Abstract
The most immature lymphoid-committed progenitors in both the bone marrow (common lymphoid progenitor) and thymus (proT1) maintain a latent granulocyte/macrophage (G/M) differentiation potential that can be initiated by signals emanating from exogenously expressed IL-2 receptors. In this study, we investigate at which developmental stage thymocytes lose this G/M differentiation potential. We demonstrate that the next maturational stage after proT1 cells (proT2), but not preT (TN3) cells, can convert cell fate from lymphoid to myeloid in response to ectopic IL-2 receptor signaling in human IL-2Rβ transgenic mice. It is significant that approximately 10% of clonogenic G/M colonies derived from proT cells of IL-2Rβ transgenic mice have DJ rearrangement specifically at the Dβ1 but not Dβ2 segment in the TCRβ locus. No TCR gene rearrangement is observed in G/M cells from nontransgenic mice, suggesting that the G/M cells we observe in this system were truly lymphoid-committed before stimulation with IL-2. In addition, Dβ1 and Dβ2 DJ rearrangement of the TCRβ gene may be differentially regulated and thus serve as markers for distinct proT cell maturational stages.
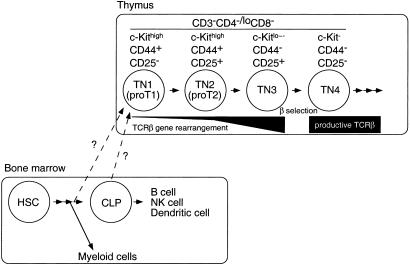
The thymus is the major site of T cell development. Immature thymocytes express neither the T cell receptor subunit CD3 nor the coreceptor molecules CD4 and CD8, and are thus referred to as triple-negative (TN) cells (1–3). These TN cells can be further subdivided into four populations based on T cell antigen receptor (TCR) gene status and expression of cell surface molecules (Fig. 1). The most immature TN cells (proT1) are defined as c-Kit (CD117)high CD44+CD25− and as a population are not yet committed to the T lineage. Cells with the proT1 phenotype as a population maintain the potential to develop into B cells, NK cells, and dendritic cells as well as T cells (4–6). In addition, few of these cells have initiated TCRβ gene rearrangement (7). Most TN thymocytes at the next maturational stage (proT2), defined as c-KithighCD44+CD25+, also maintain their TCR gene loci in the germ-line configuration, and as a population these cells have a bipotent developmental potential into T and natural killer (NK) cells (7, 8). Cells in the proT2 population can also give rise to dendritic cells (9). Both c-Kit and CD44 expression are down-regulated during progression to the next maturational stage (TN3), and these cells have now begun to rearrange their TCRβ loci. For the transition from TN3 to TN4 (CD44−CD25−) to occur, cells must undergo selection through the preT cell antigen receptor (preTCR) complex composed of a preTα chain and a productively rearranged and expressed TCRβ chain (10–13). Thus, the primary purpose of the TN phase in T cell development is likely to select for cells that have undergone productive rearrangement and expression of the β chain of the TCR.
Figure 1.
Schematic diagram of early T cell development. Immigrant cells into the thymus from the bone marrow remain to be identified. HSC, hematopoietic stem cell.
Recently, we found that the most immature lymphoid-committed cell population in the bone marrow, the common lymphoid progenitor (CLP) (14), maintains a latent granulocyte/macrophage (G/M) differentiation program (15). Lineage conversion from lymphoid to myeloid can be initiated by signaling through exogenously expressed cytokine receptors for IL-2 or G/M colony-stimulating factor (GM-CSF), which normally are not expressed on CLPs. Given that GM-CSF receptor expression can be detected in the most primitive hematopoietic precursors (hematopoietic stem cells, HSCs), but not in CLPs, we have proposed that one of the earliest events in lymphoid commitment is down-regulation of cytokine receptors that promote myeloid-lineage cell development. In addition, we have found that the most immature thymocytes, proT1 cells, also maintain a latent G/M differentiation potential. In contrast, pre-proB cells, the most immature B lineage-committed cells, are not diverted to the myeloid lineage when stimulated through reconstituted IL-2 receptors. Thus, it seems that commitment to the B lineage occurs before rearrangement of the Ig heavy chain (IgH) gene at the pre-proB cell stage (16). This result prompted us to examine at which stage T cell progenitors lose this cytokine-inducible G/M differentiation potential and fully commit to the T lineage.
In this study, we characterize the differentiation potential of CD3−CD4−/loCD8− TN cells. We demonstrate that proT1 and proT2 cells can change cell fate from lymphoid to myeloid after signaling through exogenously expressed IL-2 receptors, revealing a greater plasticity of differentiation activity than previously thought.
Methods
Mice.
Wild-type mice are C57BL/Ka-Thy1.1 (Ly5.2) and congenic C57BL/Ka-Thy1.1-Ly5.1. Human IL-2Rβ transgenic mice, originally bred on the C57BL/6 background (17), were backcrossed to C57BL/Ka-Thy1.1 through two generations. RAG-2−/− (Ly5.1) mice (11) were established by backcrossing to C57BL/Ka-Thy1.1-Ly5.1 mice. Mice were bred and maintained in the animal care facility at Stanford University School of Medicine.
Antibodies.
Antibodies used for flow cytometric analysis were as follows: FITC- or allophycocyanin (APC)-conjugated anti-Gr-1 (RB6–8C5), APC-conjugated anti-c-Kit (2B8), APC- and nonconjugated anti-CD4 (GK1.5), and Texas Red-conjugated anti-CD8α (53–6.7). These antibodies were affinity-purified from culture supernatant from hybridoma cells by protein G column chromatography (Protein G-Sepharose 4 Fast Flow, Amersham Pharmacia) and labeled in our laboratory by standard methods. Phycoerythrin (PE)-conjugated anti-CD25 was purchased from Caltag (Burlingame, CA). FITC-conjugated anti-CD44, FITC- and PE-conjugated anti-CD8α, PE- and biotin-conjugated anti-NK1.1, Cy-Chrome-conjugated anti-CD3, PE-conjugated and biotinylated anti-human IL-2Rβ, and PE-conjugated and biotinylated anti-CD19 were purchased from PharMingen. Streptavidin-PE (Caltag) and streptavidin-APC (PharMingen) were used as a secondary reagent if necessary.
Flow Cytometric Analysis and Cell Sorting.
For analysis, 1 × 106 or fewer cells were stained in 50 μl of staining medium (Hanks' Balanced Salt Solution with 2% FCS and 0.02% NaN3) with combinations of labeled antibodies (that had been individually titrated for optimal concentration) on ice for 20 min. To sort TN thymocytes, single-cell suspensions were made from thymuses of 4- to 5-week old mice. Cells were incubated on ice for 20 min with anti-mouse CD8α (clone AD4, mouse IgM) obtained from Cedarlane Laboratories. After washing with staining medium, cells were resuspended in complete medium (Iscove's MEM supplemented with 10% FCS, 50 μM 2-mercaptoethanol and antibiotics) with 10% Low-Tox-M rabbit complement (Cedarlane) and 20 μg/ml DNase (Sigma), and incubated at 37°C for 30 min with constant mixing. The cell suspension was then overlaid onto Lympholyte-M (Cedarlane) and spun at 2,000 rpm for 20 min at 20°C. The interphase containing live thymocytes was collected and washed with staining medium. Then cells were incubated on ice for 20 min with Texas Red-conjugated anti-CD8α and unconjugated anti-CD4 antibodies. After washing twice with staining medium, magnetic beads conjugated with sheep anti-rat IgG (Dynabeads M-450; Dynal, Oslo) were added and incubated on ice for 10 min. The unbound cells were collected after magnetic depletion and incubated with anti-CD44-FITC, anti-CD25-PE, anti-CD3-CyChrome, and anti-c-Kit-APC on ice for 20 min. Cells were washed and finally resuspended in staining medium containing propidium iodide. All cell sorting and flow cytometric analysis was done on a highly modified fluorescence-activated cell sorter (FACS) Vantage equipped with 488-nm argon and 597-nm dye lasers available at the Stanford shared FACS facility. Collected data were analyzed with flowjo (Tree Star, San Carlos, CA).
In Vitro Cell Culture.
One to forty cells were sorted by the automatic cell deposition unit (Becton Dickinson) on the FACS Vantage into 96-well plates containing OP9 stromal cells, and cultured for 7–14 days in complete medium in the presence of cytokines, as indicated in the figure legends. For methylcellulose colony assay, 200 cells were sorted into 35-mm dishes (Falcon 3001; Becton Dickinson) with 1 ml methylcellulose medium (MethoCult H4100, StemCell Technologies, Vancouver) containing 30% FCS, 40% Iscove's MEM, 50 μM 2-mercaptoethanol, antibiotics, and indicated cytokines. All cytokines were purchased from R&D Systems except human IL-2 (PeproTech, Rocky Hill, NJ).
Synthesized Oligos.
Oligonucleotides that were used in this study are as follows:
Dβ1.1ext: 5′-GAGGAGCAGCTTATCTGGTG-3′
Dβ1.1int: 5′-GGTAGACCTATGGGAGGGC-3′
Jβ1.7ext: 5′-AAGGGACGACTCTGTCTTAC-3′
Jβ1.7int: 5′-ACCATGGTCATCCAACACAG-3′
Dβ2.1ext: 5′-TAGGCAACCTGTGGGGAAGAAAC-3′
Dβ2.1int: 5′-GTATCACGATGTAACATTGTG-3′
Jβ2.7ext: 5′-TGAGAGCTGTCTCCTACTATC-3′
Jβ2.7int: 5′-GGAAGCGAGAGATGTGAATC-3′.
PCR Analysis of the TCRβ Genes.
Genomic DNA was isolated by incubating cells in 10 μl of 1×PCR buffer (Perkin–Elmer) containing 0.15 mg/ml proteinase K (Roche Molecular Biochemicals) at 60°C for 40 min. After incubation at 94°C for 10 min to denature proteinase K, extracted DNA was amplified by 10-cycle touchdown PCR (10 s at 94°C, 30 s at 68–63°C, 2 min at 72°C) followed by a 15-cycle PCR (10 s at 94°C, 30 s at 63°C, 2 min at 72°C) with primers (Dβ1.1ext and Jβ1.7ext, or Dβ2.1ext and Jβ2.7ext) by using a Perkin–Elmer Gene-Amp 9700. A 0.5-μl aliquot from the first amplification was subjected to the second PCR with nested primers (Dβ1.1int and Jβ1.7int, or Dβ2.1int and Jβ2.7int) for 24–30 cycles. For the analysis of TCRβ gene status in proT1 and proT2 populations, after the first 25-cycle PCR, the amplified products were separated on 1.2% agarose gel, and 0.2–2 kb DNA was excised to exclude germ-line bands. DNA was purified by spin column after melting the gel (QIAquick gel extraction kit; Qiagen, Valencia, CA), and eluted with 50 μl of elution buffer included in the kit. Purified DNA (1 μl) was used in the 24-cycle second PCR. For the analysis of TCRβ gene status in bulk populations (5 × 105 FACS-sorted cells), products after the first PCR were subjected to Southern blotting.
Results
ProT2 Cells but Not TN3 Cells in the Thymus Maintain a Latent G/M Differentiation Potential.
We have shown that CLP and proT1 cells from human IL-2Rβ transgenic mice can be induced to transdifferentiate into G/M cells by IL-2 signaling (15). CLPs maintain this latent G/M differentiation potential for 2 days after being placed in stromal cell cultures, suggesting that lymphoid precursors lose their developmental plasticity at a subsequent maturational stage. To determine the stage at which thymocytes lose IL-2-inducible G/M differentiation potential, we assayed the CD3−CD4−/loCD8− TN thymocyte populations of human IL-2Rβ transgenic mice for this activity. TN thymocytes are subdivided into four distinct populations defined by the expression of CD44, CD25, and c-Kit (CD117) (Figs. 1 and 2A). All TN cells in the thymus express the cytokine receptor common γ (γc) chain, which is an indispensable subunit for a functional IL-2 receptor complex (18, 19). The transgene is under the control of the MHC class I promoter (17), and human IL-2Rβ expression was observed in all TN cells of the thymus (Fig. 2B). Thus, all CD3−CD4−/loCD8− TN cells of the human IL-2Rβ transgenic mice express functional IL-2 receptors comprising human IL-2Rβ and mouse γc chains (20, 21). This receptor complex is responsive to human but not mouse IL-2.
Figure 2.
Expression of human IL-2Rβ transgene in CD3−CD4−/loCD8− TN thymocytes. (A) Sorting gates of subpopulations of CD3−CD4−/loCD8− TN thymocytes in this study. (B) Expression of human IL-2Rβ in CD3−CD4−/loCD8− TN thymocytes from IL-2Rβ transgenic mice (filled histogram). Background staining is also shown as a negative control (open histogram). (C Left) 2.5 × 104 proT1 (Upper) and proT2 cells (Lower) from C57BL/Ka-Thy1.1 (Ly5.2) were intravenously injected into 400 rad-irradiated RAG2−/− (Ly5.1) mice. Four weeks after injection, donor-derived cells (Ly5.2+ cells) in the spleen were analyzed by a flow cytometer. (Right) In in vitro stromal cell culture, 40 proT cells were cultured in 96-well plates on an OP9 cell layer in the presence of SIF, FL, IL-7, and human IL-2 for 7 days. Cells in positive wells were pooled and analyzed on a flow cytometer.
It is well established that proT1 cells maintain the potential to develop into B cells, NK cells, and T cells (5, 22). In keeping with this observation, we detected both T and B cell readout from proT1 cells intravenously injected into sublethally irradiated RAG2−/− mice (Fig. 2C). In contrast, proT2 cells did not give rise to B cells in in vivo reconstitution nor in in vitro stromal cell culture (Fig. 2C). Although no appreciable NK cell readout was observed from either proT1 or proT2 cells in vivo, both cells gave rise to NK1.1+ cells in vitro. Neither TN3 nor TN4 cells gave rise to B or NK cells in the stromal cell culture, but they differentiated into mature T cells in vivo (data not shown).
Next, we purified each population of TN cells by FACS from human IL-2Rβ transgenic thymuses and cultured cells in methylcellulose in the presence of a combination of cytokines sufficient for induction of G/M colonies from bone marrow cells. As we have reported, CLPs from human IL-2Rβ transgenic mice formed G/M colonies in methylcellulose only when human IL-2 was added into the culture. ProT1 cells have a basal level of myeloid cell differentiation potential (Fig. 3A and Table 1), and this activity is greatly enhanced in human IL-2Rβ transgenic proT1 cells in the presence of IL-2. ProT2 cells from IL-2Rβ transgenic mice also formed G/M colonies (Fig. 3 A and C) in the presence of IL-2. ProT2 cells show no basal G/M developmental potential, demonstrating that G/M colony formation is totally dependent on human IL-2 (Fig. 3 A and B) as for CLPs (15). In stromal cell cultures, Gr-1+Mac-1+ G/M cells were induced from proT1 and proT2 but not from TN3 or TN4 populations of human IL-2Rβ transgenic mice in the presence of human IL-2 (Fig. 3D). When the latent myeloid differentiation potential of proT1 and proT2 cells was induced, lymphoid readout frequency (B or NK cell readout in this case) from these cells in stromal cell culture was reduced (Table 1), demonstrating that myeloid cell readout occurs at the expense of lymphoid development. G/M cell readout was not observed from IL-2Rβ transgenic proT1 and proT2 cells when human IL-2 was added 4 days after initiation of the culture (Fig. 3D), suggesting that this latent G/M differentiation potential initiated by ectopic IL-2 receptor signaling is maintained in proT1 and proT2 cells for a limited time during development.
Figure 3.
A latent myeloid differentiation potential of CD3−CD4−/loCD8− TN thymocytes. (A) G/M colony formation of CD3−CD4−/loCD8− TN thymocytes derived from IL-2Rβ transgenic mice. Two hundred double-sorted cells were cultured in methylcellulose medium for 5–7 days in the presence of cytokines indicated in the figure. (B) Morphology of colonies from IL-2Rβ transgenic proT2 cells after the culture under the condition indicated in A. Only when IL-2 was added did we observe colony formation. (C) Cytospin of G/M colony-forming cells derived from IL-2Rβ transgenic proT2 cells. All colonies we confirmed by cytospin analysis were composed solely of G/M cells. (D) Stromal cell culture of CD3−CD4−/loCD8− TN thymocytes from IL-2Rβ transgenic mice. One hundred double-sorted cells were cultured on OP9 stromal cell layers in the presence of SlF, FL, IL-3, and GM-CSF. Human IL-2 was also added on either Day 0 or Day 4 of the culture as indicated in the figure. Readout populations were analyzed by a flow cytometer after 6 days of the culture. WT, wild type.
Table 1.
Limiting number of each lineage readout from proT cells in stromal cell culture
| Population | Mouse | Readout lineage, 1 in
|
||
|---|---|---|---|---|
| G/M cells | NK cells | B cells | ||
| ProT1 | WT | 500 | 22 | 150 |
| IL-2Rβ transgenic | 10 | 60 | >1,000* | |
| ProT2 | WT | ∞† | 31 | ∞† |
| IL-2Rβ transgenic | 8 | 54 | ∞† | |
One to 40 cells were clone-sorted into each well of 96-well plates layered with OP9 stromal cells and cultured for 7–10 days in the presence of SIF, FL, IL-7, and human IL-2. Positive readout was determined by microscopic observation. Outcome lineage was confirmed by flow cytometric analysis. The limiting number was obtained as described (14).
Too few positive events to determine the limiting number.
No positive readout was observed.
TCR Gene Rearrangement in G/M Colonies Derived from proT Cells of IL-2Rβ Transgenic Mice.
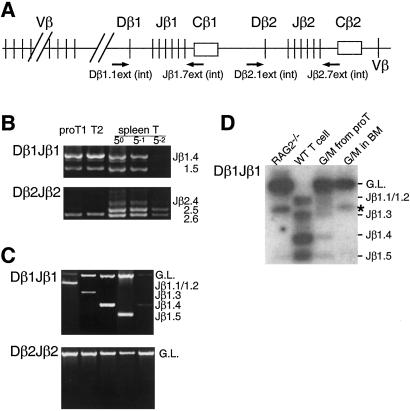
Because TCR gene rearrangement is a lymphoid-specific event (23), one may imagine that irreversible lymphoid commitment occurs at or before the initiation of TCR gene rearrangement. The TCR loci (Fig. 4A) of all proT1 and proT2 cells are in the germ-line configuration (7); however, more recent results suggest that a substantial number of proT cells have DJ rearrangement of the TCRβ genes (24, 25). In fact, DJ rearrangement of the TCRβ loci was observed in proT1 and proT2 cells by using nested PCR (Fig. 4B), although we could not detect TCRβ DJ rearrangement bands after single-round PCR by staining with ethidium bromide (data not shown), which may be due simply to the low frequency of proT cells with TCRβ DJ rearrangement. This evidence prompted us to examine TCRβ locus rearrangement status in G/M cells derived from IL-2Rβ transgenic proT cells to determine whether a correlation exists between G/M differentiation potential and TCRβ gene rearrangement. We analyzed 220 G/M colonies derived from proT1 and 126 G/M colonies derived from proT2 cells of IL-2Rβ transgenic mice for TCRβ rearrangement. Because each colony formed in methylcellulose is derived from a single cell, the TCR gene status should reflect the TCRβ gene status of the originally plated proT cell. By using genomic PCR analysis, we found that 18 colonies (8.1% of total colonies) from proT1 and 12 colonies (9.5%) from proT2 had Dβ1Jβ1 rearrangement but not Dβ2Jβ2 or V(D)Jβ rearrangement of the TCRβ loci (Fig. 4C). Obvious skewing in the usage of specific Jβ1 segments was not observed (data not shown).
Figure 4.
Rearrangement of the TCRβ gene in G/M cells derived from IL-2Rβ transgenic proT cells. (A) Schematic diagram of the TCRβ gene locus. Relative position of PCR primers used in this study is shown by arrows. (B) Rearrangement of the TCRβ gene in proT1 and proT2 populations. Genomic DNA was extracted from 5 × 103 double-sorted proT1 and proT2 cells. Dominant germ-line bands were excluded after the first PCR as described in Methods. After nested PCR, amplified products were subjected to 1.2% agarose gel electrophoresis and visualized under UV light after ethydium bromide staining. Spleen T cells (1 × 103) were used as a control. After purification from agarose gel, amplified DNA from the first PCR was serially diluted as indicated in the figure and used as the template for the second PCR. (C) Dβ1Jβ1 (Top) and Dβ2Jβ2 (Bottom) rearrangement was examined in G/M colony-forming cells derived from IL-2Rβ transgenic proT cells. Data shown are representative of five independent colonies derived from proT1 cells of IL-2Rβ transgenic mice. Because the sizes of Dβ1Jβ1.1 and Dβ1Jβ1.2 were too close to be distinguished, these two are denoted by Jβ1.1/1.2 in this study. No Dβ2Jβ2 rearrangement was observed in any of the G/M colonies analyzed. (D) TCRβ gene rearrangement in polyclonal cell populations. Double-sorted IL-2Rβ transgenic proT cells (1 × 105; both proT1 and proT2 cells) were cultured on OP9 stromal cell layers in the presence of SlF, FL, IL-3, GM-CSF, and human IL-2. After 6 days of culture, Gr-1+Mac-1+ G/M cells were sorted and analyzed for TCRβ gene rearrangement by PCR (lane 3). Gr-1+Mac-1+ G/M cells from wild-type (WT) bone marrow were also used in this analysis (lane 4). Amplified products were separated on 1.2% agarose gel and transferred to a nylon membrane. Amplified bands were detected by hybridization with 32P-labeled Dβ1.1 int and Jβ1.7 oligos followed by autoradiography. Genomic DNA from wild-type spleen T cells (lane 2) and RAG2−/− bone marrow cells (lane 1) were used as positive and negative controls, respectively. The band marked with an asterisk (*) is a PCR artifact that is also seen in RAG2−/− cells.
It has been established that TCRβ DJ rearrangement may occur in other lymphoid lineage cells, such as B cells, but not in myeloid cells (23). If this finding is true, the existence of TCRβ DJ rearrangement in G/M cells demonstrates genetically that lineage conversion from lymphoid to myeloid outcomes is induced by ectopic cytokine receptor signaling.
To confirm this assertion, we purified polyclonal Gr-1+Mac-1+ G/M cells derived from IL-2Rβ transgenic proT1 and proT2 cells in stromal cell cultures and analyzed their TCRβ gene status with PCR analysis (Fig. 4D). Bone marrow cells from RAG2−/− mice were used as background control for this assay, because they do not have any TCR gene rearrangement (11). In accordance with the results of the clonogenic analysis shown in Fig. 4C, Dβ1Jβ1 rearrangement bands were observed in the G/M cells derived from IL-2Rβ transgenic proT cells (Fig. 4D, lane 3). However, no rearrangement bands were observed in G/M cells purified from the bone marrow of wild-type C57BL/Ka-Thy1.1 mice (Fig. 4D, lane 4). We independently sorted Gr-1+Mac-1+ cells from the bone marrow of wild-type mice three times. In each case, we did not observe any TCR gene rearrangement bands in the G/M cells from the normal bone marrow, even after prolonged exposure (data not shown), demonstrating that G/M cells do not rearrange their TCR loci under normal physiological conditions. In contrast, 8–10% of proT cell-derived G/M cells have TCR gene rearrangement, which is limited to Dβ1 rearrangement. These results suggest that proT cells that have initiated Dβ1 rearrangement of the TCRβ locus maintain a latent G/M differentiation potential. However, the initiation of Dβ2 gene rearrangement seems to occur at a distinct developmental stage associated with the loss of latent G/M differentiation potential. More importantly, these results provide strong genetic evidence that lineage conversion from lymphoid to myeloid can be induced through cytokine receptor signaling.
Discussion
Hematopoietic stem cells can differentiate into all blood cells and reconstitute the hematopoietic system in vivo (26). During maturation, hematopoietic stem cells gradually lose differentiation potential as they commit to certain lineages (27). The recent identification of CLPs and common myeloid progenitors demonstrates that lymphoid and myeloid lineages are separable at the progenitor level (14, 28). Lineage commitment (at least in the hematopoietic lineage) is generally thought to be an irreversible event that is tightly regulated by lineage-specific gene expression (29). Although lineage commitment is irreversible under physiological conditions, we find a latent G/M differentiation potential in CLPs and proT cells that can be activated by signals emanating from ectopically expressed cytokine receptors (Fig. 3) (15). These data give us a more complete understanding of lymphoid lineage commitment.
One critical aspect of lineage commitment from multipotent progenitors is the expression of genes that drive this process. Although many genes play critical roles in lymphocyte development (30, 31), these genes do not necessarily prohibit differentiation programs for myeloid cell development. Our studies support a model for lymphoid commitment in which the first step is down-regulation of cytokine receptors whose signaling can initiate a G/M differentiation program (15). As the cells continue developing into lymphocytes, they gradually lose all potential of differentiating into the myeloid lineage. Specifically, we showed that CLPs, proT1 and proT2 cells, develop into G/M lineage cells when stimulated through exogenously expressed IL-2 receptors. A 2-day window occurs during which these cells are responding to lymphopoietic signals and after which these cells are no longer able to convert from their lymphoid fate to the G/M lineage in in vitro bone marrow stromal cell cultures (15). These data suggest that irreversible lymphoid commitment may occur only when cells commit to a specific single-lymphoid lineage, such as T, B, or NK cells. If this suggestion is true, one would expect that genes expressed in a lineage-restricted manner play a central role in irreversible lymphoid commitment. For the B cell lineage, the Pax5 transcription factor is a good candidate for such a commitment gene because Pax5 null mice contain B lineage-like cells that do not suppress myelomonocytic outcomes (32).
Approximately 10% of G/M colonies derived from IL-2Rβ transgenic proT cells through IL-2 receptor signaling had rearrangements in the TCRβ loci. These data clearly show that the lineage conversion detected was truly from lymphoid to myeloid outcomes, because antigen receptor gene rearrangement is a lymphoid-specific event (23). This result may have significance for other cases of recorded apparent lineage infidelities. For example, it is possible that aberrant cytokine receptor expression may be responsible for the lineage infidelity seen in acute myelogenous leukemia cells, some of which have rearrangements in TCR genes together with myeloid-specific gene expression (33). Acute myelogenous leukemia cells can also show Ig gene rearrangement (33–35). As we reported (15), G/M cells could not be induced from proB cells of human IL-2Rβ transgenic mice upon human IL-2 stimulation. It is likely that proB/preB cells can be induced to change cell fate from lymphoid to myeloid, but at this time we do not know which receptors can change cell fate in primary immature B cells. In support of our hypothesis that immature B cells also maintain a latent myeloid differentiation potential, ectopic M-CSF receptor signaling or coexpression of c-myc and v-raf can induce a morphological change of B cell lines from lymphoid to macrophages (36, 37).
One particularly important finding we describe here is that the DJβ rearrangement observed in proT-derived G/M cells is limited to the Dβ1 locus. Gene rearrangement of antigen receptors is regulated by various factors (38–40). Although the specific role of transcriptional activation in gene rearrangement is not fully understood, activation of the promoter located 5′ to Dβ1 (PDβ) is necessary for initiation of Dβ1 rearrangement. Replacement of PDβ with a tetracycline-inducible promoter initiates DJβ rearrangement in a tetracycline-dependent manner (41). In addition, mice that have a deletion of PDβ have no Dβ1 rearrangement (42). In contrast, Dβ2 rearrangement and Cβ2 germ-line transcription are not affected in this line of mice, suggesting that the promoter at the 5′ flanking region of the Dβ2 segment is independently regulated. In regard to lineage commitment, we find that proT cells clearly maintain a latent G/M differentiation activity through the stage during which Dβ1 gene rearrangement occurs. Because we failed to detect Dβ2 rearrangement in proT-derived G/M colonies, it may be that when proT cells transit to the maturational stage at which Dβ2 rearrangement occurs, they have lost G/M transdifferentiation activity. However, given that Dβ1 rearrangement is much more prevalent in proT1 and proT2 cells than Dβ2 (Fig. 4B), assuming that the amplification efficiency of both PCR reactions is comparable, we simply may not have screened enough proT-derived G/M colonies to detect Dβ2 rearrangement. If the former hypothesis holds true, it would be of significant interest to determine whether the same regulatory factors are responsible for both the initiation of Dβ2 rearrangement and irreversible T lymphocyte commitment.
To understand the plasticity of differentiation potential that we observe in CLPs and proT cells, it is necessary to elucidate the gene expression profiles of each cell population. Of central interest are the genes that are induced in lymphoid-committed progenitors during the cytokine-induced transdifferentiation process from lymphoid to myeloid lineage initiated by signaling through ectopic cytokine receptors. Thus far, we have not observed G/M lineage-specific gene expression in either CLPs or proT cells (15, 28). In depth analysis of the hierarchical relationship of genes that determine cell fate should lead us to a better understanding of normal lymphopoiesis and the aberrant process of leukemogenesis.
Acknowledgments
We thank Tasuku Honjo and Shin-Ichi Nishikawa for human IL-2Rβ transgenic mice and OP9 cells, respectively; Toshihiro Miyamoto for methylcellulose colony assay; Libuse Jerabek for thymic injection; and Amy Wagers and Susan Prohaska for critically reading the manuscript. This work was supported by U.S. Public Health Service Grants CA 42551, AI 47457, and AI 47458 (to I.L.W.). A.G.K. is a recipient of U.S. Public Health Service Training Grant CA 09302. D.C.S. is a fellow of the American Cancer Society California Division (Fellowship no. 2-42-99). M.K. was a fellow of the Irvington Institute for Immunological Research.
Abbreviations
- TN
triple negative
- CLP
common lymphoid progenitor
- G/M
granulocyte/macrophage
- TCR
T cell antigen receptor
- APC
allophycocyanin
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- FACS
fluorescence-activated cell sorter
- PE
phycoerythrin
- NK
natural killer
References
- 1.Adkins B, Mueller C, Okada C Y, Reichert R A, Weissman I L, Spangrude G J. Annu Rev Immunol. 1987;5:325–365. doi: 10.1146/annurev.iy.05.040187.001545. [DOI] [PubMed] [Google Scholar]
- 2.Shortman K, Wu L. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 3.Michie A M, Carlyle J R, Zuniga-Pflucker J C. J Immunol. 1998;160:1735–1741. [PubMed] [Google Scholar]
- 4.Godfrey D I, Kennedy J, Suda T, Zlotnik A. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 5.Matsuzaki Y, Gyotoku J, Ogawa M, Nishikawa S, Katsura Y, Gachelin G, Nakauchi H. J Exp Med. 1993;178:1283–1292. doi: 10.1084/jem.178.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardavin C, Wu L, Li C L, Shortman K. Nature (London) 1993;362:761–763. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- 7.Godfrey D I, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 8.Moore T A, Zlotnik A. Blood. 1995;86:1850–1860. [PubMed] [Google Scholar]
- 9.Wu L, Li C L, Shortman K. J Exp Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehling H J, Krotkova A, Saint-Ruf C, von Boehmer H. Nature (London) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 11.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 12.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 13.Mombaerts P, Clarke A R, Rudnicki M A, Iacomini J, Itohara S, Lafaille J J, Wang L, Ichikawa Y, Jaenisch R, Hooper M L, et al. Nature (London) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 14.Kondo M, Weissman I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 15.Kondo M, Scherer D C, Miyamoto T, King A G, Akashi K, Sugamura K, Weissman I L. Nature (London) 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 16.Allman D, Li J, Hardy R R. J Exp Med. 1999;189:735–740. doi: 10.1084/jem.189.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asano M, Ishida Y, Sabe H, Kondo M, Sugamura K, Honjo T. J Immunol. 1994;153:5373–5381. [PubMed] [Google Scholar]
- 18.Kondo M, Ohashi Y, Tada K, Nakamura M, Sugamura K. Eur J Immunol. 1994;24:2026–2030. doi: 10.1002/eji.1830240914. [DOI] [PubMed] [Google Scholar]
- 19.Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, Tanaka N, Munakata H, Nakamura M, Sugamura K. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 20.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 21.Nemoto T, Takeshita T, Ishii N, Kondo M, Higuchi M, Satomi S, Nakamura M, Mori S, Sugamura K. Eur J Immunol. 1995;25:3001–3005. doi: 10.1002/eji.1830251102. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Antica M, Johnson G R, Scollay R, Shortman K. J Exp Med. 1991;174:1617–1627. doi: 10.1084/jem.174.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlissel M S, Stanhope-Baker P. Semin Immunol. 1997;9:161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 24.Capone M, Hockett R D, Jr, Zlotnik A. Proc Natl Acad Sci USA. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak F, Tourigny M, Schatz D G, Petrie H T. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 26.Morrison S J, Uchida N, Weissman I L. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 27.Weissman I L. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 28.Akashi K, Traver D, Miyamoto T, Weissman I L. Nature (London) 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 29.Metcalf D. Ann NY Acad Sci. 1999;872:289–304. doi: 10.1111/j.1749-6632.1999.tb08473.x. [DOI] [PubMed] [Google Scholar]
- 30.Orkin S H. Nat Rev Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 31.Glimcher L H, Singh H. Cell. 1999;96:13–23. doi: 10.1016/s0092-8674(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 32.Nutt S L, Heavey B, Rolink A G, Busslinger M. Nature (London) 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 33.Cheng G Y, Minden M D, Toyonaga B, Mak T W, McCulloch E A. J Exp Med. 1986;163:414–424. doi: 10.1084/jem.163.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha K, Minden M, Hozumi N, Gelfand E W. Cancer Res. 1984;44:4658–4660. [PubMed] [Google Scholar]
- 35.Palumbo A, Minowada J, Erikson J, Croce C M, Rovera G. Blood. 1984;64:1059–1063. [PubMed] [Google Scholar]
- 36.Borzillo G V, Ashmun R A, Sherr C J. Mol Cell Biol. 1990;10:2703–2714. doi: 10.1128/mcb.10.6.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinken S P, Alexander W S, Adams J M. Cell. 1988;53:857–867. doi: 10.1016/s0092-8674(88)90309-1. [DOI] [PubMed] [Google Scholar]
- 38.Stanhope-Baker P, Hudson K M, Shaffer A L, Constantinescu A, Schlissel M S. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 39.Sleckman B P, Gorman J R, Alt F W. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 40.Hesslein D G, Schatz D G. Adv Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- 41.Sikes M L, Oltz E M. J Immunol Methods. 1999;224:25–29. doi: 10.1016/s0022-1759(99)00003-4. [DOI] [PubMed] [Google Scholar]
- 42.Whitehurst C E, Chattopadhyay S, Chen J. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]