a role for oxidative phosphorylation in driving cell proliferation (36).
Taken together, our observations suggest that accessory-subunit mutants sustain overall mitochondrial function sufficient for the larval transition into pupariation, whereas in catalytic-subunit mutants, mitochondrial function is disrupted such that transition into the pupal stage is prevented, presumably by the inability of the larva to reach the appropriate size and developmental stage. The observed differences between the phenotypes of the accessory and catalytic mutants may reflect a difference in the amount and/or the perdurance of the maternal contribution in tissues that are critical for the viability of the organism. Our observation that mtDNA is absent in the two mutant strains in the late third instar stage leaves open the possibility that mutations in the catalytic subunit gene actually cause mitochondrial dysfunction earlier than this point in development. Both differences in the relative maternal contribution and the developmental regulation of these genes (37) may account for an earlier onset of mitochondrial dysfunction.
Acknowledgments
We thank John Roote for pointing out the l (2)34De lethal complementation group as the most likely candidate for the gene encoding the accessory subunit of DNA polymerase γ and the Bloomington Stock Center at Indiana University for the fly stocks. A.R.C. is indebted to Dr. R. A. Morton and Dr. G. Singh for comments on the manuscript. This work was supported by Grant MT12700 from Canadian Institutes of Health Research (to A.R.C.) and National Institutes of Health Grants GM45295 and HL59656 (to L.S.K.).
Abbreviations
- mtDNA
mitochondrial DNA
- CNS
central nervous system
- Pol γ
mitochondrial DNA polymerase
- BrdUrd
5′-bromo-2′-deoxyuridine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Garesse R, Vallejo C G. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz De Mena I, Lefai E, Garesse R, Kaguni L S. J Biol Chem. 2000;275:13628–13636. doi: 10.1074/jbc.275.18.13628. [DOI] [PubMed] [Google Scholar]
- 3.Takata K, Yoshida H, Hirose F, Yamaguchi M, Kai M, Oshige M, Sakimoto I, Koiwai O, Sakaguchi K. Biochem Biophys Res Commun. 2001;287:474–483. doi: 10.1006/bbrc.2001.5528. [DOI] [PubMed] [Google Scholar]
- 4.Lefai E, Fernandez-Moreno M A, Kaguni L S, Garesse R. Insect Mol Biol. 2000;9:315–322. doi: 10.1046/j.1365-2583.2000.00191.x. [DOI] [PubMed] [Google Scholar]
- 5.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. J Biol Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi M, Hayashi Y, Nishimoto Y, Hirose F, Matsukage A. J Biol Chem. 1995;270:15808–15814. doi: 10.1074/jbc.270.26.15808. [DOI] [PubMed] [Google Scholar]
- 7.Ferri K F, Kroemer G. BioEssays. 2001;23:111–115. doi: 10.1002/1521-1878(200102)23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Goto Y. Neuropathology. 2000;20,Suppl.:S82–S84. doi: 10.1046/j.1440-1789.2000.00304.x. [DOI] [PubMed] [Google Scholar]
- 9.Jaarsma D, Haasdijk E D, Grashorn J A, Hawkins R, van Duijn W, Verspaget H W, London J, Holstege J C. Neurobiol Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 10.Berman S B, Hastings T G. J Neurochem. 1999;73:1127–1137. doi: 10.1046/j.1471-4159.1999.0731127.x. [DOI] [PubMed] [Google Scholar]
- 11.Van Goethem G, Dermaut B, Lofgren A, Martin J J, Van Broeckhoven C. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 12.Iyengar B, Roote J, Campos A R. Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier D, Farr C L, Poeck B, Alahari A, Vogel M, Fischer S, Kaguni L S, Schneuwly S. Mol Biol Cell. 2001;12:821–830. doi: 10.1091/mbc.12.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis D L, Farr C L, Wang Y, Lagina A T, 3rd, Kaguni L S. J Biol Chem. 1996;271:23389–23394. doi: 10.1074/jbc.271.38.23389. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Farr C L, Kaguni L S. J Biol Chem. 1997;272:13640–13646. doi: 10.1074/jbc.272.21.13640. [DOI] [PubMed] [Google Scholar]
- 16.Carrodeguas J A, Kobayashi R, Lim S E, Copeland W C, Bogenhagen D F. Mol Cell Biol. 1999;19:4039–4046. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Kaguni L S. J Biol Chem. 1999;274:28972–28977. doi: 10.1074/jbc.274.41.28972. [DOI] [PubMed] [Google Scholar]
- 18.Lim S E, Longley M J, Copeland W C. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 19.Carrodeguas J A, Bogenhagen D F. Nucleic Acids Res. 2000;28:1237–1244. doi: 10.1093/nar/28.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson A A, Tsai Y, Graves S W, Johnson K A. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 21.Fan L, Sanschagrin P C, Kaguni L S, Kuhn L A. Proc Natl Acad Sci USA. 1999;96:9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrodeguas J A, Theis K, Bogenhagen D F, Kisker C. Mol Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 23.Fan L, Kaguni L S. Biochemistry. 2001;40:4780–4791. doi: 10.1021/bi010102h. [DOI] [PubMed] [Google Scholar]
- 24.Woodruff R C, Ashburner M. Genetics. 1979;92:133–149. doi: 10.1093/genetics/92.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 26.Winberg M L, Perez S E, Steller H. Development (Cambridge, UK) 1992;115:903–911. doi: 10.1242/dev.115.4.903. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilkerson R W, Margineantu D H, Capaldi R A, Selker J M. FEBS Lett. 2000;474:1–4. doi: 10.1016/s0014-5793(00)01527-1. [DOI] [PubMed] [Google Scholar]
- 29.Wernette C M, Kaguni L S. J Biol Chem. 1986;261:14764–14770. [PubMed] [Google Scholar]
- 30.Olson M W, Wang Y, Elder R H, Kaguni L S. J Biol Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 31.Madhavan M M, Schneiderman H A. Roux's Arch Dev Biol. 1977;183:269–305. doi: 10.1007/BF00848459. [DOI] [PubMed] [Google Scholar]
- 32.Smith A V, Orr-Weaver T L. Development (Cambridge, UK) 1991;112:997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- 33.Mensua J L, Moya A. Heredity. 1983;51:347–352. doi: 10.1038/hdy.1983.39. [DOI] [PubMed] [Google Scholar]
- 34.Simpson P, Berreur P, Berreur-Bonnenfant J. J Embryol Exp Morphol. 1980;57:155–165. [PubMed] [Google Scholar]
- 35.Poodry C A, Woods D F. Roux's Arch Dev Biol. 1990;199:219–227. doi: 10.1007/BF01682081. [DOI] [PubMed] [Google Scholar]
- 36.DiGregorio P J, Ubersax J A, O'Farrell P H. J Biol Chem. 2001;276:1930–1937. doi: 10.1074/jbc.M003911200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefai E, Fernandez-Moreno M A, Alahari A, Kaguni L S, Garesse R. J Biol Chem. 2000;275:33123–33133. doi: 10.1074/jbc.M003024200. [DOI] [PubMed] [Google Scholar]